Open Access Article
Open Access ArticleTailorable synthesis of heterogeneous enzyme–copper nanobiohybrids and their application in the selective oxidation of benzene to phenol†
Noelia
Losada-García
 ,
Alba
Rodríguez-Otero
and
Jose M.
Palomo
,
Alba
Rodríguez-Otero
and
Jose M.
Palomo
 *
*
Department of Biocatalysis, Institute of Catalysis (CSIC), Marie Curie 2. Cantoblanco. Campus UAM, 28049 Madrid, Spain. E-mail: josempalomo@icp.csic.es; Fax: +34 91 585 4760
First published on 27th November 2019
Abstract
A protein-direct synthetic route to make nanobiohybrids containing copper nanoparticles (CuNPs) using an enzyme in combination with copper sulfate in aqueous media at room temperature is described. Enzyme–Cu(II) intermediates, rapidly generated in aqueous media, are transformed, induced by the enzyme structure, to facilitate the formation of a particular Cu species, tailoring the size of the nanoparticles formed depending on the experimental conditions, e.g. the pH, reducing step, amount of enzyme or incubation time. This controlled synthesis allows the preparation of tailor-made nanobiohybrids of Cu(0)NPs, Cu(0)/Cu2ONPs, Cu2ONPs or Cu3(PO4)2NPs on the multimilligram scale. The catalytic performance of the novel CuNPs nanobiohybrids was evaluated in relation to the selective C–H functionalization of benzene via direct monohydroxylation using hydrogen peroxide as a green oxidant under mild conditions, in aqueous media and at moderate temperature. Optimization of the amount of hydrogen peroxide, the co-solvent, and the temperature was performed, resulting in the production of 17 mM phenol with 83% selectivity using the Cu-CALB-PHOS-NR nanobiohybrid as a catalyst.
Introduction
Copper is a low cost and earth-abundant metal, which has been described as being very useful for different applications,1–5 especially catalysis.6–9 In terms of the different Cu compounds available, copper nanoparticles (CuNPs) have generated great interest in recent years.10 The high surface-to-volume ratio of nanomaterials compared to bulk materials generally makes them attractive candidates for application as catalysts.11 A multitude of different practical and straightforward ways of preparing Cu nanomaterials have been described.12–14 However, the use of CuNPs is restricted by the inherent instability of copper under atmospheric conditions, which makes it prone to oxidation. Many efforts to develop methods and support materials that increase the stability of CuNPs, by altering their sensitivity to oxygen, water and other chemical entities, have encouraged the exploration of Cu-based NPs with more complex structures, such as core/shell CuNPs or systems based on copper oxides.15–20The synthesis of copper and copper oxide NPs is essentially focused on four types of chemical reactions: reduction, oxidation, hydrolysis or condensation. Depending on the choice of final materials, one or a combination of the above-mentioned chemistries may be applied. The synthesis of CuNPs often entails the reduction of Cu(I) or Cu(II) sources. The synthesis of copper oxide NPs, on the other hand, basically requires hydrolysis of the precursors, followed by a dehydration process that leads to the final materials. In addition, an oxidation process (sometimes unavoidable for Cu-based NPs) can be deployed for the preparation of Cu-based NPs with higher oxidation numbers from their respective lower oxidation state precursors. In synthetic processes, the techniques that are applied provide an adequate environment and enough energy to facilitate the process of choice, while additional restrictions are imposed to modulate the stability, properties and morphology of the final NPs.21–23
Therefore, the development of methodologies to synthesize highly active, selective, stable and robust copper nanoparticles is mandatory.24 Sustainable and inexpensive systems that allow the production of a large amount of CuNPs are also desirable from an application point-of-view. Copper species—in terms of catalysis—are one of the important aspects that affect the final properties of nanocatalysts for any given reaction.
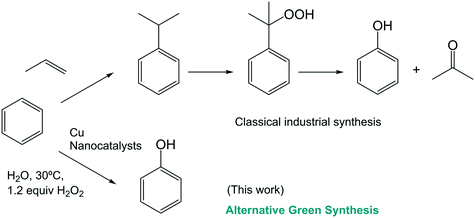
Selective catalytic oxidation of inactive aromatic C–H bonds is one of the most active research topics, but it remains a long-term challenge.25
In particular, direct catalytic oxidation in one step from benzene to phenol remains a central topic in fundamental and applied research. Phenol is one of the most important industrial chemicals,26 however, its current industrial production is still limited to the three-step cumene process (Scheme 1). These reactions are carried out at elevated temperatures and high pressure resulting in a low overall phenol yield (<5%).27
Recently, a few examples have been described in the literature of the application of Cu complexes in the monohydroxylation of benzene, although in most cases, low or moderate yields were obtained and total selectivity was not achieved.26–32
In this work, different Cu nanobiohybrids have been synthesized for the first time via an in situ controllable Cu species and nanoparticle size in a protein host network and their properties have been demonstrated using X-ray diffraction (XRD) and transmission electron microscopy (TEM).
Furthermore, some of these new Cu nanobiohybrids show catalytic activity in the direct sustainable selective conversion of benzene to phenol (Scheme 1).
