Open Access Article
Open Access ArticleSupply of methionine and arginine alters phosphorylation of mechanistic target of rapamycin (mTOR), circadian clock proteins, and α-s1-casein abundance in bovine mammary epithelial cells†
Liangyu
Hu
ab,
Yifei
Chen
a,
Ismael M.
Cortes
c,
Danielle N.
Coleman
b,
Hongyu
Dai
bd,
Yusheng
Liang
b,
Claudia
Parys
e,
Carlos
Fernandez
f,
Mengzhi
Wang
*a and
Juan J.
Loor
 *b
*b
aCollege of Animal Science and Technology, Yangzhou University, Yangzhou, 225009, P.R. China. E-mail: mengzhiwang@126.com
bDepartment of Animal Sciences and Division of Nutritional Sciences, University of Illinois, Urbana 61801, USA. E-mail: jloor@illinois.edu
cAgricultural and Animal Production Department, UAM-Xochimilco, Mexico City, Mexico 04960
dCollege of Veterinary Medicine, Nanjing Agricultural University, Nanjing, 210095, P.R. China
eEvonik Nutrition & Care GmbH, Hanau-Wolfgang, Germany
fAnimal Science Department, Universitàt Politècnica de Valencia, 46022 Valencia, Spain
First published on 16th January 2020
Abstract
Methionine (Met) and arginine (Arg) regulate casein protein abundance through alterations in activity of the mechanistic target of rapamycin complex 1 (mTORC1) signaling pathway. A potential role for the circadian clock network on the regulation of protein synthesis, partly via activity of mTORC1, has been highlighted in non-ruminants. The main objective of the study was to determine in ruminant mammary cells alterations in mRNA, protein abundance and phosphorylation status of mTORC1-related upstream targets, circadian clock proteins, and protein kinase AMP-activated catalytic subunit alpha (AMPK) in relation to α-s1-casein protein (CSN1S1) abundance in response to greater supply of Met and Arg alone or in combination. Primary bovine mammary epithelial cells (BMEC) were incubated for 12 h in a 2 × 2 arrangement of treatments with control media (ideal profile of amino acids, IPAA), or media supplemented with increased Met (incMet), Arg (incArg), or both (incMet + incArg). Data were analyzed testing the main effects of Met and Arg and their interaction. Among 7 amino acid (AA) transporters known to be mTORC1 targets, increasing supply of Arg downregulated SLC1A5, SLC3A2, SLC7A1, and SLC7A5, while increasing supply of Met upregulated SLC7A1. mRNA abundance of the cytosolic Arg sensor (CASTOR1) was lower when supply of Arg and Met alone increased. p-TSC2 (TSC complex subunit 2) was greater when the Arg supply was increased, while the phosphoralation ratio of p-AKT (AKT serine/threonine kinase 1):total (t) AKT and p-AMPK:tAMPK were lower. In spite of this, the ratio of p-mTOR:tmTOR nearly doubled with incArg but such response did not prevent a decrease in CSN1S1 abundance. The abundance of period circadian regulator 1 (PER1) protein nearly doubled with all treatments, but only incMet + incArg led to greater clock circadian regulator (CLOCK) protein abundance. Overall, data suggest that a greater supply of Met and Arg could influence CSN1S1 synthesis of BMEC through changes in the mTORC1, circadian clock, and AMPK pathways. Identifying mechanistic relationships between intracellular energy, total AA supply, and these pathways in the context of milk protein synthesis in ruminants merits further research.
Introduction
Amino acids (AA) can be used by the mammary gland as substrates to synthesize milk protein1 and as signaling factors to regulate casein protein gene transcription and translation through pathways2–4 such as the mechanistic target of rapamycin complex 1 (mTORC1). Methionine (Met) is identified frequently in dairy cows as the most-limiting AA for milk and milk protein synthesis,5,6 especially on highly corn-based diets.7 Arginine (Arg) belongs to the conditionally-essential amino acids (EAA)8 that indirectly could affect milk protein synthesis.9Providing a well-balanced supply of EAA can serve as an opportunity to increase the efficiency of metabolizable protein (MP) utilization for milk protein yield.10 Rulquin et al. (2007)11 proposed an ideal profile for intestinally-absorbed EAA in the form of percentage of MP in which a 7.3% Lys to 2.5% Met was deemed optimal. This relationship was subsequently tested by Haque et al. (2012)10 where balancing the EAA profiles was independent of MP supply levels. Nan et al. (2014)12 working with mammary cells provided data suggesting that a ratio of Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met at about 2.9
Met at about 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was optimum for enhancing casein concentration in vitro, partly via alterations in mRNA and protein abundance of mTORC1 targets. In the case of Arg, published recommendations range from 3.1%13 to 4.8%14 of MP, and Haque et al. (2013)15 subsequently concluded that compared with highest levels proposed a supply of 3.5% Arg of MP (Lys
1 was optimum for enhancing casein concentration in vitro, partly via alterations in mRNA and protein abundance of mTORC1 targets. In the case of Arg, published recommendations range from 3.1%13 to 4.8%14 of MP, and Haque et al. (2013)15 subsequently concluded that compared with highest levels proposed a supply of 3.5% Arg of MP (Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg at 2
Arg at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was not limiting for milk protein synthesis.
1) was not limiting for milk protein synthesis.
Recent in vitro studies reported a positive effect of greater Arg supply (Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg at 1
Arg at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on mRNA and protein abundance of various components of the milk protein synthesis network,16 but a negative impact of supplying Met beyond the 3
1) on mRNA and protein abundance of various components of the milk protein synthesis network,16 but a negative impact of supplying Met beyond the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of Lys
1 ratio of Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met.17 Although most published studies have focused on the optimal supply of Met and Arg with respect to the role of mTORC1 downstream factors on milk protein synthesis, recent research has identified important roles of factors upstream of mTORC1,18,19 AA transporters20 and AA sensors21,22 in the overall regulation of protein synthesis. Thus, detailed mechanisms encompassing not only mTORC1 in response to changes in AA supply merit further research.
Met.17 Although most published studies have focused on the optimal supply of Met and Arg with respect to the role of mTORC1 downstream factors on milk protein synthesis, recent research has identified important roles of factors upstream of mTORC1,18,19 AA transporters20 and AA sensors21,22 in the overall regulation of protein synthesis. Thus, detailed mechanisms encompassing not only mTORC1 in response to changes in AA supply merit further research.
The circadian clock, a mammalian internal timing system present in all organs,23 orchestrates temporal physiology to optimize the timing of metabolic processes.24–26 mTORC1 is capable of coupling environmental inputs, including AA availability, to the circadian clock and its signaling network. Data from a recent review of the non-ruminant literature underscored a reciprocal interplay between circadian clock and mTORC1 signaling,27 with a key premise being that mTORC1 activity is regulated by the circadian clock. In turn, the rhythmicity of mTORC1 and its downstream signaling components impact autonomous properties as well as core clock proteins both in suprachiasmatic nucleus (SCN) and some peripheral tissues (e.g. liver).28 Although not known, bidirectional regulation between circadian clock and mTORC1 may also be part of the mechanisms controlling milk protein synthesis in mammary gland. Thus, as underscored by the recognition that there are annual rhythms in milk yield and milk components,29 adding this dimension to research of mechanisms controlling milk protein synthesis is needed for an integrated understanding of mammary cell physiology.
In this study, we hypothesized that a greater supply of Met or Arg based on an ideal AA profile which has been used in cell culture studies from our laboratory17,30,31 stimulates casein protein abundance in bovine mammary epithelial cells (BMEC). We used mRNA and protein abundance analyses to explore the potential molecular mechanisms involved in the coordination of upstream components of mTORC1 signaling and the circadian clock network.
Materials and methods
Cell culture
The cell culture was performed using primary BMEC (kindly provided by Prof. Mengzhi Wang, Yangzhou University). The BMEC cells were allowed to grow in cell dishes (BioLite 100 mm, #130182, Thermo Scientific, Rochester, NY) at 37 °C and 5% CO2 to generate enough biological material, and were then transferred into 6-well plates (1.2 × 105 cells per well, #140675, Thermo Scientific, Waltham, MA) with basal media composed of high-glucose Dulbecco's modified Eagle's media (1, #11995, Gibco, Carlsbad, CA), 10% fetal bovine serum (#16140, Gibco, Invitrogen, Carlsbad, CA) and 1× penicillin/streptomycin (#sv30010, HyClone, South Logan, UT). Cells grew to approximately 90% confluence before imposing treatments as described previously.32Treatments and experimental design
The BMEC in 6-well plates (about 90% confluence) were serum-starved overnight and then incubated (n = 5 replicates per treatment) for 12 h with 4 modified lactogenic media as follows: media with ideal profile of AA10,15 as the control (IPAA; Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and treatment media was supplemented with increased Met (incMet, Lys
1), and treatment media was supplemented with increased Met (incMet, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.5
Met 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), Arg (incArg, Lys
1), Arg (incArg, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 1
Arg 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), or both (incMet + incArg; Lys
1), or both (incMet + incArg; Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.5
Met 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 1
Arg 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) based on the IPAA.
