 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Near-infrared fluorescent protein and bioluminescence-based probes for high-resolution in vivo optical imaging
Dhermendra K.
Tiwari
 *a,
Manisha
Tiwari
a and
Takashi
Jin
*a,
Manisha
Tiwari
a and
Takashi
Jin
 *b
*b
aDepartment of Biotechnology, Faculty of Life Sciences and Environment, Goa University, Taleigao Plateau, Goa 403206, India. E-mail: dhermendratiwari@gmail.com
bRIKEN Centre for Biosystems Dynamics Research, RIKEN, Furuedai 6-2-3, Suita, Osaka 565-0874, Japan. E-mail: tjin@riken.jp
First published on 16th June 2020
Abstract
In the last few years, high-resolution near-infrared (NIR) optical imaging has become an indispensable modality for non-invasive visualization of deep tissues both in fundamental life science and preclinical research. This is due to the high tissue permeability, low absorption and low scattering of NIR light as well as the low autofluorescence in the NIR wavelength region (700–1400 nm) in living systems. Compared to magnetic resonance imaging (MRI), X-ray computer tomography (X-ray CT), and positron emission tomography (PET), NIR optical imaging has a high spatiotemporal resolution (∼μm) enough to visualize cellular dynamics at the whole-body level. Additionally, NIR optical imaging does not require high-energy ionizing radiation, such as X-rays, that leads to serious radiation damage of living cells. Furthermore, NIR optical imaging can easily achieve molecular imaging with the aid of NIR optical probes, which specifically bind to biomarkers expressed on cell surfaces. Thus, NIR optical imaging has great potential to be used for non-invasive optical diagnostics of diseases in medical and clinical fields. For such NIR optical imaging, NIR fluorescent probes with high brightness and biocompatibility are crucial. Although a variety of NIR imaging probes based on nanoparticles such as quantum dots and dye-incorporated polymers have been developed, possible applications of these imaging probes to optical contrast agents are limited due to their cytotoxicity. In contrast, fluorescent proteins and bioluminescence-based probes are highly biocompatible and practical for biomedical applications. During the last few years, a variety of NIR optical probes based on engineered proteins have been reported for fluorescence and/or bioluminescence in vivo imaging. This review describes the recent progress on NIR fluorescent proteins and bioluminescence-based probes for high-resolution in vivo optical imaging. The review also covers several cutting-edge optical imaging techniques using NIR fluorescent proteins and bioluminescent probes.
1. Introduction
NIR optical imaging of living systems is widely used for tracking of live phenomena, detection of cancerous tumours, and secure handling of surgery.1–3 Currently, NIR optical imaging has become an indispensable modality for the non-invasive imaging of deep tissues in fundamental life science and preclinical research. Compared to other conventional imaging modalities such as MRI, X-ray CT, and PET, NIR optical imaging has a high spatiotemporal resolution (∼μm) enough to visualize molecular and cellular dynamics at the whole-body level.4,5 Recent development in NIR fluorescent proteins (FPs) and bioluminescence-based probes permits non-invasive molecular imaging in vivo, where it is difficult to achieve high-resolution in vivo molecular imaging with the conventional imaging modalities such as MRI and X-ray CT.To date, conventional FPs emitting in the visible region have been commonly used for imaging of live cells.6 However, visible-emitting FPs are not suitable for tissue imaging at the whole-body level,7,8 because of the strong tissue absorption and scattering of visible light (400–700 nm). In contrast, NIR light in the wavelength range of 700–1400 nm, the so-called “first and second NIR window” (Fig. 1a), is preferable for deep-tissue imaging.1,9–11 This is due to the high permeability, and low absorption and scattering of NIR light in living tissues in addition to the low tissue autofluorescence in the NIR wavelength region (Fig. 1b and c).10–15 These superior properties of NIR light lead to clearer deep-tissue imaging with high signal-to-noise (S/N) ratios (Fig. 1d).15
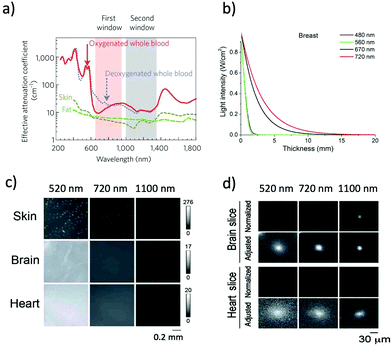 | ||
| Fig. 1 (a) The first and second NIR window ranging from 700 nm to 1400 nm shows minimal light absorption by deoxy-hemoglobin (Hb), hemoglobin (HbO2), and water. Reproduced with permission from ref. 11 (Copyright 2009, Springer-Nature). (b) Attenuation of light in breast tissue. Reproduced from ref. 14 (Copyright 2016, Creative Commons). (c) Autofluorescencee from skin, brain, and heart tissues. The images were taken at a wavelength of 520 nm, 720 nm, and 1100 nm by excitation at 482 nm, 670 nm, and 785 nm, respectively. (d) Microscopic fluorescence images of QD-doped porous beads (15 μm in diameter) and their passage images through brain and heart tissues. The images were taken at a wavelength of 520 nm, 720 nm, and 1100 nm by excitation at 482 nm, 670 nm, and 785 nm, respectively. The slice thicknesses of the brain and heart are 100 and 200 μm, respectively. Normalized images show the fluorescence intensities normalized against the intensities in the absence of tissue slices. Adjusted images show the fluorescence intensities normalized to the peak to peak intensity. Figure (c) and (d) are reproduced with permission from ref. 15 (Copyright 2014, Royal Society of Chemistry). | ||
In the last few years, a variety of NIR FPs have been developed for non-invasive in vivo imaging in animals.16–25 NIR FPs have several advantages over chemically synthesized fluorescent dyes. NIR FPs can be genetically expressed in living systems with cellular protein tags, which help in tracking the activities of a gene of interest inside living cells. So far, many effective NIR FPs have been developed from bacterial phytochromes, leading to various applications in NIR in vivo imaging. These NIR FPs are categorized into three groups, namely, dimeric NIR FPs (iRFPs and IFPs),7,17,18 monomeric NIR FPs (miRFPs, mIFP, miRFPnano and Wi-Phy),19–24 and photo-switchable NIR FPs (PAiRFPs).25 Most of the NIR FPs have high extinction coefficients. Therefore their relatively low quantum yields are compensated by their high extinction coefficients for live imaging applications. Among the available NIR-FPs, miRFP680 is the brightest FP. All NIR-FPs emit fluorescence by utilizing endogenous chromophores in eukaryotic and mammalian cells.
In addition to NIR FPs, bioluminescence-based probes can also be used for non-invasive in vivo imaging in the NIR window. Unlike NIR FPs, bioluminescent proteins do not need external excitation, leading to high signal to background imaging because of almost no tissue autofluorescence. However, only a limited number of bioluminescent proteins are available for non-invasive in vivo imaging in the NIR window. Most bioluminescent protein–substrate systems emit luminescence in the visible region.26 For example, firefly luciferase/D-luciferin and Renilla luciferase/coelenterazine (CTZ) systems emit bioluminescence at around 480 nm and 560 nm, respectively.26 However, red-shifted bioluminescent protein–substrate pairs (luciferase/luciferin) have recently been developed for in vivo imaging by modification of the chemical structures of luciferins to emit in the longer wavelength region.27–32 To date, a variety of luciferin analogues with extended π-conjugation have been synthesized to achieve bioluminescence in vivo imaging in living systems.33–35 For instance, a red-shifted luciferin analogue (AkaLumine) emits in the NIR region over 700 nm, which was used for single-cell bioluminescence imaging in freely moving animals.34 Red-shifted luciferin analogues were also used for NIR in vivo imaging of cancer cells and cancer metastasis in living mice.30,35
The use of bioluminescence resonance energy transfer (BRET) is an alternative method to achieve bioluminescence imaging beyond the visible region.36,37 BRET consists of a luciferase/luciferin system as an energy donor and a NIR fluorophore as an energy acceptor. Several types of NIR fluorophores such as organic dyes, NIR FPs, and quantum dots (QDs) have been used as energy acceptors in the BRET system.38 Compared to organic dyes and nanoparticle-based fluorophores such as QDs, NIR FPs can be genetically encoded into cells and tissues in living systems. The genetically encoded NIR FPs are relatively easy to use to label specific biomolecules and organelles to track their functional roles in living systems.