Results and discussion
Synthesis and structural characterization
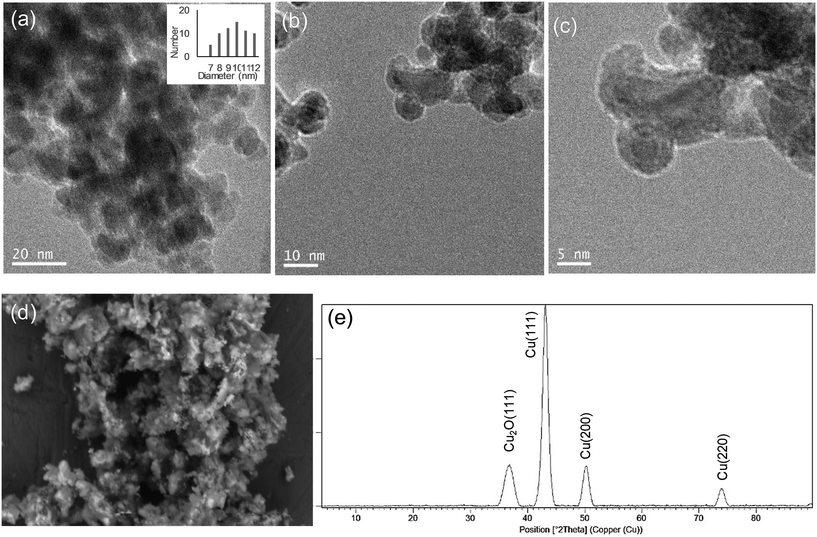
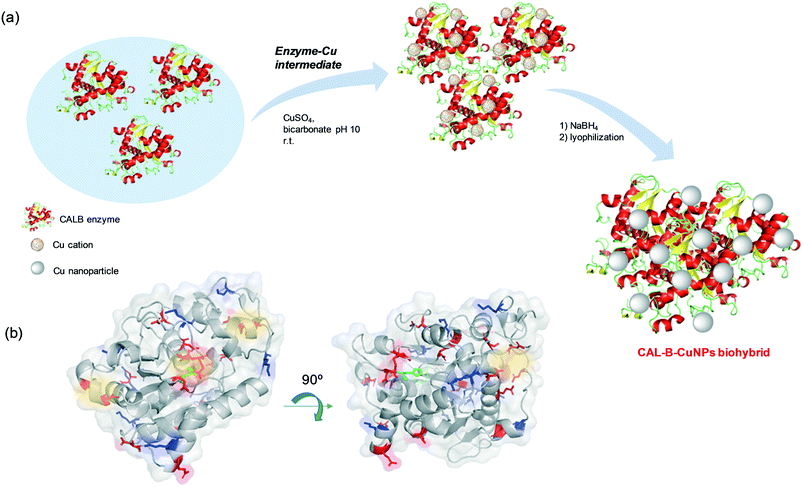
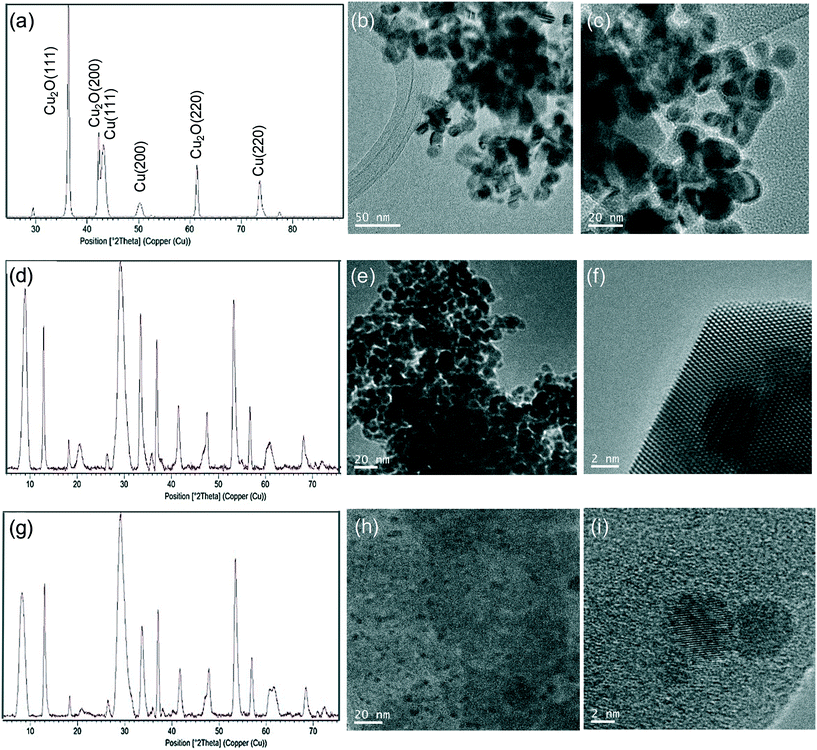
Hybrid copper nanoparticles (Cu nanobiohybrids) were synthesized via an aqueous soft-templating method, where an enzyme (commercial lipase B from Candida antarctica, CAL-B, 33 kDa, monomeric enzyme, supplied by Novozymes), copper sulfate, 100 mM of sodium bicarbonate at pH 10 and NaBH4 were used as the template and binder, Cu precursor, reaction medium and reducing agent, respectively.The first three components were combined and then the resulting solid formed was treated with the reducing agent, collected by centrifugation and washed with distilled H2O. Finally, the solid was dissolved in water, frozen and lyophilized. The Cu nanobiohybrid (Cu-CALB-BIC) morphology was initially observed via electron microscopy. Scanning electron microscopy (SEM) analysis revealed that the precipitate was made up of an aggregate with a mesoporous amorphous superstructure composed of copper atoms dispersed in an organic matrix (CAL-B) (Fig. 1).
TEM analysis showed the formation of nanoparticles with spherical morphology densely deposited throughout the hybrid composite (Fig. 1a–c). The average diameter of the nanoparticles was 9 nm (Fig. 1a). Wide-angle XRD further displayed four characteristic peaks, three of them assigned to the (111), (200) and (220) planes, respectively, of the fcc Cu lattice (matched well with JCPDS card no. 04-0836), whereas the peak at 37 degrees was assigned to the (111) plane characteristic of Cu2O (matched well with JCPDS card no. 05-0667) (Fig. 1d). Inductively coupled plasma-optical emission spectrometry (ICP-OES) analysis showed that the copper content in the nanobiohybrid was 84%.
Thus, the formation of hybrid copper nanoparticles first undergoes rapid coordination between the amino acid residues of the enzyme with the copper ions, generating enzyme–Cu(II) intermediates (Scheme 2). It is well described that copper ions have very good coordination capacity, especially with carboxylic groups (Asp, Glu) and also with amino groups, mainly those containing histidine or lysine residues. Interaction with cysteine residues may also be possible.