1) based on the IPAA.
The modified lactogenic media was prepared according to Dong et al. (2018).17,33 The 10 EAA (L-isomer, Sigma-Aldrich, St Louis, MO) were individually added into the custom high-glucose DMEM (devoid of these 10 EAA, custom made from Gibco, Carlsbad, CA). Treatment media were prepared by increasing only Met, Arg or both while keeping the concentrations of 7 other EAA (histidine, isoleucine, leucine, phenylalanine, threonine, tryptophan, valine) the same across all media (Table 1).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and treatments supplemented with increased Arg (incArg), Met (incMet), or both (incMet + incArg)
1) and treatments supplemented with increased Arg (incArg), Met (incMet), or both (incMet + incArg)
| Amino acid | Treatments | |||
|---|---|---|---|---|
| IPAAb | incArg | incMet | incMet + incArg | |
a The AA were individually added into the custom high-glucose Dulbecco's modified Eagle's media (DMEM) devoid of these 10 EAA. Concentrations of other AA in the custom high-glucose DMEM are glycine = 30 μg mL−1, L-cystine = 63 μg mL−1, L-glutamine = 584 μg mL−1, L-serine = 42 μg mL−1, and L-tyrosine = 104 μg mL−1.
b IPAA = ideal profile of AA, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9 Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys 1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2 Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; incArg = increased supply of Arg, Lys 1; incArg = increased supply of Arg, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9 Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys 1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 1 Arg 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; incMet = increased supply of Met, Lys 1; incMet = increased supply of Met, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.5 Met 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys 1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2 Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; incMet + incArg = increased supply both of Met and Arg, Lys 1; incMet + incArg = increased supply both of Met and Arg, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.5 Met 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys 1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 1 Arg 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Concentrations and ratios of other 7 AA were the same in all groups. 1. Concentrations and ratios of other 7 AA were the same in all groups.
|
||||
| Concentration (μg mL−1) | ||||
| L-Lysine | 175 | 175 | 175 | 175 |
| L-Methionine | 60 | 60 | 70 | 70 |
| L-Arginine | 84 | 175 | 84 | 175 |
| L-Histidine | 74 | 74 | 74 | 74 |
| L-Isoleucine | 121 | 121 | 121 | 121 |
| L-Leucine | 206 | 206 | 206 | 206 |
| L-Phenylalanine | 93 | 93 | 93 | 93 |
| L-Threonine | 97 | 97 | 97 | 97 |
| L-Tryptophan | 16 | 16 | 16 | 16 |
| L-Valine | 142 | 142 | 142 | 142 |
| Ratio | ||||
Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met Met |
2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg Arg |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
RNA extraction and quantitative RT-PCR analysis
After 12 h of incubations with treatments, BMEC were scraped using a cell lifter in 1 mL of QIAzol reagent (#79306, Qiagen, Valencia, CA) and total RNA was obtained using the RNeasy Mini kits (#74104, Qiagen, Valencia, CA). The RNA concentration and integrity were determined by ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples for subsequent analysis had an RNA Integrity Number (RIN) ≥8.0, and 100 ng of RNA was used for cDNA synthesis.Quantitative real-time polymerase chain reaction (qRT-PCR) was performed as described previously. The list of target genes is reported in ESI Table 1.† Briefly, 4 μL diluted cDNA was combined with 6 μL of a mixture composed of 5 μL 1× SYBR Green Master Mix (Quanta, Gaithersburg, MD), 0.4 μL each of 10 μM forward and reverse primers, and 0.2 μL RNase free water. Reaction mixtures were preheated at 95 °C for 5 min, followed by 40 cycles of melting at 95 °C for 15 s, and annealing at 60 °C for 30 s. The standard curve method and QuantStudio™ 7 Flex Real-Time PCR Software (Applied Biosystems, CA) were used for data analysis. The internal controls used for normalization of target gene abundance were glyceraldehyde-3-phosphate dehydrogenase (GADPH), ubiquitously expressed prefoldin like chaperone (UXT), and ribosomal protein S9 (RPS9). The expression of the internal control genes was not affected by treatment (P > 0.10). Normalization of the target gene abundance was performed by dividing their relative abundance by the geometric mean of the three internal control genes. Primers (ESI Table 1†) were designed and verified based on established protocols in our laboratory.34 qRT-PCR performance was reported in ESI Table 2.†
Western blotting
Complete details of these protocols are reported in our previous publications. Briefly, the BMEC were washed three times with phosphate-buffered saline, and total protein collected using ice-cold M-PER mammalian protein extraction reagent (#78501, Thermo Scientific, Waltham, MA) containing 1% halt protease and phosphatase inhibitor cocktail (#78440, Thermo Scientific). Lysates were centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C, and then the supernatants harvested for measuring and normalizing protein concentration with the BCA protein assay kit (#23227, Thermo Scientific). Proteins (20 μL of total protein) and 4× Laemmli sample buffer (#161-0747, Bio-Rad, Hercules, CA) were mixed at a ratio of 3
000g for 10 min at 4 °C, and then the supernatants harvested for measuring and normalizing protein concentration with the BCA protein assay kit (#23227, Thermo Scientific). Proteins (20 μL of total protein) and 4× Laemmli sample buffer (#161-0747, Bio-Rad, Hercules, CA) were mixed at a ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (wt
1 (wt![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) vol) and denatured at 95 °C for 10 min. Samples were subjected to 10% SDS/PAGE and then transferred to a PVDF membrane (#1620261, Bio-Rad) by using Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (#170-3940, Bio-Rad) for 25 min, and PVDF membranes were incubated with primary antibody (listed in ESI Table 3†) overnight at 4 °C after blocking for 2 h with 5% skim milk (#1706404XTU, Bio-Rad) for total proteins or 5% bovine serum albumin (#A7960, sigma, St Louis, MO) for phosphorylated proteins and casein at room temperature. The PVDF membranes were then washed 6 times (5 min per time) and incubated with secondary antibody for 1.5 h at room temperature before washing again. The visualization of PVDF membranes was performed using the ChemiDoc MP System (Bio-Rad) with Clarity Western ECL Substrate (#170-5060, Bio-Rad) reagent. Band intensities were quantified using Image Lab software (version 3.0, Bio-Rad), and phosphorylation state was calculated as the intensity ratio of phosphorylated protein to total protein (arbitrary units). The intensity of GAPDH was used to normalize target protein abundance.
vol) and denatured at 95 °C for 10 min. Samples were subjected to 10% SDS/PAGE and then transferred to a PVDF membrane (#1620261, Bio-Rad) by using Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (#170-3940, Bio-Rad) for 25 min, and PVDF membranes were incubated with primary antibody (listed in ESI Table 3†) overnight at 4 °C after blocking for 2 h with 5% skim milk (#1706404XTU, Bio-Rad) for total proteins or 5% bovine serum albumin (#A7960, sigma, St Louis, MO) for phosphorylated proteins and casein at room temperature. The PVDF membranes were then washed 6 times (5 min per time) and incubated with secondary antibody for 1.5 h at room temperature before washing again. The visualization of PVDF membranes was performed using the ChemiDoc MP System (Bio-Rad) with Clarity Western ECL Substrate (#170-5060, Bio-Rad) reagent. Band intensities were quantified using Image Lab software (version 3.0, Bio-Rad), and phosphorylation state was calculated as the intensity ratio of phosphorylated protein to total protein (arbitrary units). The intensity of GAPDH was used to normalize target protein abundance.
