High-throughput ultra-sensitive discrimination of single nucleotide polymorphism via click chemical ligation†
Qian-Yu
Zhou
a,
Xin-Ying
Zhong
a,
Ling-Li
Zhao
a,
Li-Juan
Wang
ab,
Ying-Lin
Zhou
 *a and
Xin-Xiang
Zhang
a
*a and
Xin-Xiang
Zhang
a
aBeijing National Laboratory for Molecular Sciences (BNLMS), MOE Key Laboratory of Bioorganic Chemistry and Molecular Engineering, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China. E-mail: zhouyl@pku.edu.cn
bKey Laboratory of Medical Chemistry and Molecular Diagnosis, Ministry of Education, Hebei University, Baoding 071002, Hebei, China
First published on 1st November 2019
Abstract
Single nucleotide polymorphisms (SNPs) have been proven to be important biomarkers for disease diagnosis, prognosis and disease pathogenesis. Here, taking the advantages of a self-assembled oligonucleotide sandwich structure and robust chemical reactions, we have developed a simple, high-throughput and effective colorimetric analytical technique termed CuAAC-based ligation-assisted assays (CuAAC-LA) for SNP detection using a DNA-BIND 96-well plate. With the 5′-azide and 3′-alkyne groups labelled on two oligonucleotide probes, the target DNA can direct a Cu(I)-catalyzed alkyne–azide cycloaddition (CuAAC) click reaction. Since the small difference in duplex stability caused by a single-nucleotide mismatch was amplified by the steric effects of these reactive groups for the ligation reaction of an unstable duplex, CuAAC-LA exhibited an ultra-sensitive discrimination ability for a mutant type target in the presence of large amounts of wild type targets. As low as 0.05% SNP could be clearly detected, which was better than most previously reported methods by various DNA ligases, indicating that a simple and rapid synthetic method i.e., the DNA template-directed click reaction held the potential to replace the ligase for SNP detection.
Introduction
Recently, there have been increasing number of studies focused on the identification and detection of single nucleotide polymorphisms (SNPs) associated with the disease diagnosis, prognosis and disease pathogenesis.1,2 A variety of technologies have been developed for SNP genotyping.3–9 Among these methods, ligation-based techniques10–14 have been considered as promising and powerful detection assays. The most commonly used ligation-based strategies for the detection of SNPs involve the use of DNA ligase enzymes, which are sensitive to the mismatched bases at the ligation site. Although these methods can achieve the sensitive detection of SNPs, the complexity of the operation, demand for a strict environment and high cost of DNA ligase enzymes hinder their wide use for clinical diagnosis.Over the past decade, numerous non-enzymatic template-directed chemical reactions15–18 have been developed. In contrast to the enzymatic methods, these non-enzymatic methods have the advantages of simple operations, common environments and low cost without sample preparation or target isolation.19,20 Based on the high specificity and programmability of these template-directed chemical reactions, the non-enzymatic methods have found valuable applications in many aspects. They have been used as a powerful tool to translate the instructions of nucleic acids into the direction of controlled organic synthesis, as well as small molecules and some useful materials.21–23 The DNA-templated synthesis also makes the libraries for the drug discovery based on the DNA-encoded chemistry.24,25 In the latter case, templated reactions can be used for sensing DNA or RNA in vitro.19,26 Furthermore, the cellular nucleic acids (miRNA, rRNA, and mRNA) can also be detected via the templated reactions in living cells, circulating exosomes, and tissues,20,27–29 while enzymatic methods cannot work in intact cells due to the difficulty of delivering enzymes into cells.