In this particular case, CALB does not have free cysteines (cysteine residues are involved in the formation of disulfide bridges) in its structure (Fig. S1†). However, at the pH where hybrids form, Asp, Glu and Lys could be the main binding sites of the Cu2+ ions in the protein. Take into account that, the tridimensional structure seems to have a particular area where Cu(II) ions can initially bind (Scheme 2a and Fig. S2†). Here, we can observe a rich area of carboxylic group residues and even the imidazole from the histidine. Two types of amino acid residue (Asp, His) are involved in the formation of the catalytic triad of the enzyme.
This coordination could be responsible for the loss of enzymatic catalytic activity observed (data not shown). Free lysine residues that do not form salt bridges with the corresponding Asp or Glu residues may also be involved.
Then, in situ reduction by NaBH4 with optimal reduction kinetics results in the formation of a CuNPs-enzyme nanobiohybrid.
Tailor-made synthesis of CuNPs in nanobiohybrids
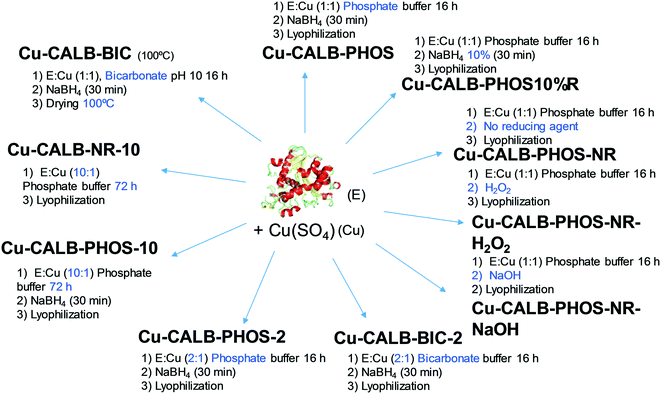
This approach has demonstrated the specific formation of particular copper nanoparticles embedded in the protein matrix, where the role of the enzyme is crucial. In this respect, considering the effect that experimental conditions can have on the protein structure, we have evaluated our method in terms of experimental conditions to control the copper species, morphology and size of Cu nanoparticles in the nanobiohybrids. A summary of the different approaches developed is described in Fig. 2.Initially, drying conditions were modified in the Cu-CALB-BIC nanobiohybrid, replacing lyophilization via treatment at 100 °C. Under these conditions, the XRD pattern of the hybrid Cu-CALB-BIC(100°C) showed the increase of Cu2O as a copper species, and a decrease in the intensity of the characteristic peaks of Cu(0) (Fig. S3†).
The pH is an important parameter for proteins, so the synthesis of the Cu nanobiohybrid was attempted at pH 7 instead of 10. For that, an aqueous sodium phosphate buffer solution was used. The synthetic protocol was repeated as described above to produce the Cu-CALB-PHOS nanobiohybrid (Fig. 2).
Wide-angle XRD measurements showed that the main Cu species was Cu(I) in the form of Cu2O, which displayed three main characteristic peaks, assigned to the (111), (200) and (220) planes (Fig. 3a). A small amount of Cu(0) was also determined. TEM experiments revealed the formation of 15 nm dispersed nanoparticles (Fig. 3b and c), slightly larger than those previously observed for Cu-CALB-BIC (Fig. 1). This nanobiohybrid presented 81% Cu content, as calculated from ICP-OES analysis. This method was also modified by using acetone in the washing step instead of water, or by heating at 80 °C instead of lyophilization to obtain the solid. Under all these conditions, the nanobiohybrids presented a mixture of different Cu species and the catalytic properties and mechanical stability of the solids were worse (data not shown). A test was also performed by applying the synthetic protocol directly in distilled water (pH of around 5.5) without using any buffer in the solution. Under these conditions, the XRD pattern showed that a mixture of Cu(I) in the form of Cu2O and Cu(0) was obtained (Fig. S4†), and ICP-OES determined a quantity of 68% copper in the sample.
The next strategy was to decrease the amount of reducing agent, NaBH4, using only 10% (w/v) of the previous amount. This simple modification resulted in significant changes in the final Cu NPs of the nanobiohybrid synthesized (Fig. 3d–f). Wide-angle XRD further displayed characteristic peaks of Cu(PO4)3, which matched well with the JCPDS card no. 01-080-0991 (Fig. 3d). TEM analysis showed the formation of spherical nanoparticles (of around 10 nm in diameter) densely deposited throughout the hybrid composite (Fig. 3e). High-resolution TEM showed the high crystallinity of the nanoparticle framework (Fig. 3f). ICP-OES determined 48% copper in the hybrid solid powder.
In addition, the Cu nanobiohybrid was also synthesized without the use of a reducing step, to obtain the so-called Cu-CALB-PHOS-NR nanobiohybrid (Fig. 2). The XRD pattern showed the same peak distribution previously observed for Cu-CALB-PHOS-10%R, corresponding to the Cu(PO4)3 species (Fig. 3g). However, in this case, TEM analysis revealed the formation of smaller crystalline spherical nanoparticles (of around 3 nm) (Fig. 3h and i). The amount of copper contained in the solid was 32%.
Other variations were made to the protocol, such as substituting the reducing step with an oxidative process via the addition of NaOH or H2O2, resulting in the synthesis of Cu-CALB-PHOS-NR-NaOH and Cu-CALB-PHOS-NR-H2O2, respectively. In this case, as in the previous one, a light blue solid was obtained instead of the typical black colour for the other nanobiohybrids. The copper species was conserved and spherical nanoparticles were also obtained of around 5–6 nm in size (Fig. S4–S6†). The amounts of copper contained in the solid were 35% and 22%, respectively.