Statistical analysis
All data were analyzed as a 2 × 2 factorial arrangement of treatments using the MIXED procedure of SAS 9.4 (SAS Institute Inc., Cary, NC). The main factors were Met and Arg, with each containing 2 levels: “yes” the AA was greater or “no” it was the same as IPAA. The model tested the effects of Met, Arg and the interaction between them. Least square means and standard errors were determined using the LSMEANS statement and means were compared using Tukey's when a significant interaction between Met and Arg was observed. Variables were assessed for normality of distribution using the Shapiro–Wilk test. mRNA abundance data were log2-scale transformed to fit normal distribution of residuals. The tables contain the log2 back-transformed means that resulted from analysis. Significance was declared at P ≤ 0.05 and tendencies as P ≤ 0.10.Results
mRNA abundance
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative mRNA abundance (log2-scale)a of AA transporters, AA sensors, and components of AMPK–SIRT1 pathway
1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative mRNA abundance (log2-scale)a of AA transporters, AA sensors, and components of AMPK–SIRT1 pathway
| Gene | Treatment | P-Value | ||||||
|---|---|---|---|---|---|---|---|---|
| IPAA | incArg | incMet | incMet + incArg | SEM | Met | Arg | Met × Arg | |
| a mRNA data were log2-scale transformed to fit normal distribution of residuals, and analyzed least squares means (LSM) and standard errors (SEM) were log2 back-transformed. | ||||||||
| Amino acid (AA) transport | ||||||||
| SLC1A1 | 0.92 | 0.94 | 0.89 | 0.92 | 0.01 | 0.098 | 0.108 | 0.544 |
| SLC1A5 | 1.17 | 0.94 | 1.16 | 0.92 | 0.03 | 0.567 | <0.0001 | 0.829 |
| SLC3A2 | 1.10 | 0.89 | 1.14 | 0.98 | 0.04 | 0.069 | <0.0001 | 0.446 |
| SLC7A1 | 0.65 | 0.57 | 0.74 | 0.66 | 0.03 | 0.003 | 0.011 | 0.789 |
| SLC7A5 | 1.25 | 1.00 | 1.25 | 0.99 | 0.04 | 0.843 | <0.0001 | 0.913 |
| SLC38A2 | 1.01 | 1.03 | 1.06 | 1.09 | 0.04 | 0.140 | 0.579 | 0.820 |
| SLC38A9 | 1.02 | 0.95 | 0.94 | 0.95 | 0.03 | 0.202 | 0.270 | 0.178 |
| AA sensors and intracellular signaling | ||||||||
| CASTOR1 | 1.24 | 1.19 | 1.04 | 0.77 | 0.10 | 0.002 | 0.051 | 0.120 |
| MIOS | 1.26 | 1.00 | 1.08 | 1.01 | 0.07 | 0.180 | 0.014 | 0.150 |
| DEPDC5 | 1.06 | 1.13 | 1.05 | 1.13 | 0.03 | 0.876 | 0.007 | 0.752 |
| RRAGA | 1.19 | 1.16 | 1.14 | 1.14 | 0.02 | 0.011 | 0.281 | 0.498 |
| WDR24 | 1.02 | 0.96 | 0.90 | 0.90 | 0.02 | <0.0001 | 0.068 | 0.083 |
| TAS1R3 | 0.44 | 0.51 | 0.46 | 0.47 | 0.03 | 0.663 | 0.186 | 0.319 |
| PIK3C3 | 0.99 | 0.96 | 0.97 | 0.98 | 0.03 | 0.985 | 0.741 | 0.546 |
| AMPK–SIRT1 pathway genes | ||||||||
| SIRT1 | 1.27 | 1.10 | 1.24 | 1.10 | 0.03 | 0.644 | <0.0001 | 0.554 |
| NAMPT | 1.12 | 1.03 | 1.10 | 0.95 | 0.02 | 0.046 | <0.0001 | 0.143 |
| STK11 | 1.07 | 1.04 | 1.08 | 1.08 | 0.03 | 0.397 | 0.702 | 0.611 |
Protein abundance and phosphorylation status
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative protein abundancea of upstream regulators of mTORC1
1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative protein abundancea of upstream regulators of mTORC1
| Protein | Treatment | P-Values | ||||||
|---|---|---|---|---|---|---|---|---|
| IPAA | incArg | incMet | incMet + incArg | SEM | Met | Arg | Met × Arg | |
| a The intensity of GAPDH (arbitrary units) was used to normalize target protein abundance (i.e. target protein abundance = total target protein intensity/GAPDH intensity). | ||||||||
| CASTOR1 | 0.14 | 0.19 | 0.19 | 0.19 | 0.02 | 0.143 | 0.204 | 0.227 |
| MIOS | 0.57 | 0.55 | 0.50 | 0.59 | 0.05 | 0.776 | 0.388 | 0.179 |
| NPRL3 | 0.61 | 0.68 | 0.58 | 0.66 | 0.07 | 0.714 | 0.283 | 0.916 |
| RRAGA | 0.80 | 0.85 | 0.80 | 0.84 | 0.06 | 0.923 | 0.500 | 0.930 |
| PIK3C3 | 0.50 | 0.43 | 0.43 | 0.44 | 0.08 | 0.686 | 0.688 | 0.584 |
| TAS1R1 | 0.46 | 0.49 | 0.43 | 0.53 | 0.05 | 0.866 | 0.170 | 0.511 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative protein abundance and/or phosphorylation statusa of α-s1-casein and components of the mTOR, insulin, and energy metabolism pathways
1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative protein abundance and/or phosphorylation statusa of α-s1-casein and components of the mTOR, insulin, and energy metabolism pathways
| Protein | Treatment | P-Value | ||||||
|---|---|---|---|---|---|---|---|---|
| IPAA | incArg | incMet | incMet + incArg | SEM | Met | Arg | Met × Arg | |
| a–cMeans within a row with different superscripts differ (Met × Arg, P < 0.05).a The intensity of GAPDH (arbitrary units) was used to normalize target protein abundance (i.e. target protein abundance = total target protein intensity/GAPDH intensity). The phosphorylation status was calculated as the intensity ratio of phosphorylated protein to total protein (arbitrary units). | ||||||||
| Total | ||||||||
| CSN1S1 | 0.35a | 0.20b | 0.24ab | 0.30ab | 0.05 | 0.916 | 0.364 | 0.034 |
| ERK | 1.49 | 1.56 | 1.51 | 1.52 | 0.08 | 0.824 | 0.622 | 0.666 |
| AKT | 0.65 | 0.52 | 0.54 | 0.61 | 0.06 | 0.850 | 0.601 | 0.067 |
| AMPK | 0.73 | 0.54 | 0.71 | 0.82 | 0.11 | 0.213 | 0.691 | 0.170 |
| TSC2 | 0.75 | 0.56 | 0.62 | 0.66 | 0.07 | 0.866 | 0.280 | 0.097 |
| mTOR | 0.48a | 0.28b | 0.32b | 0.32b | 0.03 | 0.069 | 0.004 | 0.005 |
| Phosphorylated | ||||||||
| p-ERK | 1.44 | 1.79 | 1.49 | 1.62 | 0.20 | 0.739 | 0.209 | 0.546 |
| p-AKT | 0.66 | 0.45 | 0.55 | 0.36 | 0.08 | 0.219 | 0.019 | 0.898 |
| p-AMPK | 0.50a | 0.13b | 0.37a | 0.36ab | 0.06 | 0.392 | 0.004 | 0.006 |
| p-TSC2 | 0.07 | 0.15 | 0.11 | 0.17 | 0.02 | 0.193 | 0.006 | 0.600 |
| p-mTOR | 0.42ab | 0.48a | 0.42ab | 0.29b | 0.05 | 0.039 | 0.439 | 0.042 |
| Phosphorylated:total | ||||||||
| p-ERK:tERK | 0.98 | 1.14 | 0.98 | 1.08 | 0.12 | 0.797 | 0.255 | 0.788 |
| p-AKT:tAKT | 0.99 | 0.86 | 1.06 | 0.53 | 0.17 | 0.387 | 0.049 | 0.207 |
| p-AMPK:tAMPK | 0.71 | 0.33 | 0.55 | 0.43 | 0.08 | 0.744 | 0.007 | 0.115 |
| p-TSC2:tTSC2 | 0.08 | 0.27 | 0.19 | 0.25 | 0.03 | 0.158 | 0.001 | 0.064 |
| p-mTOR:tmTOR | 0.72b | 1.51a | 1.32ab | 0.90ab | 0.20 | 0.752 | 0.236 | 0.002 |
An overall effect of Arg on p-AKT (AKT serine/threonine kinase 1) (P = 0.02), p-AKT:tAKT (P = 0.049), and p-AMPK:tAMPK (P = 0.007) was observed where abundance was decreased when Arg supply increased. No statistical difference (P![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) >
>![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05) was detected for total protein abundance of AKT, AMPK, TSC2, extracellular regulated protein kinase (ERK), as well as phosphorylation status of ERK. Representative western blots are reported in ESI Fig. 2.†
0.05) was detected for total protein abundance of AKT, AMPK, TSC2, extracellular regulated protein kinase (ERK), as well as phosphorylation status of ERK. Representative western blots are reported in ESI Fig. 2.†
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met 2.9
Met 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, Lys
1, Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg 2
Arg 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative abundance of circadian clock-related proteinsa
1), and increased supply of Arg (incArg), Met (incMet), or both (incMet + incArg) on relative abundance of circadian clock-related proteinsa
| Protein | Treatment | P-Values | ||||||
|---|---|---|---|---|---|---|---|---|
| IPAA | incArg | incMet | incMet + incArg | SEM | Met | Arg | Met × Arg | |
| a–cMeans within a row with different superscripts differ (Met × Arg, P < 0.05).a The intensity of GAPDH (arbitrary units) was used to normalize target protein abundance (i.e. target protein abundance = total target protein intensity/GAPDH intensity). | ||||||||
| CLOCK | 0.15b | 0.10c | 0.11c | 0.22a | 0.02 | 0.001 | 0.059 | <0.0001 |
| ARNTL | 0.79 | 0.74 | 0.70 | 0.67 | 0.12 | 0.481 | 0.703 | 0.922 |