Based on the above, the non-enzymatic template-directed chemical reactions show the promise to replace the DNA ligase enzymes for some applications. Most templated strand ligations, such as nucleophilic substitution, condensation, and cycloaddition, show different levels of biocompatibility, orthogonality and reaction efficiency. Taking advantages of fast reaction speed, high yield of the product and mild reaction conditions of click chemistry,30–35 we have proposed a strategy based on the DNA target-directed Cu(I)-catalyzed alkyne–azide cycloaddition (CuAAC) click reaction for multiplex SNP detection based on capillary gel electrophoresis with laser-induced fluorescence (CGE-LIF).36 SNPs can be identified at an abundance of as low as 0.5% in the presence of a wild type target. In this study, we further developed a simple, high-throughput and colorimetric analytical technique based on a click chemical ligation for the detection of SNPs similar to the enzyme-linked immunosorbent assay (ELISA), which is based on a solid-phase oligonucleotide reaction instead of immunoassay.37,38 The use of a 96-well plate for the assay procedure can easily achieve the simultaneous detection for a large number of samples using small amounts of reagents. The assay results caused by the enzymatic substrate can provide the visible signal directly or through the use of a microplate reader, which simplifies the detection process. The performance of our assay termed CuAAC-based ligation-assisted assay (CuAAC-LA) for the detection of SNPs was evaluated and high sensitivity for the detection of SNPs at abundance as low as 0.05%–0.1% in the presence of large amounts of wild type target was achieved.
Experimental section
Materials and reagents
All chemicals were of analytical grade and were used as received. NaH2PO4·2H2O, Na2HPO4·12H2O, KCl and CuSO4·5H2O were purchased from Xilong Chemical Co. Ltd (Guangdong, China). Sodium ascorbate was obtained from Aladdin (Shanghai, China). Streptavidin-horseradish peroxidase (SA-HRP) and tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) were bought from Sigma-Aldrich (St Louis, MO, USA). Bovine serum albumin (BSA) and 3,3′,5,5′-tetramethyl benzidine dihydrochloride (TMB·2HCl) were obtained from Coolaber Technology Co., Ltd (Beijing, China). DNA-BIND 96-well plates were obtained from Corning Incorporated (USA). HPLC purified oligonucleotides were acquired from Shanghai Sangon Biotech (Shanghai, China) and have the sequences in Table S1.†Procedure of CuAAC-LA for target ssDNA detection
The reaction procedure was similar to ELISA. First, the amine-modified capture probes were dissolved in a coupling buffer (0.5 M Na2HPO4-NaH2PO4, pH 8.5) and divided into the wells of a DNA-BIND 96-well plate (100 μL per well) at a concentration of 250 nM. After incubation at 4 °C overnight, the wells were washed using a washing buffer (0.1 M Na2HPO4-NaH2PO4, pH 7.4, 5 mM KCl, 0.05% Tween 20) three times. Then, the wells were blocked using blocking solutions (5% BSA in coupling buffer) by 1 h at 37 °C and washed using the washing buffer three times. Different amounts of target DNA and 50 nM P2 in the mixed solutions (2 mM THPTA, 100 μM CuSO4 and 1 mM sodium ascorbate in PBS, pH 7.4) were added to each well and incubated at 25 °C for 4 h. The wells were then washed using the washing buffer, containing 7 M urea to remove the target chains and unreacted probe chains. After washing, 0.4 μg mL−1 SA-HRP dissolved in a phosphate buffer was added into the plate and incubated at 25 °C for 0.5 h. Then, 200 μL per well TMB–H2O2 working solutions (0.12 mg mL−1 TMB·2HCl, 18 mM H2O2, 0.1 M NaH2PO4-Na2HPO4, pH 6.0) were added to start chromogenic reaction with HRP after washing. After incubating for 40 min in dark at 25 °C, the mixture was terminated by 50 μL per well 2 M H2SO4. The absorbance (A450–A620) was recorded using a Synergy H1 multi-mode microplate reader (Biotek, Winooski, USA).Procedure of CuAAC-LA for the detection of SNPs
The immobilized and blocking procedure was performed in the same way as described above. Then, 50 nM P2, target DNA mixed with mutant DNA (total amount of 200 nM) at different ratios (2 mM THPTA, 100 μM CuSO4 and 1 mM sodium ascorbate in PBS, pH 7.4) were added to start the click reaction. The washing step and chromogenic reaction were also performed in the same way as described above.Results and discussion