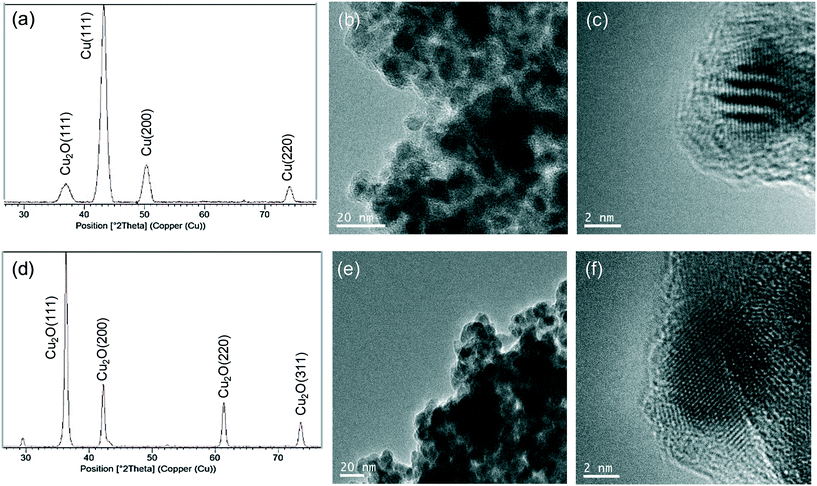
Taking into account the clear differences between the use of bicarbonate or phosphate in the synthesis, in each case a new synthetic protocol was applied where the amount of enzyme was doubled. Thus, the Cu-CALB-BIC-2 and Cu-CALB-PHOS-2 nanobiohybrids were obtained (Fig. 2).
For the first, the XRD pattern showed the same characteristic peaks of the Cu fcc as those observed in Cu-CALB-BIC (Fig. 4a). However, TEM analysis revealed the formation of very crystalline nanoparticles with a size of 6 nm in diameter (Fig. 4b and c) (instead of the 9 nm nanoparticles contained in Cu-CALB-BIC, Fig. 1). A slight increase in the content of the amount of Cu in the solid was observed.
For Cu-CALB-PHOS-2, the XRD pattern showed characteristic peaks of the fcc Cu2O (Fig. 4d) and the amount of copper contained in the solid was 61% (20% less than in Cu-CALB-PHOS). No traces of further oxidation states (CuO or Cu(OH)2) were observed even upon increasing the time of the reducing step (data not shown). The TEM images revealed the formation of crystalline nanoparticles with a diameter of 10 nm (Fig. 4e and f) (instead of the 15 nm nanoparticles containing Cu-CALB-PHOS, Fig. 3b and c).
Therefore, when using this last methodology, a novel Cu nanobiohybrid with controlled morphology, size and metal species was synthesized. This could be explained by the concept that a greater amount of protein influences the control of the reduction of copper oxide species and also influences the control of the nanoparticle growth.
A final test was performed using ten times more protein in the phosphate solution synthesis. The incubation time was also increased to 72 h. The protocol was performed with and without the reducing step, synthesizing the nanobiohybrids Cu-CALB-PHOS-10 and Cu-CALB-PHOS-NR-10, respectively. In both cases, SEM analysis revealed the formation of well-formed nanoflowers (Fig. S7 and S8†). The XRD patterns showed the presence of Cu3(PO4)2 as the main copper species, and the mass analysis revealed a 50% copper content in the solid in each case.
In all cases, the enzyme–Cu nanoparticle hybrids were synthesized in a very effective, simple and sustainable way on a multimilligram scale, easily scalable to grams.
In order to test the stability of the hybrids, they were incubated at 100 °C in water and 100 °C in the presence of mercaptoethanol and sodium dodecyl sulfate (SDS, disruption buffer) (conditions for protein denaturisation). In both cases the hybrids conserved their heterogeneous nature and copper species, confirmed by XRD (Fig. S9†).
Catalytic performance of Cu nanobiohybrids in the selective oxidation of benzene to phenol in aqueous media
The different synthesized CuNPs nanobiohybrids were evaluated as catalysts in a C–H activation process, the direct selective oxidation of benzene to phenol under mild conditions (Table 1). The reaction was first performed in aqueous media (99% water, 1% acetonitrile (ACN)) using a green oxidant (hydrogen peroxide) at 30 °C. After solubility studies, we determined that using 100 mM of benzene, 62 mM was soluble under these conditions (Table S2†), which was the concentration used for phenol yield calculation. The best results in phenol production were obtained using the Cu nanobiohybrids containing Cu(PO4)3 (entries 10–13, Table 1), with around 12 mM phenol (19% yield) with a selectivity of 77% achieved using Cu-CALB-PHOS-NR (entry 10, Table 1).| Entry | Cu nanobiohybrid | Time (h) | [Phenol]b (mM) | Yieldc (%) | Selectivityd (%) |
|---|---|---|---|---|---|
| a Benzene (1 mmol), 33% H2O2 (1.25 mmol), catalyst (5 mg), 10 mL of solution (99% water, 1% ACN), 30 °C, 24 h. b Phenol concentration produced, calculated from a quantification calibration curve (Fig. S10†). c Phenol yield was calculated using high performance liquid chromatography (HPLC). d Selectivity of phenol with respect to the formation of other byproducts (benzoquinone, hydroquinone or catechol) was determined using HPLC. | |||||
| 1 | — | 24 | 0 | — | — |
| 2 | CAL-B | 24 | 0 | — | — |
| 3 | Cu-PHOS | 24 | 0 | — | — |
| 4 | Cu-BIC | 24 | 0 | — | — |
| 5 | Cu-CALB-PHOS | 24 | <1 | <2 | — |
| 6 | Cu-CALB-BIC | 24 | <1 | <2 | — |
| 7 | Cu-CALB-PHOS-2 | 24 | 4 | 6 | >99 |
| 8 | Cu-CALB-BIC-2 | 24 | 1.5 | 2 | >99 |
| 9 | Cu-PHOSNR | 24 | 0 | — | — |
| 10 | Cu-CALB-PHOS-NR | 24 | 11.5 | 18 | 77 |
| 11 | Cu-CALB-PHOS-NR-NaOH | 24 | 12.5 | 19 | 71.4 |
| 12 | Cu-CALB-PHOS-NR-H 2 O 2 | 24 | 8 | 13.1 | 55.4 |
| 13 | Cu-CALB-PHOS10%R | 24 | 10 | 15.6 | 77.1 |
A plausible mechanism for the reaction could be similar to that recently reported,28 which first involves the degradation of hydrogen peroxide via a Fenton-like reaction to produce a hydroxyl radical and Cu(III), which in the presence of hydrogen peroxide produces a HO2˙ radical with the regeneration of Cu(II). The following hydrogen abstraction from H2O2 by the HO2˙ adduct produces phenol and H2O.