| CRY1 | 0.37 | 0.31 | 0.34 | 0.37 | 0.03 | 0.644 | 0.636 | 0.167 |
| PER1 | 0.27b | 0.51a | 0.46a | 0.40a | 0.05 | 0.037 | 0.003 | 0.008 |
| PER2 | 0.30 | 0.24 | 0.34 | 0.17 | 0.04 | 0.820 | 0.016 | 0.211 |
Discussion
Without supplemental rumen-protected Met, the ratio of Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met often ranges between 3.6
Met often ranges between 3.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3.8
1 and 3.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in corn silage-based diets.6,35 The currently-accepted IPAA ratio to maximize milk protein content and yield in established lactation is approximately 2.9
1 in corn silage-based diets.6,35 The currently-accepted IPAA ratio to maximize milk protein content and yield in established lactation is approximately 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met and is achieved by feeding rumen-protected Met.10–12,36 Increasing the post-ruminal supply of Met in dairy cows to levels in IPAA by feeding rumen-protected supplements has positive effects on production performance,6 milk protein content,37,38 and other physiological functions such as the immune response.39,40 As for Arg, compared with other EAA, there are inconsistent recommendations for optimal requirements.15 However, previous work indicated that a 2
Met and is achieved by feeding rumen-protected Met.10–12,36 Increasing the post-ruminal supply of Met in dairy cows to levels in IPAA by feeding rumen-protected supplements has positive effects on production performance,6 milk protein content,37,38 and other physiological functions such as the immune response.39,40 As for Arg, compared with other EAA, there are inconsistent recommendations for optimal requirements.15 However, previous work indicated that a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg would not be detrimental to milk protein synthesis.13–15
Arg would not be detrimental to milk protein synthesis.13–15
The present study aimed to mimic feeding conditions that could potentially be achieved in the field in order to modify the ratio of EAA available to mammary cells to test the effects of increased supply of Met (Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met = 2.5
Met = 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and Arg (Lys
1) and Arg (Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg = 1
Arg = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) using their profiles in the IPAA as a guide. Thus, the levels of Met and Arg used in the present study were in accordance with our previous work.16,17 Within those in vitro studies, although there was an overall positive effect on mammary cell metabolism, the negative effects on phosphorylation of 4E binding protein 1 (EIF4EBP1), eukaryotic elongation factor 2 (EEF2),16 mTOR,17 and mRNA abundance of β-casein and AA transporters17 were partly consistent with the present results. Similarly, despite using EAA profiles similar to those in IPAA, previous in vivo studies also reported results contrary to ours, for instance phosphorylation ratio of ribosomal protein S6 (RPS6) was lower at a Lys
1) using their profiles in the IPAA as a guide. Thus, the levels of Met and Arg used in the present study were in accordance with our previous work.16,17 Within those in vitro studies, although there was an overall positive effect on mammary cell metabolism, the negative effects on phosphorylation of 4E binding protein 1 (EIF4EBP1), eukaryotic elongation factor 2 (EEF2),16 mTOR,17 and mRNA abundance of β-casein and AA transporters17 were partly consistent with the present results. Similarly, despite using EAA profiles similar to those in IPAA, previous in vivo studies also reported results contrary to ours, for instance phosphorylation ratio of ribosomal protein S6 (RPS6) was lower at a Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met = 2.9
Met = 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 141 and milk protein synthesis or the efficiency of N utilization was not modified at an Lys
141 and milk protein synthesis or the efficiency of N utilization was not modified at an Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg = 2
Arg = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.15 Thus, it seems plausible that the decrease in CSN1S1 abundance observed in the present study in response to 2.5
1.15 Thus, it seems plausible that the decrease in CSN1S1 abundance observed in the present study in response to 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met and 1
Met and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg resembles the in vivo response. Clearly, from a practical production standpoint, the focus should be in reaching at the very least the levels of Met and Arg that are in the IPAA. However, depending on stage of lactation, e.g. early versus late lactation, additional supply of Met and Arg could benefit health and production performance.
Arg resembles the in vivo response. Clearly, from a practical production standpoint, the focus should be in reaching at the very least the levels of Met and Arg that are in the IPAA. However, depending on stage of lactation, e.g. early versus late lactation, additional supply of Met and Arg could benefit health and production performance.
AA transport and sensing
The availability of AA is instrumental to the efficiency of milk protein synthesis,42,43 with Met and Arg being two of the most-limiting and the most-extracted from blood by bovine mammary gland. Due to selective barrier function,44 AA require membrane-spanning AA transporters (AAT) for entry into and out of cells. The stage of lactation in dairy cows34 and AA profiles in the MP17 have been suggested to elicit some direct or indirect control over the transcription of AAT in mammary cells.Among 7 AAT known to be mTORC1 targets, the opposite response in SLC7A1 mRNA abundance (Arg and Lys transporter) elicited with incMet or incArg suggested a different sensitivity to additional supply of Arg and Met. The fact that SLC7A1 mRNA abundance was lower with greater Arg supply agrees with previous studies in ruminants45 and non-ruminants.46 The down-regulation in mRNA abundance of SLC7A5, SLC3A2, and SLC1A5 in response to greater Arg supply suggested a negative feedback on the uptake of AA when other EAA are constant, possibly because cells were trying to deal with too high concentrations of Met or Arg. We also recently detected similar responses with increased supply of Met in MAC-T cells.17
An important consideration to keep in mind is the fact that SLC3A2 and SLC7A5 counter-transport cationic AA out of mammary cells, suggesting the possibility that beyond a certain extracellular concentration the AAT mechanisms might become saturated. More importantly, because SLC7A5, SLC3A2 and SLC1A5 have a reciprocal regulatory connection with mTORC147 and are stimulated via mTOR activity when exogenous AA supplementation increases,48 it seems plausible that greater supply of Met and Arg to BMEC could influence milk protein synthesis potentially via regulating AA transport through its effect on transcription of those AAT.
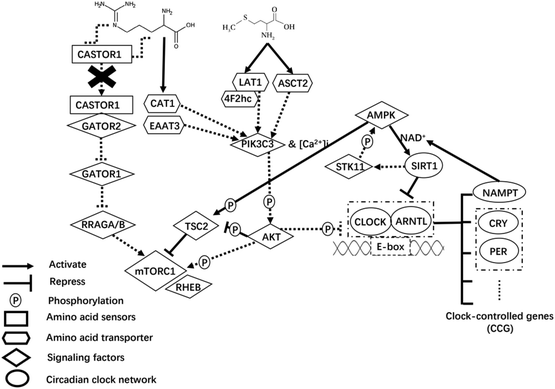
In addition to AAT-mediated regulation on milk protein synthesis,49 AA play a regulatory role by inducing changes in upstream mTORC1 signaling sensing components.50 Of note, the cytosolic Arg sensors for mTORC1 (i.e., CASTOR proteins) link this AA with the mTORC1 pathway.51 In fact, Arg disrupts the interaction of CASTOR dimers with the GATOR2 complex, presumably allowing free GATOR2 to inhibit the GATOR1 complex.52 The GATOR2 complex protein WDR24 is particularly important in sensing AA and activating mTORC1.53 Activation of GATOR2 further activates mTORC1 through the active Rag GTPases54 that aid in localizing mTORC1 to lysosomes where the protein Ras homolog, mTORC1 binding (RHEB) is located55 (Fig. 1).