Principle of CuAAC-LA for SNP detection
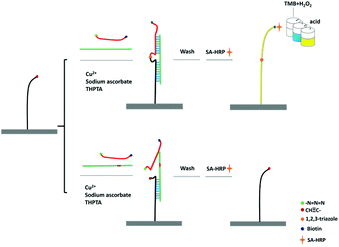
The principle of CuAAC-LA for the detection of SNPs has been shown in Scheme 1. 5′-Amine-modified capture probe P1 was immobilized on the 96-well plate surface with the reactive N-hydroxysuccinimide esters (NHS group). In the simultaneous presence of perfectly matched target DNA (T) and a signal probe P2, the specific hybridization of P1 and P2 to T formed the sandwich DNA structure. Then, the chemical ligation has occurred through CuAAC reaction since the alkynyl group labelled on P1 and the azide group labelled on P2 were induced to approach each other via hybridization reaction. A single-base mismatched target (M) cannot form the stable double structures due to the thermodynamic difference, and combined with the steric effects of these reactive groups, the significant ligation between P1 and P2 may not happen. The system was washed using the washing buffer containing urea, which was used for the destabilization of double chains to make sure the signal only came from the ligated P1/P2 chain.As the signal probe is modified with the biotin, which can easily react with SA-HRP. Based on the highly catalytic peroxidase activity of HRP, the signal produced by the addition of TMB–H2O2 solution can be visually determined directly or read using a microplate reader.
The feasibility of the CuAAC-LA
The performances of probe P1 and probe P2 in the presence of T and M (the sequences are listed in Table S1†) and in the absence of T were investigated. As shown in Fig. 1A, in the absence of any targets or in the presence of M, the immobilized P1 and signal P2 could not be ligated by the CuAAC reaction. However, in the presence of T, a significant signal could be detected, indicating that the click reaction had occurred. Another control experiment was performed to confirm that the signal was produced by a click reaction between P1 and P2 as follow. The hybridization system in the presence of P1, P2 and target without a catalyst was washed using the washing buffer containing urea. The weak signal (in the second column shown in Fig. 1A), similar to those in the absence of T or in the presence of M, indicated that the washing ability of the washing buffer was strong enough to eliminate the effects of hybridization, and the absence of catalysts could not lead to the ligation between P1 and P2. Simultaneously, the colour changes between the T and M can be clearly observed by naked eyes (Fig. 1B). Therefore, T and M can be easily discriminated by the assay.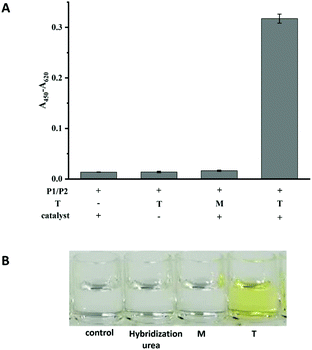 | ||
| Fig. 1 The colorimetric responses (A) and photograph (B) for the feasibility of the CuAAC-LA based SNP detection. Experimental conditions: 250 nM P1, 50 nM P2, 10 nM T or M4. | ||
Optimization of experimental conditions
Several reaction conditions, which might influence the discrimination ability of SNPs, were investigated to obtain optimal conditions for CuAAC-LA. The quantity of immobilized P1 was primary to form the sandwich DNA structure. At a constant concentration of P2, the effects of various quantities of P1 were investigated. As shown in Fig. S1,† it was found that the ligation efficiency gradually increased with the increase in the concentration of P1. However, the excess surface density might raise the possibility of free collisions of two probes and get a higher background signal. Considering the appropriate signal difference and reagent consumption, the 250 nM per well P1 was used for the immobilization process. Similarly, the concentration of P2 was optimized at the constant concentration of P1 (Fig. S2†). The probe concentration of 50 nM was used for the reaction.The reaction temperature of the assay could affect the sensitivity of the ligation efficiency (Fig. S3†). On increasing the reaction temperature, the ligation efficiency gradually decreased. The sensitivity of the assay was best at 25 °C for the formation of a stable sandwich DNA structure. The reaction time (Fig. S4†) was also related to the ligation efficiency. Considering the signal difference, the reaction time of 4 h was used for the assay.