Owing to the development of NIR FPs and bioluminescence-based probes, a variety of high-resolution in vivo imaging techniques in the NIR region have been developed. These include wide field/confocal NIR fluorescence microscopy, super-resolution microscopy, and light sheet fluorescence microscopy. These imaging techniques have been applied to high-resolution in vivo imaging to visualize the three-dimensional structure of molecular and cellular dynamics at a sub-micrometer scale. In this review, we present the recent progress on NIR FPs and bioluminescence-based probes as well as high-resolution in vivo imaging techniques.
2. NIR FP-based probes for in vivo imaging
In the past two decades, genetically encoded FPs have been widely used for visualization of a variety of biological functions such as calcium ion fluctuation, protein–protein interactions, and cell-cycle study in living cells.6 Recently, several types of NIR FPs have been developed for their applications in non-invasive in vivo imaging.16 Compared to visible light, NIR light (wavelength: 700–1400 nm) is relatively permeable in living tissues because of its low absorption and low scattering in tissues. In addition, tissue autofluorescence in the NIR region is very weak compared to that in the visible region (wavelength: 400–700 nm) (Fig. 1c). By combining NIR FPs with advanced imaging techniques such as two-photon microscopy, non-invasive deep-tissue imaging enables a high spatial resolution of ∼μm at millimeter depths. NIR FPs for in vivo imaging can be categorized into three groups, dimeric NIR proteins, monomeric NIR proteins, and photo-switchable NIR proteins (Tables 1 and 2).| NIR-FPs | Characteristics | |||||||
|---|---|---|---|---|---|---|---|---|
| Excitation (nm) | Emission (nm) | QY (%) | Dimeric state | Extinction coefficient (M−1 cm−1) | Molecular brightness vs. miRFP720 (%) | Brightness in mammalian cells vs. miRFP720 (%) | Ref. | |
| a An exogenous supply of biliverdin chromophore is required for fluorescence emission in eukaryotic and mammalian cells. | ||||||||
| emiRFP670 | 642 | 670 | 14 | Monomeric | 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
204.7 | 117 | 42 |
| miRFP670 | 642 | 670 | 14 | Monomeric | 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
204.7 | 67.9 | 19 |
| miRFP670-2 | 643 | 670 | 13.6 | Monomeric | 103![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
234.3 | 80.8 | 42 |
| miRFP670nano | 645 | 670 | 10.8 | Monomeric | 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
171.9 | 67.9 | 23 |
| miRFP680 | 661 | 680 | 14.5 | Monomeric | 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
228 | 156.7 | 42 |
| miRFP682 | 663 | 682 | 11.2 | Monomeric | 91![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
170.5 | 101.8 | 42 |
| iRFP702 | 673 | 702 | 8.2 | Dimeric | 93![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
56 | 17 | |
| miRFP702 | 673 | 702 | 8.1 | Monomeric | 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
119.2 | 40.6 | 20 |
| emiRFP703 | 674 | 703 | 8.6 | Monomeric | 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
130.8 | 77.6 | 42 |
| miRFP703 | 674 | 703 | 8.6 | Monomeric | 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
130.8 | 49.8 | 42 |
| miRFP709 | 683 | 709 | 5.4 | Monomeric | 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
79.8 | 29 | 19 |
| mIFP | 683 | 705 (704) | 6.9 (8.4) | Monomeric | 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (82 900 (82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) 000) |
114.6 | 17.6 | 22 |
| IFP1.4a | 684 | 708 | 7 | Dimeric | 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
NA | NA | 18 |
| IFP2.0a | 688 (690) | 709 (711) | 6.8 (8.1) | Dimeric | 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 (98 900 (98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) 000) |
NA | NA | 18 |
| iRFP713 (akaiRFP) | 690 | 713 | 6.3 | Dimeric | 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
NA | 90 | 7 |
| miRFP713 | 690 | 713 | 7 | Monomeric | 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
115.9 | 94.9 | 42 |
| Wi-Phya | 701 | 719 | 4.7 | Monomeric | 92![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 991 991 |
NA | NA | 24 |
| miRFP720 | 702 | 720 | 6.1 | Monomeric | 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
100 | 100 | 21 |
| iRFP720 | 702 | 720 | 6 | Dimeric | 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
NA | 96 | 17 |
| Properties | NIR-FP | |
|---|---|---|
| PAiRFP1 | PAiRFP2 | |
| Excitation-max (nm) | 373/659 (relaxed) | 383/657 (relaxed) |
| 390/690 (photoactivatable) | 389/692 (photoactivatable) | |
| Emission-max (nm) | 703 (relaxed) | 708 (relaxed) |
| 717 (photoactivatable) | 719 (photoactivatable) | |
| QY (%) | 4.8 | 4.7 |
| Dimeric state | Monomeric | Monomeric |
| Molecular brightness | ND | ND |
| Extinction coefficient (M−1 cm−1) | 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 (relaxed) 700 (relaxed) |
39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (relaxed) 500 (relaxed) |
67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (excited) 100 (excited) |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 (excited) 600 (excited) |
|
| Half time for conversion | 58 minutes | 233 minutes |
| Photoconversion | Reversible | Reversible |
| Fate of conversion | Weakly fluorescent to highly fluorescent | Weakly fluorescent to highly fluorescent |
| Ref. | 25 | 25 |
2.1. Dimeric NIR FPs
More than twenty NIR FPs are now available. These NIR FPs can be selected based on the user's choice and the type of biological imaging applications. Typical NIR FPs are summarised in Table 1. Among them, iRFP720 is the most far-red emitting dimeric FP (34.6 kDa) which emits in the conventional NIR region (Exmax: 702 nm and Emmax: 720 nm).17 This is a dimeric NIR FP based on iRFP713, which is derived from the RpBphP2 phytochrome photoreceptor of Rhodopseudomonas palustris39 (Fig. 2). iRFP720 requires the cofactor biliverdin for fluorescence emission. Because of its low acid sensitivity, it can also be utilized for specific applications where the cellular environment is acidic.17 iRFP720 works well with multicolor in vivo imaging with other iRFP variants fused with several cellular protein tags.17 However, this protein has limited use for in vivo tracking and protein dynamics study due to its dimeric nature. Another variant, iRFP702, has among the highest quantum yields (QY = 8.2%);17,18 iRFP720 gives superior brightness in in vivo imaging owing to the NIR-shifted emission, weak autofluorescence, low scattering and better tissue penetration of the NIR light.40,41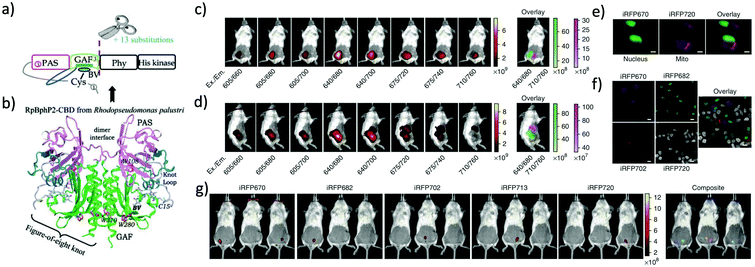 | ||
| Fig. 2 (a) iRFP is engineered from R. palustris bacterial phytochrome RpBphP2 by removal of the PHY and His kinase domains, and consists only of the CBD comprising the PAS and GAF domains, which differs from the CBD of wildtype RpBphP2 by 13 amino acid substitutions. (b) The 3D structure of the CBD of bacterial phytochrome RpBphP2. Figure (a) and (b) are reproduced with permission from ref. 39 (Copyright 2014, FEBS Journal). (c) and (d) Multicolor imaging of a living mouse with two injected tumors expressing iRFP670 (left tumor) and iRFP720 (right tumor). (e) Microscopy of HeLa cells coexpressing iRFP670 and iRFP720 in the nucleus and mitochondria (Mito). Pseudocolor images (iRFP670 channel in green and iRFP720 channel in magenta) and the overlay are shown. Scale bars, 10 μm. (f) Confocal microscopy with spectral detection and linear unmixing of MTLn3 cells expressing iRFP670, iRFP682, iRFP702 and iRFP720. The unmixed channels and the overlay are shown in pseudocolors. Scale bars, 20 μm. (g) Separate detection of five types of tumors expressing iRFPs in living mice. Mice were injected with two types of MTLn3 cells each, from left to right in each image: iRFP670, iRFP682, iRFP702, iRFP713 and iRFP720. Figures (c)–(g) are reproduced with permission from ref. 17 (Copyright 2013, Springer-Nature). | ||
iRFP720 can also be used as a BRET acceptor for BRET-based in vivo deep-tissue imaging. Verkhusha et al. engineered two novel chimeric probes based on RLuc8 luciferase fused iRFP670 and iRFP720 NIR FPs.14 Due to intramolecular BRET between RLuc8 and iRFP, the chimeric probes show NIR bioluminescence with maxima at 670 nm and 720 nm. These BRET probes enable combined multi-color bioluminescence imaging of the iRFPs, leading to BRET-based highly sensitive NIR detection of cancer cells as well as continuous spatiotemporal analysis of metastatic cells in living mice.