In the present study, despite the lack of differences at the protein level, the lower mRNA abundance of CASTOR1 with increased supply of Arg and Met was consistent with greater phosphorylation status of mTOR, likely because less CASTOR1 interacting with GATOR2 alleviates their negative regulation on mTOR activity.56 Greater supply of Arg was associated with the dissociation of CASTOR1 from GATOR2, which might explain the lower abundance of CASTOR1. Because the protein abundance of CASTOR1, MIOS (subunit of GATOR2), and RRAGA did not differ due to Arg or Met alone or in combination, the observed differences at the mRNA abundance level are difficult to explain. However, the differences in p-mTOR:tmTOR and total CSN1S1 abundance indicate that greater activation of mTOR alone might be insufficient to enhance CSN1S1 synthesis. Whether the other 3 casein isoforms would respond in a similar fashion is unknown.
Upstream signaling from mTORC1
In non-ruminants, a large array of factors required for protein synthesis affects the mTORC1 signaling pathway via interacting with the tuberous sclerosis (TSC1/TSC2) complex57 in which TSC2 acts as a GTPase-activating protein towards RHEB to impact mTORC1 activity.58 Our theoretical model (Fig. 1) mainly focuses on two signaling pathways upstream of mTORC1 with TSC2 being a key integration point to link regulators that coordinate nutrient signals (AA availability), cellular energy status, and protein synthesis.Availability of AA could feed into the core PIK3C3-AKT pathway that assembles AAT and targets TSC2 functioning downstream of AKT and upstream of mTORC1.59 Upon activation of PIK3C3, AKT further activates mTORC1 through phosphorylating and inactivating TSC2 on Thr14,60,62 or directly phosphorylates mTORC1.61 The lack of statistically significant effect on mRNA and protein abundance of PIK3C3 and the fact that most AAT for Arg and Met measured in the present study were downregulated with the treatments suggests that intracellular AA availability might not have been enough. Other signaling events leading from activation of PIK3C3 to downstream changes in cell physiology, particularly the response of intracellular [Ca2+]i or insulin, merit detailed research.
The lower phosphorylation ratio of p-AKT:tAKT (i.e. less active AKT) with greater supply of Arg (particularly when combined with greater Met) might partly explain the greater p-TSC2 in the present study, i.e., less repression of AKT on TSC2. However, the fact that increased Arg supply also nearly doubled the p-mTOR:tmTOR ratio seems contradictory. As Sekulić et al. (2000)61 reported that phosphorylation of mTOR by AKT had no subsequent influence on downstream targets (e.g. RPS6K1 and EIF4EBP1), whether the greater p-mTOR:tmTOR might be the result of other kinases remains to be determined. The phosphorylation ratio changes in mTOR were due primarily to a reduction in total mTOR. In that context, too high concentrations of Met and Arg relative to the ideal AA profile might explain the decrease in CSN1S1 and total mTOR protein detected in the present study.
Because the response on p-AMPK:tAMPK ratio due to incArg was similar to p-AKT:tAKT, we speculate that intracellular glucose supply and its metabolism was sufficient to support utilization for anabolic purposes. The kinase AMPK, a conserved cellular energy sensor,62 phosphorylates and activates TSC2 that is required to inhibit mTORC163 and, hence, serves as a signaling nexus for energy homeostasis and nutrient-sensing that inhibits protein synthesis, the most energy-demanding anabolic process.64 Although we did not measure cellular energy status per se, the greater p-TSC2 with increased supply of Arg seems to confirm that activation of AMPK was not the sole regulator of TSC2. There might be other kinases specific to mammary cells that regulate this protein. For instance, ERK-dependent phosphorylation leads to dissociation of the TSC1–TSC2 complex, and markedly impairs the ability of TSC2 to inhibit mTOR signaling.65 However, contrary to previous work66 the possibility exists that ERK-TSC2 signaling was not necessary for controlling p-mTOR in response to increased supply Arg or Met. It is also noteworthy that the addition of Met to the incArg treatment (i.e. incArg + incMet) was needed to return CSN1S1 abundance to those of the control or incMet, i.e. in spite of achieving the greatest p-mTOR:tmTOR incArg led to lower CSN1S1 abundance. That response agrees with a recent report evaluating dosage of Met on CSN1S1 abundance in mammary cells.67
Circadian clock network
Circadian cyclic signals are generated by a series of well-defined oscillators based on transcription-translation feedback loops68 (TTFL, Fig. 1). At the center of this molecular machinery, the CLOCK and ARNTL proteins act as transcriptional activators of CLOCK-controlled genes (CCG), which encode repressors PER1 and CRY1.69,70 It is noteworthy to point out that metabolic regulation is not a mere output function of the circadian system, but nutrient or energy levels signal back to cellular clock network to reinforce circadian rhythmicity and to adapt physiology to temporal needs.71 Some signaling molecules, surrounding the core clock network, coordinate cellular nutrient and energy metabolism in mammals,72,73 of which AKT and AMPK were the main emphasis in the present study. Those are at the crossroads of upstream mTORC1 signaling and the circadian clock network.Besides phosphorylating mTORC1, the protein AKT can lengthen the circadian period when activated.74 In that context, AKT might be a point through which changes in protein synthetic metabolism impact the functioning of circadian clock. This would occur via phosphorylating CLOCK (Ser845) and ARNTL (Ser42), which subsequently influences their nuclear/cytoplasmic shuttling.75,76 In the present study, the lack of statistical changes in ARNTL protein abundance due to AA treatments was inconsistent with previous work where AKT deficiency decreased ARNTL abundance.77 Lower p-AKT protein with increased supply of Arg could alleviate inhibition on clock function, but the opposite response of CLOCK protein abundance with increased Met or Arg alone or in combination might have occurred because AKT might not be the sole regulator of CLOCK and ARNTL in mammary cells. The alteration of CLOCK and ARNTL abundance with increased supply of Met and Arg might occur mainly through other pathways. Future work should investigate the effects of Met and Arg on other regulators of CLOCK and ARNTL.
The AMPK protein transmits energy-dependent signals to the CLOCK partly through coordinating with SIRT1 and NAMPT to inhibit transcriptional activity of CLOCK–ARNTL heterodimers. A role for SIRT1 on the AMPK-mediated responses in mammary cells78 cannot be discounted. Active AMPK enhances SIRT1 activity by increasing cellular NAD+ levels through the upregulation of NAMPT, a rate-limiting enzyme in NAD+ biosynthesis.79–81 There is, in fact, a mutualistic interaction between activated SIRT1 and AMPK, because the former can activate AMPK via the enzyme STK11.82,83 In the context of the present study, the lower p-AMPK:tAMPK with increased Arg supply might have been responsible for the lower mRNA abundance of SIRT1 and NAMPT. Activated SIRT1 counteracts CLOCK and ARNTL function through its histone deacetylase activity, which inhibits transcriptional activity of CCG.84,85 This role of SIRT1 is particularly relevant because an important function of CLOCK–ARNTL heterodimers is to activate NAMPT.86,87 Therefore, SIRT1 seems to sit at the center of a loop coordinating the activity of AMPK and CLOCK–ARNTL complexes. Thus, lower expression of SIRT1 in this study may partly explain the greater abundance of CLOCK and PER1 proteins with greater supply of Met or Arg.
Overall, the aforementioned link between mTORC1-mediated protein synthesis and circadian clock feedback loops might partly occur through the AMPK protein, which coordinates intracellular energy levels. Because protein synthesis is closely associated with intracellular energy metabolism, it is reasonable to foresee future studies in which energy supply to the mammary gland (e.g. glucose) can be manipulated while holding EAA profiles constant to better study the role of AMPK in milk protein synthesis and the circadian clock network.