Other Cu nanobiohybrids exhibited very low catalytic activity, with yields lower than 10%, although in the case of the Cu-CALB-PHOS-2 nanobiohybrid, 4 mM of phenol was produced with excellent selectivity of >99% (entry 7, Table 1).
The enzyme without metal and the hybrid formed without the enzyme (Cu-PHOS, Cu-BIC and Cu-PHOSNR) did not show any catalytic activity (Table 1).
Therefore, considering these results, the Cu-CALB-PHOS-NR nanobiohybrid was selected for further experimental studies.
Firstly, the effect of benzene concentration on the phenol yield was evaluated (Table 2). As described above, the solubility of benzene at 50 and 200 mM in water containing 1% ACN was also studied (Table S2†). Considering the corresponding soluble amount of benzene, we observed that at lower benzene concentration, Cu-CALB-PHOS-NR produced a higher amount of phenol (31% yield), although with lower selectivity (67%). The Cu catalyst in the presence of double the amount of benzene (although a slightly higher soluble amount of 70 mM) showed similar activity and slightly less selectivity (Table 2). Therefore, we selected this as the initial amount of benzene for further experiments.
| [C6H6]initialb (mM) | Time (h) | [Phenol]c (mM) | Yieldd (%) | Selectivitye (%) |
|---|---|---|---|---|
| a 33% H2O2 (1.25 mmol), catalyst (5 mg), 10 mL of solution (99% water, 1% ACN), 30 °C, 24 h. b Amount of benzene. c Phenol concentration produced, calculated from a quantification calibration curve (Fig. S10†). d The phenol yield was calculated using HPLC. e Selectivity of phenol with respect to the formation of other byproducts was determined using HPLC. | ||||
| 50 | 24 | 6.3 | 31.3 | 67.1 |
| 100 | 24 | 11.3 | 18 | 76.5 |
| 200 | 24 | 13.5 | 19.1 | 73.6 |
The next parameter to study was the amount of acetonitrile as a co-solvent in the aqueous media (Table 3). The catalytic efficiency and selectivity of Cu-CALB-PHOS-NR was found to be affected in the presence of different amounts of ACN. Also, the solubility of benzene in water increased with increasing amounts of acetonitrile, so a benzene solubility study was performed (Table S3†).
| Co-solventb (%) | Time (h) | [Phenol]c (mM) | Yieldd (%) | Selectivitye (%) |
|---|---|---|---|---|
| a Benzene (1 mmol), 33% H2O2 (1.25 mmol), catalyst (5 mg), 10 mL of solution (water/ACN), 30 °C, 24 h. b Amount of acetonitrile as a co-solvent in water (v/v). c Phenol concentration produced, calculated from a quantification calibration curve (Fig. S10†). d The phenol yield was calculated using HPLC. e Selectivity of phenol with respect to the formation of other byproducts (benzoquinone and hydroquinone) was determined using HPLC. | ||||
| 1 | 24 | 11.5 | 18 | 76.5 |
| 10 | 24 | 14.5 | 23.4 | 80 |
| 20 | 24 | 17.2 | 24 | 83 |
| 50 | 24 | 3.5 | 4 | 26 |
| 100 | 24 | — | — | — |
The addition of 10% acetonitrile as a co-solvent in aqueous media barely increased the solubility of the benzene, however Cu-CALB-PHOS-NR showed higher activity and selectivity under these conditions, also producing 15 mM phenol at 80% selectivity. Even an increased amount of up to 20% acetonitrile improved the catalytic efficiency of the Cu nanocatalyst, with a 24% yield of phenol (17.2 mM) and 83% selectivity (Table 3).
Surprisingly, Cu-CALB-PHOS-NR showed very low activity and selectivity in the presence of 50% acetonitrile or it no activity when the reaction was performed directly in acetonitrile.
Thus, 20% acetonitrile in water was selected as a reaction medium. In order to improve upon the best results achieved, different reaction parameters were modified, such as the amount of oxidant, temperature or reaction time (Table 4).
| Entry | H2O2b (eq.) | Co-solventc (%) | T (°C) | Time (h) | [Phenol]d (mM) | Yielde (%) | Selectivityf (%) |
|---|---|---|---|---|---|---|---|
| a Benzene (1 mmol), catalyst (5 mg), 10 mL of water/acetonitrile. b Amount of H2O2 (eq.). c Amount of acetonitrile as a co-solvent in water (v/v). d Phenol concentration produced, calculated from a quantification calibration curve (Fig. S10†). e The phenol yield was calculated using HPLC. f Selectivity of phenol with respect to the formation of other byproducts (benzoquinone and hydroquinone) was determined using HPLC. | |||||||
| 1 | 1.25 | 20 | 30 | 24 | 17.2 | 24 | 83 |
| 2 | 5 | 20 | 30 | 24 | 3 | 3 | 60 |
| 3 | 10 | 20 | 30 | 24 | 1.3 | 1.8 | >99 |
| 4 | 1.25 | 50 | 30 | 24 | 3.5 | 4 | 26 |
| 5 | 5 | 50 | 30 | 24 | 9 | 10.5 | 84.4 |
| 6 | 10 | 50 | 30 | 24 | 3.3 | 4 | 60 |
| 7 | 1.25 | 20 | 4 | 3 | — | — | — |
| 8 | 1.25 | 20 | 30 | 3 | 17 | 23.7 | 83 |
| 9 | 1.25 | 20 | 40 | 3 | 15.3 | 21.5 | 81 |
| 10 | 2 | 20 | 30 | 3 | 17.8 | 21 | 75 |
| 11 | 4 | 20 | 30 | 3 | 9.5 | 11.2 | 65.8 |
Considering the possibility of the need for more oxidant in order to improve the conversion, we tested the addition of more equivalents of hydrogen peroxide. This study showed that the increase in the amount of H2O2 to 5–10 eq. in the reaction using aqueous media containing 20% can has a very negative effect on the conversion, resulting in a very low yield of phenol (Table 4, entries 2–3). At this point, we also evaluated the effect on the amount of oxidant in the reaction using water/ACN 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as a solvent (Table 4, entries 4–6). In this case, the activity and selectivity of Cu-CALB-PHOS-NR improved by more than the double when 5 eq. of H2O2 were used in the reaction. Surprisingly, the selectively greatly improved by more than 4 times from 26 to >84% (Table 4, entries 4–5). However, the addition of extra hydrogen peroxide (10 eq.) cause a decreased in the activity and selectivity of the catalyst compared with the results using 5 eq. of oxidant (Table 4, entry 6).