IFP1.4 and IFP2.0 are other dimeric NIR FPs with an emission maximum of 708 nm and 711 nm, respectively. IRF1.4 is a 34.8 kDa protein derived from a chromophore-binding domain (DrCBD) of Deinococcus radiodurans.8 However, an exogenous supply of biliverdin cofactor is required to achieve fluorescence imaging of these NIR-FPs in mammalian cells. IFP2.0, a brighter version of IFP1.4, was developed after insertion of 11 mutations in cDNA of IFP1.4.18 In some cases, dimeric NIR FPs are preferred over monomer NIR FPs due to the superior brightness of dimeric NIR FPs. They work well with many but not all cellular tags as fusion partners. However, they have limited use in live-cell dynamics and tracking applications due to their dimeric nature.
2.2. Monomeric NIR FPs
The development of monomeric NIR FPs makes in vivo live cell imaging and dynamics study more easy with lower laser power.1,10,12 Monomeric NIR FP variants have nearly similar photophysical properties to dimeric NIR FPs with slightly compromised brightness (Table 1). However, they are preferred over dimer NIR FPs for live-cell study and animal imaging with other cellular fusion protein tags because of their monomeric nature. Also, because of their monomeric nature, the localization efficiency of these NIR FPs is better than that of dimer NIR FPs.The first set of bright monomeric NIR FPs engineered from bacterial phytochromes are mIFP22 and miRFPs (miRFP670, miRFP670-2, emiRFP670, miRFP680, miRFP682, miRFP703, emiRFP703, miRFP709, miRFP713 and miRFP720)19,21,42 (Fig. 3). They work as a good fusion partner with several cellular protein tags like collagen, H2B, vimentin, tubulin, and actin and in free form in a living system without showing any aggregation. The molecular weight of mIFPs and miRFPs is 35.1 kDa and 34.6 kDa, respectively. Among them, miRFP680 has the highest QY and superior molecular brightness; this protein expressed very well with many cellular tag-proteins and miRFP720. Other bright and photostable NIR-FPs such as emiRFP670 and emiRFP703 with enhanced photophysical characteristics have been successfully utilized for single and multiplex non-invasive whole body imaging of mice.42 However, miRFP713 and miRFP720 are more preferable for deep-tissue in vivo imaging because of their NIR-shifted emission maximum (Emmax: 713 nm and Emmax: 720 nm, respectively). These proteins can be used for dual-colour multiplex imaging with other spectrally far NIR-FP bright variants of miRFPs like miRFP 680, miRFP670 and emiRFP670 for dual-colour in vivo imaging.
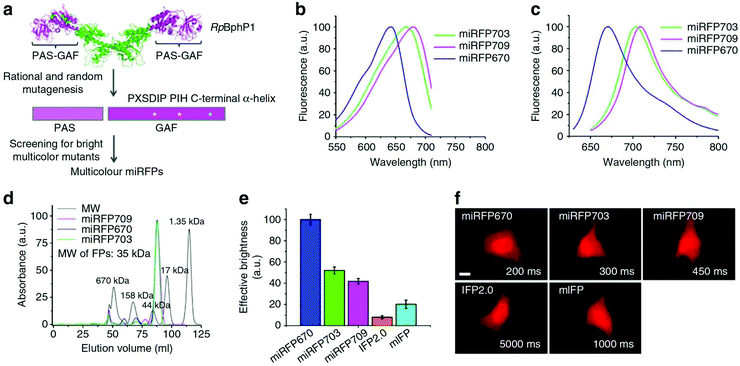 | ||
| Fig. 3 (a) Schematics of directed molecular evolution resulting in three monomeric miRFPs. The chromophore-binding PAS-GAF domains, which are not involved in dimerization of RpBphP1, were used as a starting point. To exclude formation of even weak dimers, residues in the C-terminal a-helix in the GAF domain were mutated. To obtain spectrally distinct variants, residues 201 and 202 in the –PXSDIP– motif and residue 253 in the –PIH– motif in the GAF domain were mutated. (b) Fluorescence excitation spectra of engineered miRFP670, miRFP703 and miRFP709. (c) Fluorescence emission spectra of miRFPs. (d) Size-exclusion chromatography of miRFPs. (e) Brightness of live HeLa cells transiently transfected with several BphP-based NIR FPs. (f) Representative fluorescence images of several BphP-based NIR FPs in live HeLa cells. Reproduced from ref. 19 (Copyright 2016, Creative Commons). | ||
Among the monomeric NIR FPs, miRFP670nano is the latest and smallest NIR FP (17 kDa). This protein has an emission maximum at 670 nm upon excitation at 645 nm with a 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 extinction coefficient.19 The monomeric protein miRFP670nano has nearly half the molecular weight of miRFP670, which makes it a better fusion partner candidate with other cellular proteins. In mammalian cells, miRFP670nano exhibits 2.8- and 1.3-fold higher photostability than miRFP670 and miRFP703. It shows precise expression with several fusion tag cellular proteins and behaves as a perfect fluorescence resonance energy transfer (FRET) donor with miRFP720 for in vitro and in vivo imaging.19
000 extinction coefficient.19 The monomeric protein miRFP670nano has nearly half the molecular weight of miRFP670, which makes it a better fusion partner candidate with other cellular proteins. In mammalian cells, miRFP670nano exhibits 2.8- and 1.3-fold higher photostability than miRFP670 and miRFP703. It shows precise expression with several fusion tag cellular proteins and behaves as a perfect fluorescence resonance energy transfer (FRET) donor with miRFP720 for in vitro and in vivo imaging.19
NIR FPs also prove to be useful in super-resolution imaging as they generate low autofluorescence signals in living cells. Structured illumination microscopy with miRFP703 and other variants provides superior spatial resolution compared to wide-field fluorescence imaging for tubulin, H2B and various other cellular tags.9 The enhanced brightness and photostability of the newly developed emiRFP670 and emiRFP703 worked perfectly well in single and dual-colour STED imaging with an 11–36 fold decrease in the STED illumination dose in mammalian cells compared to their previously existing monomer NIR-FPs.42
2.3. Photo-switchable NIR FPs
Photo-switchable FPs allow illuminating only the area of interest, which drastically improves the spatial and temporal resolution and image quality. PAiRFP1 and PAiRFP2 are the only photo-switchable NIR FPs emitting above 700 nm (Table 2).25 They are capable of undergoing multiple dark–bright cycles upon excitation with NIR light before bleaching.25 The emission peak of PAiRFPs is almost 50 nm red-shifted relative to that of the conventional red-shifted photo-switchable GFP-like protein, PSmOrange, leading to the use of PAiRFPs for dual-colour imaging.PAiRFPs are developed from a bacterial phytochrome photoreceptor (BphP) known as AtBphP2,43,44 which utilizes heme-derived biliverdin (ubiquitous in mammalian tissues) as the chromophore. Initially, weakly fluorescent PAiRFPs undergo photoconversion into a highly fluorescent state with an excitation/emission wavelength of 690 nm/717 nm. They are not as bright as the other monomeric NIR FPs but can be used in protein tracking with localized excitation and super-resolution imaging. Verkhusha et al. reported that whole-body imaging of mice using PAiRFPs gave two times higher signals than iRFP because of their photo-conversion properties.25 Photo-switchable FPs have been widely used for various 2D and 3D super-resolution imaging in photoactivated localization microscopy (PALM), reversible saturable optical fluorescence transition (RESOLFT), structured illumination microscopy (SIM) and light sheet fluorescence microscopy (LSFM).45 Photo-switchable NIR FPs could have the potential to be used for in vivo super-resolution imaging with superior signal-to-noise ratios.