Conclusions
Increasing the supply of Met and Arg to reach ratios of 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met and 1
Met and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Lys
1 Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg while maintaining concentrations of other EAA constant induces alterations in mRNA abundance of AA transporters and activity of upstream AA-sensing components in the mTORC1 and circadian clock signaling pathways. The decrease of α-s1-casein protein abundance might indicate a response of cells to a high concentration of AA. From a practical standpoint, targeting the supply of Met and Arg to the ideal ratio of Lys
Arg while maintaining concentrations of other EAA constant induces alterations in mRNA abundance of AA transporters and activity of upstream AA-sensing components in the mTORC1 and circadian clock signaling pathways. The decrease of α-s1-casein protein abundance might indicate a response of cells to a high concentration of AA. From a practical standpoint, targeting the supply of Met and Arg to the ideal ratio of Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Met and Lys
Met and Lys![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg (i.e. IPAA) during established lactation should help optimize casein protein synthesis. However, recent data underscore the potential for enhancing the supply of Met and Arg beyond those in the IPAA as a way to enhance health and production performance, i.e. these EAA might exert a functional role beyond serving solely as precursors for protein synthesis. Considering that protein synthesis is one of the most energy-consuming cellular processes and intracellular energy levels link mTORC1-mediated protein synthesis and circadian clock feedback loops, future studies should control energy status and total AA supply to explore further the underlying mechanistic relationships in the context of milk protein synthesis.
Arg (i.e. IPAA) during established lactation should help optimize casein protein synthesis. However, recent data underscore the potential for enhancing the supply of Met and Arg beyond those in the IPAA as a way to enhance health and production performance, i.e. these EAA might exert a functional role beyond serving solely as precursors for protein synthesis. Considering that protein synthesis is one of the most energy-consuming cellular processes and intracellular energy levels link mTORC1-mediated protein synthesis and circadian clock feedback loops, future studies should control energy status and total AA supply to explore further the underlying mechanistic relationships in the context of milk protein synthesis.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
L. Hu was recipient of a 2017 Yangzhou University International Academic Exchange award and a Postgraduate Research & Practice Innovation Program of Jiangsu Province (CX137) to train at University of Illinois. L. Hu and M. Wang were supported by project from Natural Science Foundation of China (31672446). H. Dai and Y. Liang received scholarships from China Scholarship Council (Beijing, China) to undertake PhD training at University of Illinois.References
- A. Rius, J. Appuhamy, J. Cyriac, D. Kirovski, O. Becvar, J. Escobar, M. McGilliard, B. Bequette, R. Akers and M. Hanigan, Regulation of protein synthesis in mammary glands of lactating dairy cows by starch and amino acids, J. Dairy Sci., 2010, 93, 3114–3127 CrossRef CAS PubMed.
- Y. Moshel, R. E. Rhoads and I. Barash, Role of amino acids in translational mechanisms governing milk protein synthesis in murine and ruminant mammary epithelial cells, J. Cell. Biochem., 2006, 98, 685–700 CrossRef CAS PubMed.
- P. Nicklin, P. Bergman, B. Zhang, E. Triantafellow, H. Wang, B. Nyfeler, H. Yang, M. Hild, C. Kung and C. Wilson, Bidirectional transport of amino acids regulates mTOR and autophagy, Cell, 2009, 136, 521–534 CrossRef CAS PubMed.
- J. Appuhamy, W. Nayananjalie, E. England, D. Gerrard, R. Akers and M. Hanigan, Effects of AMP-activated protein kinase (AMPK) signaling and essential amino acids on mammalian target of rapamycin (mTOR) signaling and protein synthesis rates in mammary cells, J. Dairy Sci., 2014, 97, 419–429 CrossRef CAS.
- M. Hanigan, L. Crompton, B. Bequette, J. Mills and J. France, Modelling mammary metabolism in the dairy cow to predict milk constituent yield, with emphasis on amino acid metabolism and milk protein production: model evaluation, J. Theor. Biol., 2002, 217, 311–330 CrossRef CAS PubMed.
- J. S. Osorio, P. Ji, J. K. Drackley, D. Luchini and J. J. Loor, Supplemental Smartamine M or MetaSmart during the transition period benefits postpartal cow performance and blood neutrophil function, J. Dairy Sci., 2013, 96, 6248–6263 CrossRef CAS PubMed.
- T. L. Weekes, P. H. Luimes and J. P. Cant, Responses to amino acid imbalances and deficiencies in lactating dairy cows, J. Dairy Sci., 2006, 89, 2177–2187 CrossRef CAS PubMed.
- G. Wu, F. W. Bazer, T. A. Davis, S. W. Kim, L. Peng, J. M. Rhoads, M. C. Satterfield, S. B. Smith, T. E. Spencer and Y. Yin, Arginine metabolism and nutrition in growth, health and disease, Amino Acids, 2009, 37, 153–168 CrossRef CAS PubMed.
- T. B. Mepham, Amino acid utilization by lactating mammary gland, J. Dairy Sci., 1982, 65, 287–298 CrossRef CAS PubMed.
- M. Haque, H. Rulquin, A. Andrade, P. Faverdin, J. L. Peyraud and S. Lemosquet, Milk protein synthesis in response to the provision of an “ideal” amino acid profile at 2 levels of metabolizable protein supply in dairy cows, J. Dairy Sci., 2012, 95, 5876–5887 CrossRef CAS PubMed.
- H. Rulquin, G. Raggio, H. Lapierre and S. Lemosquet, Energy and Protein Metabolism and Nutrition, Wageningen Academic Publishers, Wageningen, 2007 Search PubMed.
- X. Nan, D. Bu, X. Li, J. Wang, H. Wei, H. Hu, L. Zhou and J. J. Loor, Ratio of lysine to methionine alters expression of genes involved in milk protein transcription and translation and mTOR phosphorylation in bovine mammary cells, Physiol. Genomics, 2014, 46, 268–275 CrossRef CAS PubMed.
- D. L. Fraser, E. R. Ørskov, F. G. Whitelaw and M. F. Franklin, Limiting amino acids in dairy cows given casein as the sole source of protein, Livest. Prod. Sci., 1991, 28, 235–252 CrossRef.
- L. Doepel, D. Pacheco, J. J. Kennelly, M. D. Hanigan, I. F. López and H. Lapierre, Milk protein synthesis as a function of amino acid supply, J. Dairy Sci., 2004, 87, 1279–1297 CrossRef CAS.
- M. N. Haque, H. Rulquin and S. Lemosquet, Milk protein responses in dairy cows to changes in postruminal supplies of arginine, isoleucine, and valine, J. Dairy Sci., 2013, 96, 420–430 CrossRef CAS PubMed.
- A. Salama, M. Duque, L. Wang, K. Shahzad, M. Olivera and J. J. Loor, Enhanced supply of methionine or arginine alters mechanistic target of rapamycin signaling proteins, messenger RNA, and microRNA abundance in heat-stressed bovine mammary epithelial cells in vitro, J. Dairy Sci., 2019, 102, 2469–2480 CrossRef CAS PubMed.
- X. Dong, Z. Zhou, B. Saremi, A. Helmbrecht, Z. Wang and J. J. Loor, Varying the ratio of Lys: Met while maintaining the ratios of Thr: Phe, Lys: Thr, Lys: His, and Lys: Val alters mammary cellular metabolites, mammalian target of rapamycin signaling, and gene transcription, J. Dairy Sci., 2018, 101, 1708–1718 CrossRef CAS PubMed.
- L. Deng, C. Jiang, L. Chen, J. Jin, J. Wei, L. Zhao, M. Chen, W. Pan, Y. Xu and H. Chu, The ubiquitination of RagA GTPase by RNF152 negatively regulates mTORC1 activation, Mol. Cell., 2015, 58, 804–818 CrossRef CAS PubMed.
- B. Carroll, D. Maetzel, O. D. Maddocks, G. Otten, M. Ratcliff, G. R. Smith, E. A. Dunlop, J. F. Passos, O. R. Davies and R. Jaenisch, Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity, eLife, 2016, 5, e11058 CrossRef PubMed.
- R. L. Wolfson and D. M. Sabatini, The dawn of the age of amino acid sensors for the mTORC1 pathway, Cell Metab., 2017, 26, 301–309 CrossRef CAS PubMed.
- S. Wang, Z. Y. Tsun, R. L. Wolfson, K. Shen, G. A. Wyant, M. E. Plovanich, E. D. Yuan, T. D. Jones, L. Chantranupong and W. Comb, Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1, Science, 2015, 347, 188–194 CrossRef CAS PubMed.
- R. L. Wolfson, L. Chantranupong, R. A. Saxton, K. Shen, S. M. Scaria, J. R. Cantor and D. M. Sabatini, Sestrin2 is a leucine sensor for the mTORC1 pathway, Science, 2016, 351, 43–48 CrossRef CAS PubMed.