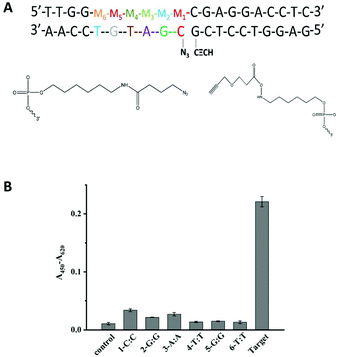
Another important factor is the discrimination ability caused by different positions of a single-base mismatch at the template DNA. In general, the short probes showed a high sequence specificity.9,39 Based on this, the designed probes in our methods were both 10 nt probes to afford high sequence specificity. The discrimination ability caused by different positions of a single-base mismatch at the template DNA was tested. Several DNA targets with a single-base mismatch at the specific positions M1 to M6 were designed (Fig. 2A). As shown in Fig. 2B, the relatively low click ligation efficiency was caused by the mismatched base at position M4, M5 or M6. Since the formation of the sandwich DNA structure was the premise that the ligation reaction could have occurred, the point mutation of centre position at P2 made the unstable duplex between target and P2, leading to the relative low click ligation efficiency. This was consistent with the perspective that the point mutation falls at the centre position of short probes made with the high sequence specificity.39 Simultaneously, the small difference in the duplex stability caused by a single-nucleotide mismatch may further be amplified by the steric effects of these reactive groups for the ligation reaction of an unstable duplex. Therefore, the single-base mismatch at M4 was selected for the further detection of SNPs.
Analytical performance of CuAAC-LA
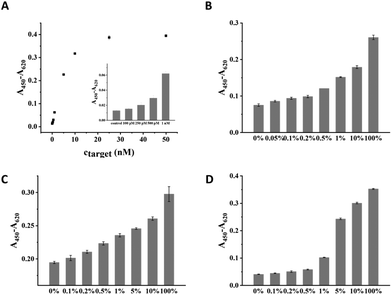
Under the optimal condition, the sensitivity of CuAAC-LA for the target DNA detection was investigated, which was directly related to the discrimination of SNPs. Different amounts of T mixed with a certain amount of P2 and catalyst were added into wells to form a sandwich DNA structure with P1 and ligated by the catalyst. As the concentration of T increased, the absorption values (A450–A620) of the CuAAC products increased based on the HRP–TMB–H2O2 colorimetric detection. As shown in Fig. 3A, 100 pM was the lowest detectable concentration for target DNA.To demonstrate the sensitivity of CuAAC-LA for the detection of SNPs, we used this method to detect different base-mismatched types. The low abundance of T mixed with different amounts of different M (M4, M7 or M8) at different ratios (0%, 0.05%, 0.1%, 0.2%, 0.5%, 1%, 5%, 10% and 100%). Then, the designed probes P1 and P2 were used to detect the mixture samples. As shown in Fig. 3B, for A > T substitution (T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) T), the target T could be significantly detected at abundance as low as 0.05% in the presence of a large number of M4. Similarly, as low as 0.1% of the T
T), the target T could be significantly detected at abundance as low as 0.05% in the presence of a large number of M4. Similarly, as low as 0.1% of the T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G (stable single mismatched base pair) and T
G (stable single mismatched base pair) and T![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C mismatches in the probe/target hybrids were also identified (Fig. 3C and D).
C mismatches in the probe/target hybrids were also identified (Fig. 3C and D).
From the above results, CuAAC-LA exhibited high sensitivity for the detection of SNPs. The results might be derived from the specificity of hybridization and effective ligation reaction. The mismatched bases of the centre position made the high sequence specificity. Since the structures of probes/mismatched target hybrids were unstable and combined with the steric effects of these reactive groups for ligation reaction, the high discrimination ability was achieved. Furthermore, the efficient ligation reaction of CuAAC-LA can not only capture the perfect matched targets from a large number of interference chains but also ensures the high sensitivity to detect the target DNA. The sensitivity of CuAAC-LA for the detection of SNPs is superior to the one based on click reaction by CGE-LIF.36 The reason might be mainly due to the differences in sensitivity for the detection of target DNA. Compared to the sensitivity for DNA detection by CGE-LIF, the sensitivity for DNA detection in this study was improved 5 times, which indicated the target DNA that could be detected was supposed to be more under the same concentration of interference chains. The performances of some current approaches are summarized in Table S2† for the detection of SNPs. The sensitivity for the detection of SNPs obtained by our methods is better than most previously reported methods by various DNA ligases. Indicated as a simple and rapid synthetic method, the DNA template-directed click reaction holds the potential to replace the ligase for the detection of SNPs. At the same time, the sensitivity of the CuAAC-LA is also comparable to the best performance detection methods.