NIR FPs with two-photon excitation have proven to give better signal-to-noise contrast and tissue scattering for in vivo and deep-tissue non-invasive imaging. Several NIR-FPs have been applied in two-photon imaging of primary neuronal cell culture, mouse brain, and brain slices of mice and monkeys.46–50 The Verkhusa group conducted two colour imaging of EGFP and iRFP680 by low-dose 880 nm two-photon excitation (6.5 mW).46 They achieved sub-cellular resolution in vivo imaging of neurons with a penetration depth as deep as 285 μm from the brain surface. NIR-FP based sensors have been recently developed for high-resolution in vivo neuroimaging using two-photon excitation in various animal models.
The Betzig group achieved wavefront sensing and visualization of the mouse cortex up to a 700 μm depth using two-photon excitation with iRFP713 NIR-FP.47 Other groups developed NIR-FP based voltage and calcium sensors using mIFP and miRFPs to monitor brain and neuronal activity in mice, zebrafish, C. elegans and Xaenopus laevis.48–50 Two-photon neuronal calcium imaging and sensing have been reported by utilizing NIR genetically encoded calcium ion indicators (GECIs) namely, NIR-GECO1 and NIR-GECO2, based on mIFP NIR-FPs.48,49 The Campbell group reported multiplex calcium imaging by utilizing 1250 nm two-photon excitation of NIR-GECO1 together with other previously developed calcium sensors such as GCaMP6f and RCaMP1.07 in live brain slices of mice.49 The same group reported other improved GECI variants such as NIR-GECO2 and NIR-GECO2G based on miRFP703.50 Bright NIR-FPs (e.g. miRFP680, emiRFP670 and miRFP720) will be good candidates for deeper-tissue calcium and voltage sensors with the two-photon excitation technique.
The major advantage of NIR-FPs over chemical dyes is genetic encodability with the fusion tag protein of interest. This helps in allowing live in vivo imaging of the whole animal. Several such studies have been done in the past to develop transgenic animals such as mice, rats and fruit flies, expressing NIR-FPs. The Miwa group developed the first iRFP based transgenic animal where they showed iRFP expression in almost every organ such as the liver, kidney, lungs, heart, brain, spleen, pancreas, bone, testis, thymus and adipose tissues.51 In two separate studies, miRFP713 was used to develop a transgenic cre-dependent inducible mouse model. In the first study, tumor development and in vivo recombinase activity were explored.52 They investigated live tumor progression, metastasis events and recombination events upon conditional activation of a cre-inducible system expressing miRFP713. In another study, a miRFP713 based mouse transgenic model was used to image a neuronal network in vivo without the supply of exogeneous chromophore biliverdin.53
A transgenic mouse model expressing Ucp1-iRFP720 was developed by the Hisatake group to study the role of uncoupling protein 1 (UCP1) activity in thermogenesis control in non-invasive in vivo systems.54 Uncoupling protein 1 (UCP1) is a mitochondrially expressed protein helping to uncouple the electron transport chain from ATP synthesis to produce heat. The mouse model developed in this study is useful for the in vivo study of pathways where adipocyte induction and non-shivering thermogenesis are involved.54
Gu et al. developed a two cell homologous recombination (2C-HR-CRISPR) method utilizing histone2B-miRFP703 NIR-FP to demonstrate the CRISPR-cas9 gene editing method for the generation of a transgenic mouse line and achieved three color imaging with mCherry, Halo-tag and miRFP703.55 McDole et al. achieved the fine detail of the developing heart of a mouse embryo with light-sheet microscopy by utilizing a histone2B-miRFP703 NIR-FP expressing transgenic mouse.56 They utilized a transgenic mouse embryo expressing histone2B-miRFP703, where they were able to achieve cellular level resolution up to a 600 μm depth covering the entire linear heart tube, head-fold and foregut pocket region.56 These studies have demonstrated the efficient use of NIR-FPs for non-invasive deep-tissue imaging and successful expression in transgenic animal systems.
3. Bioluminescence based probes for in vivo imaging
Compared to fluorescence imaging, bioluminescence in vivo imaging has a superior advantage in that the background signals due to tissue autofluorescence are almost zero, because external excitation is not needed in bioluminescence imaging. In addition, bioluminescence imaging permits long-term observation of living cells without irradiation using excitation light, leading to no photobleaching and photocytotoxicity. There are several types of bioluminescent proteins (luciferases) derived from fireflies,57–60 beetles,61–64 bacteria,65–68 and various marine species,69–83 which can be applied to in vivo imaging. Although bioluminescence imaging has such superior properties over fluorescence imaging, the observation wavelengths in bioluminescence imaging have been limited to the visible region.26 Recently, bioluminescence imaging probes based on red-shifted bioluminescent protein–substrate pairs and BRET systems have been developed for the non-invasive visualization of deep-tissues in the NIR region (Table 3).| Type | Luciferase | Substrate | BRET acceptor | λ em,max (nm) | Ref. |
|---|---|---|---|---|---|
| (1) Luciferase–substrate pairs | Firefly luciferase (FLuc) | Luciferin analog | 675, 706, 800, 667 | 28, 29, 31 and 32 | |
| FLuc | AlaLumine | 675 | 30 and 35 | ||
| Synthetic enzyme, Akaluc | AlaLumine | 650 | 34 | ||
| FLuc | Se-luciferin | 600 | 81 | ||
| NanoLuc luciferase | Coeleneterazine (CTZ) analog | 589 | 82 | ||
| Click beetle luciferase | Naphthyl luciferin | 758 | 83 | ||
| (2) BRET probes | |||||
| (a) Luciferase–FP | Renilla luciferase (RLuc8) | CTZ derivatives | iRFP670, iRFP720 | 670, 720 | 14 |
| FLuc | Luciferin | mCherry | 575 | 84 | |
| RLuc8.6 | CTZ | mKusabira Orange | 570 | 85 | |
| RLuc8.6 | CTZ | Turbo FP635 | 635 | 117 | |
| (b) Luciferase–dye | FLuc | Luciferin | Alexa Fluor680, 750 | 705, 785 | 86 |
| RLuc8.6SG | Cy5-CTZ | Cy5 | 684 | 87 | |
| RLuc8 | CTZ | NIR 775 | 775 | 88 | |
| RLuc8.6 | CTZ | Alexa Fluor680 | 700 | 89 | |
| Firefly luciferase | Cy7-luciferin | Cy7 | 800 | 27 | |
| Cypridina luciferase (CLuc) | Cypridina luciferin | HiLite Fluor647 | 670 | 36 | |
| (c) Luminol–dye | Luminol | Cyanine dye | 670 | 90 | |
| Luminol | Choline | 725, 800 | 91 | ||
| (d) Luciferase–QD | RLuc8, RLuc9 | CTZ | QD655, QD705, QD800, QD625 | 655, 705, 800, 625 | 92–98, 102, 103 and 109 |
| RLuc8 | CTZ | PbS QD | 950 | 100 | |
| RLuc8 | CTZ | CdTe/CdS QD | 650 | 104 | |
| RLuc | CTZ | CdSeTe/CdS QD | 830 | 106, 112, 113 and 115 | |
| CLuc | Cypridina luciferin | QD705 | 705 | 101 | |
| FLuc | Luciferin | CdSe/CdS Qrod, QD556 | 675, 556 | 105, 107, 108 and 111 | |
| Qrod615, 650 | 615, 650 | ||||
| NanoLuc | Furimazine | QD705 | 705 | 110 | |
| (e) Luminol–QD | Luminol | CdHgTe/CdS QD | 754 | 99 |
3.1. Red-shifted bioluminescent protein–substrate pairs
Despite the high sensitivity of bioluminescence imaging, its potential use in bioimaging is mostly restricted to the visible region less than 600 nm. Visible-emission bioluminescence is strongly absorbed and scattered in biological samples, leading to the attenuation of the bioluminescence signals. To overcome this drawback, red-shifted bioluminescent protein–substrate pairs have been developed for bioluminescence in vivo imaging, where the chemical structure of bioluminescent substrates (luciferins) has been modified to emit at longer wavelengths.27–38Conley et al. reported a selenium analogue of firefly D-luciferin with red-shifted bioluminescence emission over 600 nm.81 The emission of the selenium analogue gives a 40 nm red-shift compared to the parent D-luciferin, enabling in vivo imaging of tumor cells, which express luciferin in nude mice. Shakhimin et al. reported coelenterazine analogues with red-shifted bioluminescence with NanoLuc for deep-tissue imaging.82 The direct attachment of an aryl moiety to the imidazopyrazinone core of furimazine at the C8 position provides a significant red-shift of the emission maximum (480 nm) of coelenterazine. The modified NanoLuc substrates (coelenterazine analogues) are capable of efficient BRET to far-red and NIR fluorophores.