1 as a solvent (Table 4, entries 4–6). In this case, the activity and selectivity of Cu-CALB-PHOS-NR improved by more than the double when 5 eq. of H2O2 were used in the reaction. Surprisingly, the selectively greatly improved by more than 4 times from 26 to >84% (Table 4, entries 4–5). However, the addition of extra hydrogen peroxide (10 eq.) cause a decreased in the activity and selectivity of the catalyst compared with the results using 5 eq. of oxidant (Table 4, entry 6).
Considering the results, until now the reaction had been stopped only after 24 h, so we evaluated the results after shorter times. So, the reaction was performed using 1.25 eq. H2O2 and 20% ACN in water at 30 °C in the presence of Cu-CALB-PHOS-NR as a catalyst, and the results were evaluated after 3 h. The yield of phenol and selectivity achieved by the catalyst were the same as those achieved in 24 h (Table 4, entry 8), with a TOF value of 2.12 h−1.
Finally, to demonstrate the heterogeneity of the catalyst, a hot filtration experiment was performed as described in the experimental section for Cu-CALB-PHOS-NR under these reactions conditions, with all of the solid material being recovered.
In order to evaluate the effect of the temperature on the efficiency of the Cu catalyst, the reaction was performed at 4 and 40 °C (Table 4, entries 7 and 9). No reaction was observed at 3 h at 4 °C, whereas an increase in the temperature caused a slight decrease in the activity and selectivity of the catalyst (Table 4, entry 9).
Finally, and seeing the results obtained with the effect of the addition of more hydrogen peroxide, we tested the effect of more oxidant, but in low amounts (2 or 4 eq.), in the conversion and selectivity after 3 h of incubation. No improvements were observed in any case (Table 4, entries 10–11).
To complete the study, optimization of the phenol reaction with nanobiohydrid Cu-CALB-PHOS-2 was performed (Table 5), because although the catalyst showed a low yield, it exhibited excellent selectivity of >99% (Table 1).
| Entry | H2O2b (eq.) | T (°C) | Co-solventc (%) | Time (h) | C (%) | [Phenol]e (mM) | Yieldf (%) | Selectivityg (%) |
|---|---|---|---|---|---|---|---|---|
| a Benzene (1 mmol), catalyst (5 mg), 10 mL of aqueous media, 24 h. b Amount of H2O2 (eq.). c Amount of acetonitrile as a co-solvent in water (v/v). d C: conversion of benzene. e Phenol concentration produced, calculated from a quantification calibration curve (Fig. S10†). f The phenol yield was calculated using HPLC. g The selectivity of phenol with respect to the formation of other byproducts was determined using HPLC. | ||||||||
| 1 | 1.25 | 30 | 1 | 24 | 4 | 4 | 6 | >99 |
| 2 | 1.25 | 30 | 20 | 24 | 10 | 2 | 3 | 21.5 |
| 3 | 1.25 | 40 | 20 | 24 | 23 | 13.5 | 19 | 58.5 |
| 4 | 2 | 30 | 20 | 24 | 28 | 16 | 22.2 | 57.7 |
| 5 | 2 | 40 | 20 | 24 | 25 | 13.8 | 19.2 | 55 |
First, the reaction was attempted using 20% co-solvent in aqueous media. However, the activity, and especially the selectivity, of the catalyst were drastically effected by the increase in the solvent in the reaction solution (Table 5, entry 2), with a phenol yield of 3% and 10% conversion. The low selectivity, 21.5%, reflects the ability of the catalyst to carry out further oxidation, and it is characterized by the formation of a black liquid corresponding to the oxidation of the previous catechol formed. Under these conditions, the increase in temperature of 10 °C improved the activity of Cu-CALB-PHOS-2, producing a 19% yield of phenol and increasing the selectivity from 21 to 58.5% (Table 5, entry 3). The addition of almost double the amount of hydrogen peroxide did not improve the efficiency of the catalyst, either at 30 or 40 °C (Table 5, entries 4–5).
Conclusions
A novel protein-direct synthetic route has been developed to make highly active, selective, stable and robust nanobiohybrids containing copper nanoparticles (CuNPs) using an enzyme in combination with copper sulfate in aqueous media at room temperature. Enzyme–Cu(II) intermediates, rapidly generated in aqueous media, are transformed, induced by the enzyme structure, to facilitate the formation of a particular Cu species, tailoring the size of the nanoparticles formed depending on the experimental conditions, e.g., the pH, reducing step, amount of enzyme or incubation time.Using this cost-effective and sustainable technology, it has been possible to prepare tailor-made Cu nanobiohybrids, containing Cu(0), Cu(I) or Cu3(PO4)2 nanoparticles on the multimilligram scale. Furthermore, the enzyme plays an important role in the final nanoparticle formation, also allowing the good dispersion of the nanoparticles, and determining their size, which is demonstrated in the decreasing of the nanoparticle size upon the use of a greater amount of protein.
These novel CuNP-CALB hybrids were tested as catalysts for the C–H functionalization of benzene, via selective monohydroxylation in the presence of hydrogen peroxide in aqueous media and at moderate temperature. Of the different Cu nanobiohybrids, Cu-CALB-PHOS-NR (Cu3(PO4)2 NPs) showed the best catalytic performance, producing 17.2 mM phenol at 24% conversion of benzene with a selectivity of 83% in 20% acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80% water as a solvent using 1.25 eq. of hydrogen peroxide at 30 °C. This result for a very difficult reaction opens up the possibility of the further application of these Cu nanohybrids in other types of chemical reactions, such as reductions, C–C bond formation, etc., to expand their applicability to Cu catalysis.