Conclusions
In conclusion, we developed a simple, high-throughput and automated colorimetric analytical technique for the detection of SNPs. Taking the advantage of a self-assembled oligonucleotide sandwich structure and robust chemical reactions, an effective analytical technique termed CuAAC-LA, which was similar to ELISA, was fabricated. CuAAC-LA exhibited ultra-sensitive discrimination ability for the mutant type target in the presence of large amounts of wild type targets. As low as 0.05% SNP could be detected via this assay. We expect its ability to detect other types of SNPs only by designing probes for capturing the target DNA and the high-throughput detection method that enables an efficient and multiplex SNP detection.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21675004, 21575005 and 21775006).Notes and references
- A. A. Al Olama, Z. Kote-Jarai, G. G. Giles, M. Guy, J. Morrison, G. Severi, D. A. Leongamornlert, M. Tymrakiewicz, S. Jhavar, E. Saunders, J. L. Hopper, M. C. Southey, K. R. Muir, D. R. English, D. P. Dearnaley, A. T. Ardern-Jones, A. L. Hall, L. T. O'Brien, R. A. Wilkinson, E. Sawyer, A. Lophatananon, A. Horwich, R. A. Huddart, V. S. Khoo, C. C. Parker, C. J. Woodhouse, A. Thompson, T. Christmas, C. Ogden, C. Cooper, J. L. Donovan, F. C. Hamdy, D. E. Neal, R. A. Eeles and D. F. Easton, Nat. Genet., 2009, 41, 1058–1060 CrossRef CAS.
- G. Siravegna and A. Bardelli, Mol. Oncol., 2016, 10, 475–480 CrossRef CAS.
- M. W. Schmitt, S. R. Kennedy, J. J. Salk, E. J. Fox, J. B. Hiatt and L. A. Loeb, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14508–14513 CrossRef CAS.
- C. A. Milbury, J. Li and G. M. Makrigiorgos, Clin. Chem., 2009, 55, 632–640 CrossRef CAS.
- T. Wu, X. Xiao, Z. Zhang and M. Zhao, Chem. Sci., 2015, 6, 1206–1211 RSC.
- X. Xiao, T. Wu, F. Gu and M. Zhao, Chem. Sci., 2016, 7, 2051–2057 RSC.
- N. Wang, D. M. Kong and H. X. Shen, Chem. Commun., 2011, 47, 1728–1730 RSC.
- S. Hu, N. Li and F. Liu, Chem. Commun., 2018, 54, 3223–3226 RSC.
- Z. Li, X. Zhou, L. Li, S. Liu, C. Wang, L. Li, C. Yu and X. Su, Anal. Chem., 2018, 90, 6804–6810 CrossRef CAS.
- C. H. Chung and J. H. Kim, Analyst, 2018, 143, 3544–3548 RSC.
- J. H. Kim, Analyst, 2016, 141, 6381–6386 RSC.
- Z. Chen, L. Miao, Y. Liu, T. Dong, X. Ma, X. Guan, G. Zhou and B. Zou, Chem. Commun., 2017, 53, 12922–12925 RSC.
- Y. Sun, X. Lu, F. Su, L. Wang, C. Liu, X. Duan and Z. Li, Biosens. Bioelectron., 2015, 74, 705–710 CrossRef CAS.
- C. Shen, B. Shen, F. Mo, X. Zhou, X. Duan, X. Wei, J. Li, Y. Duan, W. Cheng and S. Ding, Sens. Actuators, B, 2018, 273, 377–383 CrossRef CAS.
- A. Kern and O. Seitz, Chem. Sci., 2015, 6, 724–728 RSC.