A recently developed luciferin analogue, AkaLumine, can emit in the NIR region over 700 nm (Fig. 4).30,32 AkaLumine is a D-luciferin analogue with an extended π-conjugation system. This red-shifted luciferin analogue has shown the capability of its NIR bioluminescence for highly sensitive deep-tissue imaging in mice.30 Recently, the Miyawaki group reported a bioluminescence deep-tissue imaging system AkaBLi (Akaluc/Akalumine), which is 100 to 1000 times brighter than conventional systems.34 They achieved non-invasive bioluminescence visualization of single cells deep inside freely moving animals, mice, and marmosets. Hall et al. reported a click beetle luciferase mutant utilizing two naphthyl–luciferin substrates to produce NIR emission (730 nm and 743 nm) for performing deep tissue multispectral tomography in mice.83
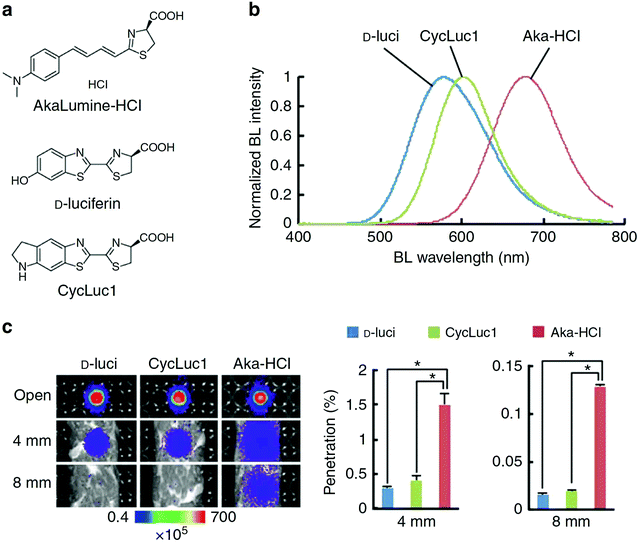 | ||
| Fig. 4 Properties of AkaLumine–HCl. (a) Chemical structures of AkaLumine–HCl, D-luciferin and CycLuc1. (b) Bioluminescence emission spectra of D-luciferin (D-luci), CycLuc1 or AkaLumine–HCl (Aka–HCl). (c) Biological tissue penetration efficiency of bioluminescence generated by D-luci, CycLuc1 or Aka–HCl. The penetration efficiency indicates the relative bioluminescence intensities of the wells covered with a biological tissue (4 or 8 mm thick) versus open wells (open). n = 3, *P < 0.05 (t-test). Reproduced from ref. 30 (Copyright 2016, Creative Commons). | ||
3.2. BRET systems
The use of BRET is an alternative method to achieve bioluminescence imaging at longer wavelengths. BRET consists of a luciferase–luciferin system as a resonance energy donor and a fluorescent acceptor as a resonance energy acceptor. BRET is a special case of resonance energy transfer, where the donor is a luciferase/luciferin system. As BRET acceptors, fluorescent materials such as FPs,14,84,85 organic dyes,35,86–91 and QDs92–115 have been employed (Fig. 5a–c).114 BRET occurs by non-radiative dipole–dipole coupling, and not by electron transfer. The efficiency of BRET is inversely proportional to the sixth power of the distance between the donor and accepter dipole:116| E = 1/[1 + (r/Ro)6] | (1) |
| Ro = 0.21[κ2QDn−4J(λ)]1/6 | (2) |
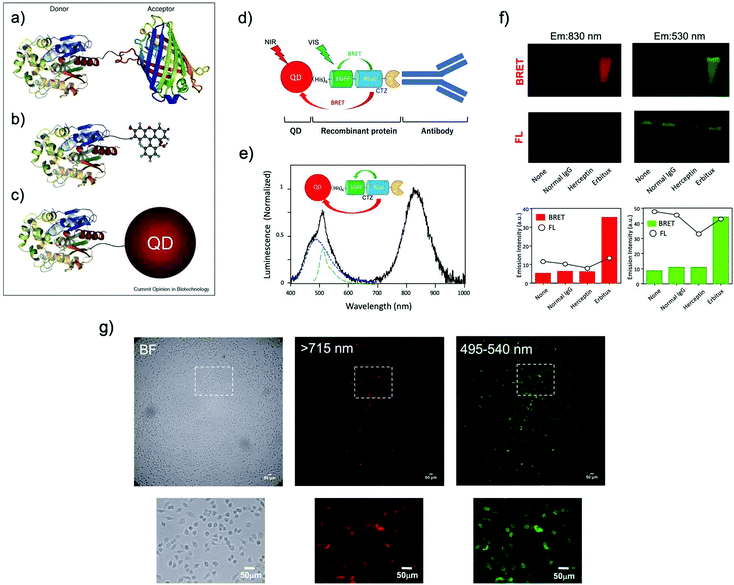 | ||
| Fig. 5 BRET systems consisting of a light-emitting protein and a fluorophore (acceptor) that can be (a) a FP, (b) a small organic dye, or (c) a quantum dot (QD). Reproduced with permission from ref. 114 (Copyright 2009, Elsevier). (d) A schematic structure of a BRET-based dual-color molecular-imaging probe. The probe consists of a NIR-emitting QD, fused protein (His-EGFP-RLuc-GB1), and monoclonal antibody. BRET is expected to occur from coelenterazine (CTZ) to EGFP and to the QD, leading to visible and NIR dual-color bioluminescence emission. (e) Bioluminescence spectra for the conjugate of a NIR-emitting QD and His-EGFP-RLuc-GB1. (f) Skin cancer cells (A431, ∼106 cells) treated with QD-His-EGFP-RLuc-GB1 (None), normal human IgG/QD-His-EGFP-RLuc-GB1 (Normal IgG), Herceptin/QD-His-EGFP-RLuc-GB1 (Herceptin), and Erbitux/QD-His-EGFP-RLuc-GB1 (Erbitux). The lower graphs show the emission intensity of BRET and FL for the above images. (g) Bright field (BF) and bioluminescence images of KLP-4 cells treated with Herceptin and His-EGFP-RLuc-GB1 conjugated QDs. Reproduced with permission from ref. 115 (Copyright 2019, Royal Society of Chemistry). | ||
Ohmiya et al. reported a BRET system using the conjugate between Cypridina luciferase (CLuc) and commercially available cyanine dye HiLyte Fluor 647.36 This BRET system showed a bimodal bioluminescence spectrum, attributable to intra-molecular BRET, having emission peaks at 460 nm and at 645 nm. They obtained high-resolution BRET images of Dlk-1-expressing Huh-7 cells, and monitored the accumulation of the BRET probe in tumor-bearing mice. Recently, Kizaka-Kondoh et al. reported an Alexa Fluor 680 conjugated BRET-probe for the detection of a ubiquitin-protease system regulated hypoxia-inducible factor.89 They achieved BRET-based NIR imaging of hypoxic liver metastasis in nude mice. Rao et al. also developed a BRET-based NIR imaging system that contains a fluorescent phthalocyanine dye, NIR775, as a resonance energy acceptor.88 They achieved BRET-based in vivo lymph-node mapping and tumor imaging. For BRET-based NIR imaging, Suzuki et al. reported a unique luciferase substrate consisting of a coelenterazine–cyanine-5 dye (CTZ–Cy5) conjugate. They developed BRET based NIR imaging using the CTZ–Cy5 substrate as a through-bond energy transfer cassette.87
In most of the BRET-based QD probes, luciferase was directly conjugated to the surface of QDs using chemical coupling methods. Since chemical coupling often results in the decrease in the enzymatic activities of luciferases, mild conjugation methods using tag technology such as Halo-tag,94 GST-tag,106 and His-tag108 are desirable. Recently, Jin et al. reported a facile method for preparing RLuc-EGFP fused protein conjugated NIR-QDs for dual-color molecular imaging of cancer cells (Fig. 5d–g).115 The major advantage in the use of QDs as BRET acceptors is the bright emission from the QDs because of the high extinction coefficients and large Stokes shifts. Although most QDs contain heavy metals with cytotoxicity, the improvement of the biocompatibility of QDs is necessary to develop the biological applications of QD-based BRET probes for NIR in vivo imaging.