80% water as a solvent using 1.25 eq. of hydrogen peroxide at 30 °C. This result for a very difficult reaction opens up the possibility of the further application of these Cu nanohybrids in other types of chemical reactions, such as reductions, C–C bond formation, etc., to expand their applicability to Cu catalysis.
Experimental section
General
Lipase B from Candida antarctica (CAL-B) solution was purchased from Novozymes (Copenhagen, Denmark). Copper(II) sulfate pentahydrate [Cu2SO4·5H2O] and hydrogen peroxide (33%) were obtained from Panreac (Barcelona, Spain). P-Nitrophenol, p-nitrophenyl propionate, sodium bicarbonate, sodium phosphate, sodium borohydride, phenol, benzoquinone, hydroquinone and catechol were purchased from Sigma-Aldrich (St. Louis, MO, USA). HPLC grade acetonitrile was obtained from Scharlab (Barcelona, Spain). ICP-OES was performed on a OPTIMA 2100 DV instrument (PerkinElmer, Waltham, MA, USA). X-ray diffraction (XRD) patterns were obtained using a Texture Analysis D8 Advance Diffractometer (Bruker, Billerica, MA, USA) with CuKα radiation. Transmission electron microscopy (TEM) and high-resolution TEM microscopy (HRTEM) images were obtained on a 2100F microscope (JEOL, Tokyo, Japan) equipped with an EDX detector INCA x-sight (Oxford Instruments, Abingdon, UK). Interplanar spacing in the nanostructures was calculated via inversed Fourier transform using the GATAN digital micrograph program (Corporate Headquarters, Pleasanton, CA, USA). Scanning electron microscopy (SEM) was performed on a TM-1000 microscope (Hitachi, Tokyo, Japan). To recover the nanobiohybrids, a Biocen 22 R (Orto-Alresa, Ajalvir, Spain) refrigerated centrifuge was used. Spectrophotometric analyses were run on a V-730 spectrophotometer (JASCO, Tokyo, Japan). A HPLC pump PU-4180 (JASCO, Tokyo, Japan) was used. Analyses were run at 25 °C using an RI-4030 refractive index detector (JASCO, Tokyo, Japan).Another variation of the phosphate protocol was used, in which the reduction step was not performed, to obtain Cu-CALB-PHOS-NR. In addition to the last variation, an oxidation step was also performed instead of a reduction step via the addition of either 6 mL of a 500 mM solution of sodium hydroxide (NaOH) for 30 min or 6 mL of a 0.1 M solution of hydrogen peroxide (H2O2) for 30 min (60 μL of the H2O2 stock solution in 6 mL of distilled water), to obtain Cu-CALB-PHOS-NRNaOH and Cu-CALB-PHOS-NRH2O2, respectively. Another one of the variations was the reduction of sodium borohydride at 10%, and the addition of 6 mL of water containing NaBH4 (30 mg), to obtain Cu-CAL-B-PHOS10%R.
Different modifications of the protocol were made by changing the amount of enzyme. Initially, different nanobiohybrids were prepared using the same protocol as those of the bicarbonate or phosphate methods using double the amount of enzyme (3.6 mL CALB solution instead of 1.8 mL), to obtain Cu-CALB-PHOS-2 and Cu-CALB-BIC-2. Also using the phosphate method (with or without the reducing step), an alternative protocol was performed using 18 mL of enzyme (10 times more than in the original protocol described above) and 72 h as a reaction time to give Cu-CALB-PHOS-10 and Cu-CALB-PHOS-NR-10.
Characterization of the different Cu nanobiohybrids was performed via XRD, ICP-OES (Table S1†), TEM, HTEM and SEM analysis.
The reaction procedure was monitored by HPLC analysis of reaction samples withdrawn at different times. The analytical conditions involved the use of a Kromasil-C8 (150 × 4.6 mm and 5 μm Ø), at a flow of 1.0 mL min−1; and a mobile phase: 50% (v/v) ACN in MilliQ water, using a refractive index detector (JASCO). Before injection, 30 μl of sample was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in acetonitrile. Under these conditions the retention time of benzene was 7.10 min, phenol was 3.20 min, benzoquinone 2.70 min, hydroquinone was 2.3 min, catechol was 1.9 min, and H2O2 was 1.4 min. Pure commercial samples of the different substrates were used as standards. The yields were obtained by extrapolating the values through a calibration curve of phenol as a reaction product (R2 = 0.9901) and also considering the difference of a non-reactive amount of benzene as a substrate.
1 in acetonitrile. Under these conditions the retention time of benzene was 7.10 min, phenol was 3.20 min, benzoquinone 2.70 min, hydroquinone was 2.3 min, catechol was 1.9 min, and H2O2 was 1.4 min. Pure commercial samples of the different substrates were used as standards. The yields were obtained by extrapolating the values through a calibration curve of phenol as a reaction product (R2 = 0.9901) and also considering the difference of a non-reactive amount of benzene as a substrate.
Hot filtration catalyst experiment
To demonstrate the heterogeneity of the catalyst, a hot filtration experiment was performed. After 1 h reaction in the following conditions: 20% ACN/80% water, 1.25 eq. hydrogen peroxide, 30 °C, Cu-CALB-PHOS-NR catalyst was centrifuged and separate of the reaction. A small amount of water was added to the solid and this was incubated at 50 °C for 30 min. Then, the catalyst was centrifuged and all the solid was recovered. The supernatant of the reaction was analyzed by ICP-OES and no traces of copper was found.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Spanish Government, the Spanish National Research Council (CSIC), and SAMSUNG via the GRO PROGRAM 2017. The authors would like to thank to the COST Action CA15106 (CHAOS). We also thank Dr. Martinez from Novozymes.Notes and references
- G. Grass, C. Rensing and M. Solioz, Appl. Environ. Microbiol., 2011, 77, 1541–1547 CrossRef CAS PubMed
.