- T. M. Casey, J. Crodian, E. Erickson, K. K. Kuropatwinski, A. S. Gleiberman and M. P. Antoch, Tissue-specific changes in molecular clocks during the transition from pregnancy to lactation in mice, Biol. Reprod., 2014, 90, 127 Search PubMed.
- K. A. Lamia, S. Kai-Florian and C. J. Weitz, Physiological significance of a peripheral tissue circadian clock, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15172–15177 CrossRef CAS PubMed.
- H. Cho, X. Zhao, M. Hatori, T. Y. Ruth, G. D. Barish, M. T. Lam, L. W. Chong, L. DiTacchio, A. R. Atkins and C. K. Glass, Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β, Nature, 2012, 485, 123–127 CrossRef CAS PubMed.
- C. Vollmers, R. Schmitz, J. Nathanson, G. Yeo, J. Ecker and S. Panda, Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome, Cell Metab., 2012, 16, 833–845 CrossRef CAS PubMed.
- R. Cao, mTOR Signaling, Translational control, and the circadian clock, Front. Genet., 2018, 9, 367 CrossRef PubMed.
- C. Ramanathan, N. D. Kathale, D. Liu, C. Lee, D. A. Freeman, J. B. Hogenesch, R. Cao and A. C. Liu, mTOR signaling regulates central and peripheral circadian clock function, PLoS Genet., 2018, 14, e1007369 CrossRef PubMed.
- I. Salfer, C. D. Dechow and K. J. Harvatine, Annual rhythms of milk and milk fat and protein production in dairy cattle in the United States, J. Dairy Sci., 2019, 102, 742–753 CrossRef CAS PubMed.
- S. Li, A. Hosseini, M. Danes, C. Jacometo, J. Liu and J. J. Loor, Essential amino acid ratios and mTOR affect lipogenic gene networks and miRNA expression in bovine mammary epithelial cells, J. Anim. Sci. Biotechnol., 2016, 7, 44 CrossRef PubMed.
- M. Vailati-Riboni, T. Xu, B. Qadir, R. Bucktrout, C. Parys and J. J. Loor, In vitro methionine supplementation during lipopolysaccharide stimulation modulates immunometabolic gene network expression in isolated polymorphonuclear cells from lactating Holstein cows, J. Dairy Sci., 2019, 102, 8343–8351 CrossRef CAS PubMed.
- A. K. G. Kadegowda, M. Bionaz, L. S. Piperova, R. A. Erdman and J. J. Loor, Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents, J. Dairy Sci., 2009, 92, 4276–4289 CrossRef CAS PubMed.
- X. Dong, Z. Zhou, L. Wang, B. Saremi, A. Helmbrecht, Z. Wang and J. J. Loor, Increasing the availability of threonine, isoleucine, valine, and leucine relative to lysine while maintaining an ideal ratio of lysine: methionine alters mammary cellular metabolites, mammalian target of rapamycin signaling, and gene transcription, J. Dairy Sci., 2018, 101, 5502–5514 CrossRef CAS PubMed.
- M. Bionaz and J. J. Loor, Gene networks driving bovine mammary protein synthesis during the lactation cycle, Bioinf. Biol. Insights, 2011, 5, 83–89 CAS.
- Z. Zhou, M. Vailati-Riboni, E. Trevisi, J. K. Drackley, D. Luchini and J. J. Loor, Better postpartal performance in dairy cows supplemented with rumen-protected methionine compared with choline during the peripartal period, J. Dairy Sci., 2016, 99, 8716–8732 CrossRef CAS.
- National Research Council, Nutrient Requirements of Dairy Cattle, National Academies Press, Washington, 2001 Search PubMed.
- F. Batistel, J. Arroyo, A. Bellingeri, L. Wang, B. Saremi, C. Parys, E. Trevisi, F. Cardoso and J. J. Loor, Ethyl-cellulose rumen-protected methionine enhances performance during the periparturient period and early lactation in Holstein dairy cows, J. Dairy Sci., 2017, 100, 7455–7467 CrossRef CAS PubMed.
- F. Batistel, J. Arroyo, C. Garces, E. Trevisi, C. Parys, M. Ballou, F. Cardoso and J. J. Loor, Ethyl-cellulose rumen-protected methionine alleviates inflammation and oxidative stress and improves neutrophil function during the periparturient period and early lactation in Holstein dairy cows, J. Dairy Sci., 2018, 101, 480–490 CrossRef CAS PubMed.
- Z. Zhou, F. Ferdous, P. Montagner, D. Luchini, M. Corrêa and J. J. Loor, Methionine and choline supply during the peripartal period alter polymorphonuclear leukocyte immune response and immunometabolic gene expression in Holstein cows, J. Dairy Sci., 2018, 101, 10374–10382 CrossRef CAS.
- M. Vailati-Riboni, Z. Zhou, C. Jacometo, A. Minuti, E. Trevisi, D. Luchini and J. J. Loor, Supplementation with rumen-protected methionine or choline during the transition period influences whole-blood immune response in periparturient dairy cows, J. Dairy Sci., 2017, 100, 3958–3968 CrossRef CAS PubMed.
- Y. Ma, F. Batistel, T. Xu, L. Han, R. Bucktrout, Y. Liang, D. Coleman, C. Parys and J. J. Loor, Phosphorylation of AKT serine/threonine kinase and abundance of milk protein synthesis gene networks in mammary tissue in response to supply of methionine in periparturient Holstein cows, J. Dairy Sci., 2019, 102, 4264–4274 CrossRef CAS PubMed.
- C. K. Reynolds, D. L. Harmon and M. J. Cecava, Absorption and delivery of nutrients for milk protein synthesis by portal-drained viscera, J. Dairy Sci., 1994, 77, 2787–2808 CrossRef CAS.
- B. J. Bequette, F. R. Backwell and L. A. Crompton, Current concepts of amino acid and protein metabolism in the mammary gland of the lactating ruminant, J. Dairy Sci., 1998, 81, 2540–2559 CrossRef CAS.
- P. M. Taylor, Role of amino acid transporters in amino acid sensing, Am. J. Clin. Nutr., 2014, 99, 223S–230S CrossRef CAS PubMed.
- H. Dai, D. Coleman, L. Hu, I. Martinez-Cortés, M. Wang, C. Parys, X. Shen and J. Loor, Methionine and arginine supplementation alter inflammatory and oxidative stress responses during lipopolysaccharide challenge in bovine mammary epithelial cells in vitro, J. Dairy Sci., 2020, 103, 676–689 CrossRef CAS.
- M. Hatzoglou, J. Fernandez, I. Yaman and E. Closs, Regulation of cationic amino acid transport: the story of the CAT-1 transporter, Annu. Rev. Nutr., 2015, 24, 377–399 CrossRef.
- B. C. Fuchs and B. P. Bode, Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime?, Semin. Cancer Biol., 2005, 15, 254–266 CrossRef CAS PubMed.
- L. Zheng, W. Zhang, Y. Zhou, F. Li, H. Wei and J. Peng, Recent advances in understanding amino acid sensing mechanisms that Regulate mTORC1, Int. J. Mol. Sci., 2016, 17, 1636 CrossRef PubMed.
- A. Suryawan and T. A. Davis, Regulation of protein synthesis by amino acids in muscle of neonates, Front. Biosci., 2011, 16, 1445–1460 CrossRef CAS PubMed.
- J. Averous, S. Lambert-Langlais, F. Mesclon, V. Carraro, L. Parry, C. Jousse, A. Bruhat, A. C. Maurin, P. Pierre and C. G. Proud, GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism, Sci. Rep., 2016, 6, 27698 CrossRef CAS PubMed.
- J. Xia, R. Wang, T. Zhang and J. Ding, Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling, Cell Discovery, 2016, 2, 16035 CrossRef CAS PubMed.
- L. Chantranupong, S. Scaria, R. Saxton, M. Gygi, K. Shen, G. Wyant, T. Wang, J. W. Harper, S. Gygi and D. Sabatini, The CASTOR proteins are arginine sensors for the mTORC1 pathway, Cell, 2016, 165, 153–164 CrossRef CAS PubMed.
- W. Cai, Y. Wei, M. Jarnik, J. Reich and M. A. Lilly, The GATOR2 component Wdr24 regulates TORC1 activity and lysosome function, PLoS Genet., 2016, 12, e1006036 CrossRef.
- K. Shen, R. K. Huang, E. J. Brignole, K. J. Condon, M. L. Valenstein, L. Chantranupong, A. Bomaliyamu, A. Choe, C. Hong and Z. Yu, Architecture of the human GATOR1 and GATOR1−Rag GTPases complexes, Nature, 2018, 556, 64–69 CrossRef CAS PubMed.