4. In vivo NIR imaging systems
In in vivo NIR imaging systems, imaging microscopes are also important as well as optical NIR probes to achieve high-resolution imaging. Although many types of in vivo imaging instruments are commercially available, most of the NIR imaging instruments are the type of macro fluorescence imaging systems, where the spatial resolution is not enough to visualize molecular and cellular dynamics in living tissues. In addition, the excitation and emission wavelengths for NIR imaging are mostly restricted to less than 900 nm (the first NIR window). In this section, we summarize in vivo NIR imaging systems consisting of wide field, confocal and super-resolution microscopes for the high-resolution visualization of living tissues at the cellular level.4.1. Wide field epi-NIR fluorescence microscopy
In most of the commercially available in vivo imaging systems, the NIR wavelength region of 700 to 900 nm (first NIR optical window wavelength) has been employed. This is because the conventional NIR photo-detectors (silicon CCD cameras) are sensitive in the first NIR region, and many kinds of NIR emitting probes (e.g. Indocyanine green, Cy 7, and CdSeTe QDs) are easily available. Although NIR fluorescence imaging in the first NIR optical window is useful for the non-invasive visualization of organs and tissues, its spatial resolution is not enough to observe cellular dynamics. Recently, NIR optical imaging in the second NIR window (1000–1400 nm) as well as in the first NIR window (700–900 nm) has attracted much attention to achieve better spatiotemporal-resolution imaging for deep tissue in small animals.11,119Dai et al. have reported a high-resolution epi-microscopic system for NIR imaging of brain imaging cerebral vessels (Fig. 6a).120–125 High-magnification intravital imaging of cerebral vessels was carried out in epifluorescence mode with an 808 nm diode laser (RMPC lasers, 160 mW) as an excitation source and two objective lenses (4× and 10×) (Fig. 6b).120 A mouse with the scalp hair removed was intravenously injected with a solution of single walled carbon nanotubes (SWNTs) and placed in a home-made stereotactic platform fixed on a motorized 3D translational stage that allowed for the digital position adjustment and readout of the mouse relative to the objective. The emitted fluorescence was filtered through a 1000 nm long-pass filter, a 1300 nm long-pass filter and a 1400 nm short-pass filter to ensure only photons in the range of 1300–1400 nm.
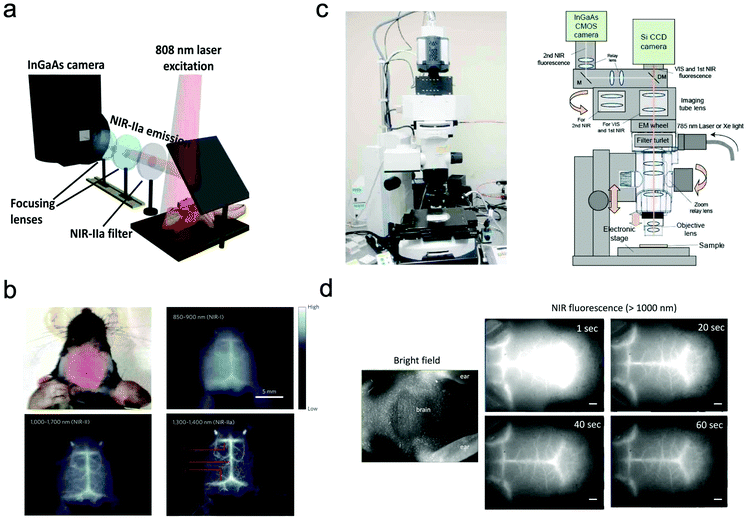 | ||
| Fig. 6 (a and b) NIR fluorescence imaging system for non-invasive through-scalp and through-skull brain vascular imaging. NIR-II emission: 1300–1400 nm. NIR fluorescence imaging of mouse brain vasculatures with SWNT-IRDye800 in different NIR subregions. Reproduced with permission from ref. 120 (Copyright 2014, Springer-Nature). (c) Upright fluorescence microscope system for in vivo imaging in the vis, first NIR, and second NIR region (400–1400 nm). (d) NIR fluorescence images of cerebral blood vessels in a nude mouse. NIR fluorescence images were taken 1 s, 20 s, 40 s, and 60 s post-injection of PbS QDs (200 μL, 2 μM) through a tail vein. Reproduced from ref. 127 (Copyright 2018, Creative Commons). | ||
Jin et al. have also reported a wide field epi-NIR fluorescence imaging system which can be used in both the first and second NIR window (Fig. 6c).15,126,127 In this imaging system, solid lasers for 645, 785, and 978 nm excitation, and emission filters of 1100 ± 25 nm, 1300 ± 25 nm, and 1500 ± 25 nm are equipped with the imaging system. A Si EM-CCD camera was employed for VIS and NIR fluorescence imaging in the first NIR window and an InGaAs CMOS camera for NIR fluorescence imaging in the second NIR window. By using the epi-NIR fluorescence microscope, non-invasive imaging of cerebral blood vasculatures in a nude mouse was easily achieved (Fig. 6d).127
4.2. Confocal NIR fluorescence microscopy
In order to achieve three-dimensional NIR fluorescence imaging with a high-spatial resolution, one-photon confocal imaging in the second NIR window has been recently reported by the Dai group (Fig. 7).128,129 Confocal imaging collects fluorescence signals from a small confocal volume, leading to high-resolution images with three-dimensional structures. They have developed a one-photon confocal NIR imaging microscope with ∼1 mm depth and high spatial resolution (sub ∼10 μm). By using the confocal NIR microscope, they achieved three dimensional images of blood vasculatures in fixed mouse brain tissue ∼5–7 μm in diameter at depth up to ∼1.3 mm. Dai et al. also achieved three-dimensional molecular imaging of targeted organs with high-performance hormone conjugated NIR dyes and quantum dots.129 Recently, Boghossian et al. reported a spinning-disc confocal laser microscope (SDCLM) in the second NIR window.130 They developed the SDCLM for high-speed optical sectioning of biological tissues in the second NIR window. The SDCLM achieved a lateral resolution of 0.5 ± 0.1 μm and an axial resolution of 0.6 ± 0.1 μm, showing a ∼17% and ∼45% enhancement in lateral and axial resolution, respectively.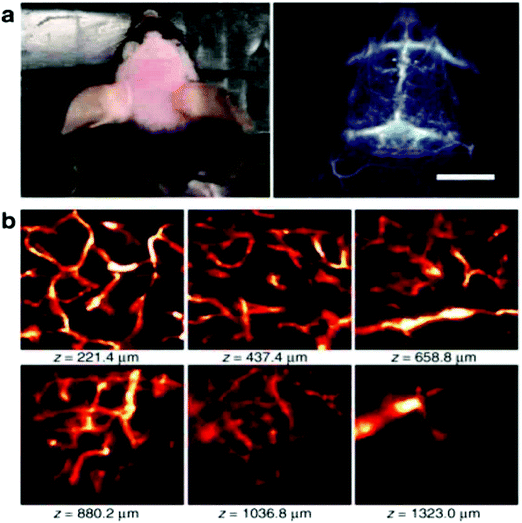 | ||
Fig. 7 (a) Photo and wide-field NIR-II epi-fluorescence imaging of brain in a mouse injected with p-FE (808![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm excitation, emission >1200 nm excitation, emission >1200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm) with an exposure time of 5 nm) with an exposure time of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ms. (b) Ex vivo confocal imaging of brain in a mouse injected with p-FE (785 ms. (b) Ex vivo confocal imaging of brain in a mouse injected with p-FE (785![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm excitation, emission >1100 nm excitation, emission >1100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm, laser power ∼30 nm, laser power ∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mW, PMT voltage ∼500 mW, PMT voltage ∼500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) V). Reproduced from ref. 128 (Copyright 2018, Creative Commons). V). Reproduced from ref. 128 (Copyright 2018, Creative Commons). | ||
4.3. Super-resolution microscopy