- M. Röthlingshöfer, K. Gorska and N. Winssinger, J. Am. Chem. Soc., 2011, 133, 18110–118113 CrossRef.
- E. M. Harcourt and E. T. Kool, Nucleic. Acids Res., 2012, 40, e65–e65 CrossRef CAS.
- D. Al Sulaiman, J. Y. H. Chang and S. Ladame, Angew. Chem., Int. Ed., 2017, 56, 5247–5251 CrossRef CAS.
- W. A. Velema and E. T. Kool, J. Am. Chem. Soc., 2017, 139, 5405–5411 CrossRef CAS.
- H. Wu, B. T. Cisneros, C. M. Cole and N. K. Devaraj, J. Am. Chem. Soc., 2014, 136, 17942–17945 CrossRef CAS.
- R. E. Kleiner, Y. Brudno, M. E. Birnbaum and D. R. Liu, J. Am. Chem. Soc., 2008, 130, 4646–4659 CrossRef CAS.
- C. S. Andersen, H. Yan and K. V. Gothelf, Angew. Chem., Int. Ed., 2008, 47, 5569–5572 CrossRef CAS.
- R. M. Franzini and E. T. Kool, J. Am. Chem. Soc., 2009, 131, 16021–16023 CrossRef CAS.
- D. L. Usanov, A. I. Chan, J. P. Maianti and D. R. Liu, Nat. Chem., 2018, 10, 704–714 CrossRef CAS.
- C. Cao, P. Zhao, Z. Li, Z. Chen, Y. Huang, Y. Bai and X. Li, Chem. Commun., 2014, 50, 10997–10999 RSC.
- H. Maruyama, R. Oikawa, M. Hayakawa, S. Takamori, Y. Kimura, N. Abe, G. Tsuji, A. Matsuda, S. Shuto and Y. Ito, Nucleic Acids Res., 2017, 45, 7042–7048 CrossRef CAS.
- Z. Pianowski, K. Gorska, L. Oswald, C. A. Merten and N. Winssinger, J. Am. Chem. Soc., 2009, 131, 6492–6497 CrossRef CAS.
- G. A. Metcalf, A. Shibakawa, H. Patel, A. Sita-Lumsden, A. Zivi, N. Rama, C. L. Bevan and S. Ladame, Anal. Chem., 2016, 88, 8091–8098 CrossRef CAS.
- L. Holtzer, I. Oleinich, M. Anzola, E. Lindberg, K. K. Sadhu, M. Gonzalez-Gaitan and N. Winssinger, ACS Cent. Sci., 2016, 2, 394–400 CrossRef CAS PubMed.
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS.
- R. Kumar, A. El-Sagheer, J. Tumpane, P. Lincoln, L. M. Wilhelmsson and T. Brown, J. Am. Chem. Soc., 2007, 129, 6859–6864 CrossRef CAS.
- A. H. El-Sagheer, A. P. Sanzone, R. Gao, A. Tavassoli and T. Brown, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11338–11343 CrossRef CAS.
- M. Kukwikila, N. Gale, A. H. El-Sagheer, T. Brown and A. Tavassoli, Nat. Chem., 2017, 9, 1089–1098 CrossRef CAS.
- E. A. Osman, T. Gadzikwa and J. M. Gibbs, ChemBioChem, 2018, 19, 2081–2087 CrossRef CAS.
- Q. Y. Zhou, F. Yuan, X. H. Zhang, Y. L. Zhou and X. X. Zhang, Chem. Sci., 2018, 9, 3335–3340 RSC.
- S. Cai, C. Lau and J. Lu, Anal. Chem., 2011, 83, 5844–5850 CrossRef CAS.
- J. Nie, Y. Deng, Q.-P. Deng, D.-W. Zhang, Y.-L. Zhou and X.-X. Zhang, Talanta, 2013, 106, 309–314 CrossRef CAS.
- Y. Z. Xu, N. B. Karalkar and E. T. Kool, Nat. Biotechnol., 2001, 19, 148–152 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9an01672d |
| This journal is © The Royal Society of Chemistry 2020 |