- L. Tamayo, M. Azócar, M. Kogan, A. Riveros and M. Páez, Mater. Sci. Eng. C Mater. Biol. Appl., 2016, 69, 1391–1409 CrossRef CAS
.
- J. Singh, T. Dutta, K.-H. Kim, M. Rawat, P. Samddar and P. Kumar, J. Nanobiotechnol., 2018, 16, 84 CrossRef CAS PubMed
.
- H. Xie, T. Wang, J. Liang, Q. Li and S. Sun, Nano Today, 2018, 21, 41–54 CrossRef CAS
.
- Z. Jia, Y. Li, Z. Zuo, H. Liu, C. Huang and Y. Li, Acc. Chem. Res., 2017, 50, 2470–2478 CrossRef CAS
.
- H. Chen, J. Motuzas, W. Martens and J. C. Diniz da Costa, Appl. Catal., B, 2018, 221, 691–700 CrossRef CAS
.
- K. Zhao, L. Duan, S. Xu, J. Jiang, Y. Fu and Z. Gu, Chem, 2018, 4, 599–612 CAS
.
- M. D. Marcinkowski, M. T. Darby, J. Liu, J. M. Wimble, F. R. Lucci, S. Lee, A. Michaelides, M. Flytzani-Stephanopoulos, M. Stamatakis and E. C. H. Sykes, Nat. Chem., 2018, 10, 325–332 CrossRef CAS
.
- X.-X. Guo, D.-W. Gu and Z. Wu, Chem. Rev., 2015, 115, 1622–1651 CrossRef CAS
.
- A. Gawande, M. B. Goswami, F.-X. Felpin, T. Asefa, X. Huang, R. Silva, X. Zou, R. Zboril and R. S. Varma, Chem. Rev., 2016, 116, 3722–3811 CrossRef
.
- S. Shylesh, V. Schnemann and W. R. Thiel, Angew. Chem., Int. Ed., 2010, 49, 342–359 Search PubMed
.
- F. Zaera, Chem. Soc. Rev., 2013, 42, 2746–2762 RSC
.
- G. Li, X. H. Li and Z. J. Zhang, Prog. Chem., 2011, 23, 1644–1656 CAS
.
- J. Mondal, A. Biswas, S. Chiba and Y. Zhao, Sci. Rep., 2015, 5, 8294 CrossRef CAS
.
- A. P. LaGrow, L. Sinatra, A. Elshewy, K. W. Huang, K. Katsiev, A. R. Kirmani, A. Amassian, D. H. Anjum and O. M. Bakr, J. Phys. Chem. C, 2014, 118, 19374–19379 CrossRef CAS
.
- A. X. Wang, D. Q. Chu, L. M. Wang, B. G. Mao, H. M. Sun and Z. C. Ma, RSC Adv., 2014, 4, 7545–7548 RSC
.
- S. R. Ye, A. R. Rathmell, I. E. Stewart, Y. C. Ha, A. R. Wilson, Z. F. Chen and B. J. Wiley, Chem. Commun., 2014, 50, 2562–2564 RSC
.
- F. Cui, Y. Yu, L. Dou, J. Sun, Q. Yang, C. Schildknecht, K. Schierle-Arndt and P. Yang, Nano Lett., 2015, 15, 7610–7615 CrossRef CAS
.
- J. C. Yu, F. G. Zhao, W. Shao, C. W. Ge and W. S. Li, Nanoscale, 2015, 7, 8811–8818 RSC
.
- M. A. Bhosale, T. Sasaki and B. M. Bhanage, Catal. Sci. Technol., 2014, 4, 4274–4280 RSC
.
- G. G. Jang, C. B. Jacobs, R. G. Gresback, I. N. Ivanov, H. M. Meyer, M. Kidder, P. C. Joshi, G. E. Jellison, T. J. Phelps, D. E. Graham and J. W. Moon, J. Mater. Chem. C, 2015, 3, 644–650 RSC
.
- R. Sankar, R. Maheswari, S. Karthik, K. S. Shivashangari and V. Ravikumar, Mater. Sci. Eng. C Mater. Biol. Appl., 2014, 44, 234–239 CrossRef CAS
.
- G. Jayakumarai, C. Gokulpriya, R. Sudhapriya, G. Sharmila and C. Muthukumaran, Appl. Nanosci., 2015, 5, 1017–1021 CrossRef CAS
.
- M. A. Ben Aissa, B. Tremblay, A. Andrieux-Ledier, E. Maisonhaute, N. Raouafi and A. Courty, Nanoscale, 2015, 7, 3189–3195 RSC
.
- S. S. Acharyya, S. Ghosh, R. Tiwari, C. Pendem, T. Sasaki and R. Bal, ACS Catal., 2015, 5, 2850–2858 CrossRef CAS
.
- R. Bal, M. Tada, T. Sasaki and Y. Iwasawa, Angew. Chem., Int. Ed., 2006, 45, 448–452 CrossRef CAS PubMed
.
- R. J. I. Schmidt, Appl. Catal., A, 2005, 280, 89–103 CrossRef CAS
.
- J. W. Han, J. Jung, Y.-M. Lee, W. Nam and S. Fukuzumi, Chem. Sci., 2017, 8, 7119 RSC
.
- J. Kumari, S. M. Mobin, S. Mukhopadhyay and K. M. Vyas, Inorg. Chem. Commun., 2019, 105, 217–220 CrossRef CAS
.
- L. Zhang, S. Qiu, G. Jiang, G. Jiang and R. Tang, Asian J. Org. Chem., 2018, 7, 165–170 CrossRef CAS
.
- W. H. Wanna, R. Ramu, D. Janmanchi, Y.-F. Tsai, N. Thiyagarajan and S. S. Yu, J. Catal., 2019, 370, 332–346 CrossRef CAS
.
- S. Farahmand, M. Ghiaci and J. S. Razavizadeh, Inorg. Chim. Acta, 2019, 484, 174–179 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cy02091h |
| This journal is © The Royal Society of Chemistry 2020 |