The controlled photoblinking of a fluorophore is produced with a specific wavelength of light either by switching them on–off or bleaching a subset of fluorophores so that only a few labeled fluorophores emit in each frame.131–137 This method is efficient in achieving a lateral resolution up to 10–20 nm, much superior to the diffraction limit in other optical microscopies. Several visible and NIR photo-switchable fluorophores are used to attain live-cell nanoscopy and in vivo super-resolution imaging; these can be repeatedly or irreversibly switchable between dark and bright states.133–135 However, organic dyes labeled with a suitable tag could also be used for various applications in living systems.132,136,137 In the past, several approaches using localization precision-based imaging techniques have been developed for live imaging like PALM, PALM with independent running acquisition, STORM and direct STORM, etc.132,134–137 In the latest development, combining these microscopy techniques with light-sheet microscopy drastically improved the resolution for thick samples and small animal in vivo imaging. The below equation explains the localization precision efficiency determination as described earlier:131,133,136
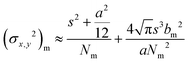 | (3) |
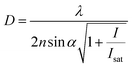 | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α is the numerical aperture, I is the applied intensity of the STED or RESOLFT pulse, and Isat is the STED intensity that gives 50% depletion of the emission.138 STED usually uses very strong laser power, which drastically affects live-cell imaging and bleaches fluorophores in parallel. Hence, STED applications have limited use in short time live-cell imaging.
α is the numerical aperture, I is the applied intensity of the STED or RESOLFT pulse, and Isat is the STED intensity that gives 50% depletion of the emission.138 STED usually uses very strong laser power, which drastically affects live-cell imaging and bleaches fluorophores in parallel. Hence, STED applications have limited use in short time live-cell imaging.
The concept of RESOLFT emerged from STED after replacing conventional fluorophores with photo-switchable fluorophores. This improvement drastically reduces the excitation and STED beam several fold to achieve long-time super-resolution imaging of live systems. RESOLFT is an extended advancement of STED microscopy using a special fluorophore, where the on–off states are controlled by two different wavelengths to achieve super-resolution up to 10–20 nm.140–144 RESOLFT has the unique advantage of reducing the laser power several fold (<10−6 times) in live-cell imaging.140,143,144 Recently, NIR FPs such as emiRFP670 and emiRFP703 achieved 50–70 nm spatial resolution in STED imaging of various cellular structures in live-cell imaging (Fig. 8).42 Development of bright photo-switchable NIR FPs will provide opportunity to users to reduce the laser power to extend the imaging time and resolution.
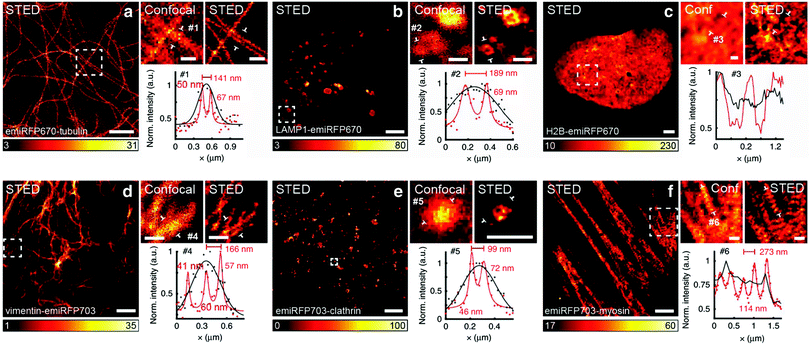 | ||
Fig. 8 Live-cell STED NIR imaging of emiRFP670 and emiRFP703 NIR-FPs with various cellular tags. (a) Confocal and STED images of HeLa cells expressing emiRFP670-tubulin. (b) Confocal and STED images of U2OS cells expressing LAMP1-emiRFP670. (c) Confocal and STED images of U2OS cells expressing H2B-emiRFP670. (d) Confocal and STED images of HeLa cells expressing vimentin-emiRFP703 NIR FP. (e) Confocal and STED images of HeLa cells expressing emiRFP703-clathrin NIR FP. (f) Confocal and STED images of U2OS cells expressing emiRFP703-myosin NIR FP. A Gaussian was used to fit the confocal data (black solid line) and a Lorentzian to fit the STED data (red solid line). Scale bar: 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm for the full images and 500 μm for the full images and 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm for the inserts. Reproduced from ref. 42 (Copyright 2018, Creative Commons). nm for the inserts. Reproduced from ref. 42 (Copyright 2018, Creative Commons). | ||
Live imaging of dendrites and dendritic spines in zebra-fish brain and mice has been achieved by SIM using green FP (GFP).149 The combination of NIR photo-switchable FPs with SSIM will further improve the resolution of such imaging conditions as proven earlier. In another study, high resolution of the neuronal structure of mouse brain was achieved using patterned illumination-based microscopy in two-photon excitation.150 This could further be improved by replacing fluorescein with NIR probes. However, the only limitation now is the availability of super-bright NIR FPs and NIR dyes. Recently, a NIR, small size and highly photostable dye, HD-Br, was successfully employed for three-dimensional super-resolution imaging of liver organoids and in Caenorhabditis elegance using SIM.151
4.4. Light sheet fluorescence microscopy (LSFM)
LSFM is widely used for 3D deep-tissue imaging. Most of the above discussed super-resolution microscopy techniques were used for thick samples, where the excitation laser light illuminates the sample in all planes (wide-field, PALM, STED and SSIM). In contrast, in LSFM imaging, a nanometer-thin sheet of light is used to excite an optically sectioned object perpendicular to the direction of observation. Dai et al. reported amazing resolution in NIR-LSFM imaging with 1300–1700 nm excitation and emission, respectively (Fig. 9).152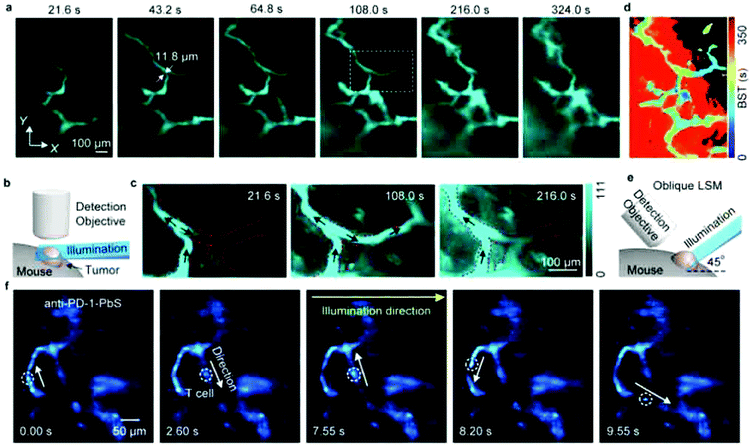 | ||
| Fig. 9 Non-invasive in vivo NIR-II light sheet microscopy of tumors on mice. (a) Time-course LSM of tumor vasculatures at a fixed illumination plane below the top of a xenograft MC38 tumor on a mouse ear at Z ∼ 300 μm after intravenous injection of a NIR probe, p-FE (excitation: 785 nm, emission: 1000–1200 nm). A 4× detection objective and a 5× illumination objective in a normal, non-oblique configuration of (b) were used. (c) Abnormal blood flows in tumor vessels, showing on–off intermittency and direction reversal in the rectangular highlighted region in (a) and gradual extravasation into the tumor space. Similar results for n = 3 (C57BL/6, female, 6 weeks old). Black arrows represent the flow direction. (d) A BST map showing highly heterogeneous blood perfusion in tumor vessels and slow, inhomogeneous extravasation behavior into the tumor space (C57BL/6, female, 6 weeks old; similar results for n = 2). (e) Schematic illustration of in vivo oblique NIR-II LSM with illumination and detection at 45° to the mouse body. (f) Time-course recording of PD-1+ cells (white circles) in a CT26 tumor labeled with anti-PD-1-CS QDs 2 h after intravenous injection of anti-PD-1-CS QDs at 20 fps by oblique LSM. Reproduced with permission from ref. 152 (Copyright 2019, Springer-Nature). | ||