- Y. Sancak, T. R. Peterson, Y. D. Shaul, R. A. Lindquist, C. C. Thoreen, L. Bar-Peled and D. M. Sabatini, The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1, Science, 2008, 320, 1496–1501 CrossRef CAS PubMed.
- R. A. Saxton, L. Chantranupong, K. E. Knockenhauer, T. U. Schwartz and D. M. Sabatini, Mechanism of arginine sensing by CASTOR1 upstream of mTORC1, Nature, 2016, 536, 229–233 CrossRef CAS.
- K. Inoki, H. Ouyang, T. Zhu, C. Lindvall, Y. Wang, X. Zhang, Q. Yang, C. Bennett, Y. Harada and K. Stankunas, TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth, Cell, 2006, 126, 955–968 CrossRef CAS PubMed.
- J. Huang and B. D. Manning, The TSC1−TSC2 complex: a molecular switchboard controlling cell growth, Biochem. J., 2008, 412, 179–190 CrossRef CAS PubMed.
- S. Menon, C. C. Dibble, G. Talbott, G. Hoxhaj, A. J. Valvezan, H. Takahashi, L. C. Cantley and B. D. Manning, Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome, Cell, 2014, 156, 771–785 CrossRef CAS PubMed.
- B. D. Manning, A. R. Tee, M. N. Logsdon, J. Blenis and L. C. Cantley, Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway, Mol. Cell, 2002, 10, 151–162 CrossRef CAS.
- A. Sekulić, C. C. Hudson, J. L. Homme, P. Yin, D. M. Otterness, L. M. Karnitz and R. T. Abraham, A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells, Cancer Res., 2000, 60, 3504–3513 Search PubMed.
- B. B. Kahn, T. Alquier, D. Carling and D. G. Hardie, AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism, Cell Metab., 2005, 1, 15–25 CrossRef CAS.
- K. Inoki, T. Zhu and K.-L. Guan, TSC2 mediates cellular energy response to control cell growth and survival, Cell, 2003, 115, 577–590 CrossRef CAS PubMed.
- S. K. Hindupur, G. Asier and M. N. Hall, The Opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control, Cold Spring Harbor Perspect. Med., 2015, 7, a019141 CrossRef PubMed.
- L. Ma, Z. Chen, H. Erdjument-Bromage, P. Tempst and P. P. Pandolfi, Phosphorylation and functional inactivation of TSC2 by Erk: Implications for tuberous sclerosisand cancer pathogenesis, Cell, 2005, 121, 179–193 CrossRef CAS PubMed.
- M. Rolfe, L. E. McLEOD, P. F. Pratt and C. G. Proud, Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2), Biochem. J., 2005, 388, 973–984 CrossRef CAS PubMed.
- J. X. Yang, C. H. Wang, Q. B. Xu, F. Q. Zhao, J. X. Liu and H. Y. Liu, Methionyl-Methionine promotes α-s1 casein synthesis in bovine mammary gland explants by enhancing intracellular substrate availability and activating JAK2-STAT5 and mTOR-mediated signaling pathways, J. Nutr., 2015, 145, 1748–1753 CrossRef CAS PubMed.
- C. H. Ko and J. S. Takahashi, Molecular components of the mammalian circadian clock, Hum. Mol. Genet., 2006, 15, R271–R277 CrossRef CAS.
- K. Yagita, F. Tamanini, G. T. Van der Horst and H. Okamura, Molecular mechanisms of the biological clock in cultured fibroblasts, Science, 2001, 292, 278–281 CrossRef CAS PubMed.
- N. Preitner, F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht and U. Schibler, The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator, Cell, 2002, 110, 251–260 CrossRef CAS.
- H. Reinke and G. Asher, Crosstalk between metabolism and circadian clocks, Nat. Rev. Mol. Cell Biol., 2019, 20, 227–241 CrossRef CAS PubMed.
- A. Balsalobre, S. A. Brown, L. Marcacci, F. Tronche, C. Kellendonk, H. M. Reichardt, G. Schütz and U. Schibler, Resetting of circadian time in peripheral tissues by glucocorticoid signaling, Science, 2000, 289, 2344–2347 CrossRef CAS PubMed.
- K. A. Lamia, U. M. Sachdeva, L. DiTacchio, E. C. Williams, J. G. Alvarez, D. F. Egan, D. S. Vasquez, H. Juguilon, S. Panda and R. J. Shaw, AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation, Science, 2009, 326, 437–440 CrossRef CAS PubMed.
- X. Zheng and A. Sehgal, AKT and TOR signaling set the pace of the circadian pacemaker, Curr. Biol., 2010, 20, 1203–1208 CrossRef CAS PubMed.
- R. Ye, C. P. Selby, Y. Y. Chiou, I. Ozkan-Dagliyan, S. Gaddameedhi and A. Sancar, Dual modes of CLOCK: BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock, Genet. Dev., 2014, 28, 1989–1998 CrossRef CAS PubMed.
- A. K. Luciano, W. Zhou, J. M. Santana, C. Kyriakides, H. Velazquez and W. C. Sessa, CLOCK phosphorylation by AKT regulates its nuclear accumulation and circadian gene expression in peripheral tissues, J. Biol. Chem., 2018, 293, 9126–9136 CrossRef CAS PubMed.
- F. Dang, X. Sun, X. Ma, R. Wu, D. Zhang, Y. Chen, Q. Xu, Y. Wu and Y. Liu, Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock, Nat. Commun., 2016, 7, 12696 CrossRef CAS PubMed.
- J. W. Mcfadden and B. A. Corl, Activation of AMP-activated protein kinase (AMPK) inhibits fatty acid synthesis in bovine mammary epithelial cells, Biochem. Biophys. Res. Commun., 2009, 390, 388–393 CrossRef CAS PubMed.
- M. Fulco, Y. Cen, P. Zhao, E. P. Hoffman, M. W. Mcburney, A. A. Sauve and V. Sartorelli, Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of nampt, Dev. Cell, 2008, 14, 661–673 CrossRef CAS PubMed.
- C. Cantó, Z. Gerhart-Hines, J. N. Feige, M. Lagouge, L. Noriega, J. C. Milne, P. J. Elliott, P. Puigserver and J. Auwerx, AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity, Nature, 2009, 458, 1056–1060 CrossRef PubMed.
- S. R. Costford, S. Bajpeyi, M. Pasarica, D. C. Albarado, S. C. Thomas, H. Xie, T. S. Church, S. A. Jubrias, K. E. Conley and S. R. Smith, Skeletal muscle NAMPT is induced by exercise in humans, Am. J. Physiol. Endocrinol. Metab., 2010, 298, E117–E126 CrossRef CAS PubMed.
- X. Hou, S. Xu, K. A. Maitland-Toolan, K. Sato, B. Jiang, Y. Ido, F. Lan, K. Walsh, M. Wierzbicki and T. J. Verbeuren, SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase, J. Biol. Chem., 2008, 283, 20015–20026 CrossRef CAS PubMed.
- F. Lan, J. M. Cacicedo, N. Ruderman and Y. Ido, SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1 possible role in AMP-activated protein kinase activation, J. Biol. Chem., 2008, 283, 27628–27635 CrossRef CAS PubMed.
- G. Asher, D. Gatfield, M. Stratmann, H. Reinke, C. Dibner, F. Kreppel, R. Mostoslavsky, F. W. Alt and U. Schibler, SIRT1 regulates circadian clock gene expression through PER2 deacetylation, Cell, 2008, 134, 317–328 CrossRef CAS PubMed.
- Y. Nakahata, M. Kaluzova, B. Grimaldi, S. Sahar, J. Hirayama, D. Chen, L. P. Guarente and P. Sassone-Corsi, The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control, Cell, 2008, 134, 329–340 CrossRef CAS PubMed.
- Y. Nakahata, S. Sahar, G. Astarita, M. Kaluzova and P. Sassone-Corsi, Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1, Science, 2009, 324, 654–657 CrossRef CAS PubMed.
- K. M. Ramsey, J. Yoshino, C. S. Brace, D. Abrassart, Y. Kobayashi, B. Marcheva, H.-K. Hong, J. L. Chong, E. D. Buhr and C. Lee, Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis, Science, 2009, 324, 651–654 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9fo02379h |
| This journal is © The Royal Society of Chemistry 2020 |