Several versions of LSFM have been invented with improved resolution in both 2D and 3D imaging. Bessel beam plane illumination microscopy (BBPIM) is the advancement in this line developed by the Eric Betzig group, which used nanometer-thin light sheet illumination.153 The resolution of BBPIM could be further improved using photo-controllable or photo-switchable fluorophores in both the visible and NIR range. BBPIM combined with a structured illumination approach proved to achieve excellent resolution (both x–y and x–z direction) with reduced phototoxicity. This new nanoscopy approach is known as lattice light sheet nanoscopy (LLSN).154 Both BBPIM and LLSN are considered as great imaging tools for 3D imaging with very low photo-cytotoxicity.
5. Conclusion and perspective
In this review, we present NIR FPs and bioluminescence-based NIR probes with their application to high-resolution in vivo imaging, which has been developed in the last decade. In living systems, there are strong tissue absorption, scattering, and autofluorescence in the visible region. Thus, the use of NIR-emitting optical probes is crucial to achieve high-resolution in vivo imaging. The combination of NIR-emitting optical probes with high-resolution imaging systems enables the non-invasive visualization of living tissues at the cellular resolution level.Among the NIR FPs designed from the bacterial phytochrome photoreceptor, miRFP680 is the brightest NIR-FP in mammalian cells.42 The brightness of miRFP613 and miRFP720 is approximately ∼30% lower than miRFP680, however their ∼33 nm and ∼40 nm red-shifted emission spectra would be advantageous for in vivo imaging and multiplex imaging with other spectrally distinct NIR-FPs. Another recently developed NIR-FP, miRFP670nano, has nearly similar brightness to miRFP720 in mammalian cells but its monomeric nature and smallest size are the advantage for use as a fusion tag for live in vivo imaging.23 Considering their monomeric nature and brightness, these NIR-FPs are suitable for high-resolution in vivo imaging.
These proteins can also be used for dual-color and FRET imaging in deep-tissues. Photo-switchable NIR FPs PAmIRFP1 and PAmiRFP2 are unique FPs, and their emission can be controlled by switching on/off with light in a localized region, which is useful for super-resolution imaging. The use of photo-switchable FPs for in vivo imaging avoids the excitation of the out-of-focus area, leading to the significant improvement of signal-to-noise ratios by a factor of several fold. After the expansion of NIR photo-switchable FPs, they could also be used with other switchable NIR proteins to develop FRET pairs to improve signal-to noise ratios for the accuracy of NIR based FRET imaging.
Red-shifted bioluminescent protein–substrate pairs as well as NIR FPs can also be used for high-resolution in vivo imaging. Luciferin analogues modified with a longer π-conjugation such as AkaLumine enable high-resolution deep-tissue imaging at the cellular level. The use of BRET is an alternative method to achieve in vivo imaging in the NIR region. BRET-based molecular imaging has a higher detection sensitivity compared to fluorescence-based molecular imaging due to its low background signals. However, NIR bioluminescent proteins are still limited with only a few good NIR emitting bioluminescent substrates like AkaLumine, which is available to use alone or as a BRET acceptor for deep-tissue imaging.
NIR FP-based fluorescence imaging and bioluminescence-based imaging are promising modalities for high-resolution in vivo imaging. These imaging techniques can be applied to non-invasive visualization of cancer metastasis, neovascularization, immune cell response and stem cell implantation both in basic life science and preclinical research. Although several types of NIR FPs and red-shifted bioluminescent protein–substrate pairs have emerged in recent years, there is a need to focus more on the development of bright and stable ones for high-resolution in vivo imaging applications. Through the development of bright NIR molecular imaging probes, NIR optical imaging would greatly contribute to optical diagnostics of diseases in biomedical and clinical fields.155–159
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Setsuko Tsuboi and Sayumi Yamada of RIKEN for their help in the preparation of the manuscript. DKT thanks Department of Biotechnology, Ministry of Science and Technology, India, for a Ramalingaswami fellowship, and University Grant Commision, India, for financial support from the faculty position in Goa University. Manisha Tiwari thanks Department of Biotechnology, Ministry of Science and Technology, India, for a BioCARE women scientist fellowship. TJ is thankful for the financial support from the Ministry of Education, Science, Sports, and Culture of Japan (Grant-in-Aid for Scientific Research (B) 19H04459 to TJ).References
- J. V. Frangioni, In vivo near-infrared fluorescence imaging, Curr. Opin. Chem. Biol., 2003, 7, 626–634 CrossRef CAS PubMed.
- S. A. Hilderbrand and R. Weissleder, Near-infrared fluorescence: application to in vivo molecular imaging, Curr. Opin. Chem. Biol., 2010, 14, 71–79 CrossRef CAS PubMed.
- R. R. Zhang, A. B. Schroeder, J. J. Grudzinski, E. L. Rosenthal, J. M. Warram, A. N. Pinchuk, K. W. Eliceiri, J. S. Kuo and J. P. Weichert, Beyond the margins: real-time detection of cancer using targeted fluorophores, Nat. Rev. Clin. Oncol., 2017, 14, 347–364 CrossRef CAS PubMed.
- A. R. Kherlopian, T. Song, Q. Duan, M. A. Neimark, M. J. Po, J. K. Gohagan and A. F. Laine, A review of imaging techniques for systems biology, BMC Syst. Biol., 2008, 2, 74 CrossRef PubMed.
- R. A. de Kemp, F. H. Epstein, C. Catana, B. M. W. Tsui and E. L. Ritman, Small-animal molecular imaging methods, J. Nucl. Med., 2010, 51, 18S–32S CrossRef CAS PubMed.
- D. M. Chudakov, M. V. Matz, S. Lukyanov and K. A. Lukyanov, Fluorescent proteins and their applications in imaging living cells and tissues, Physiol. Rev., 2010, 90, 1103–1163 CrossRef CAS PubMed.
- G. S. Filonov, K. D. Piatkevich, L.-M. Ting, J. Zhang, K. Kim and V. V. Verkhusha, Bright and stable near-infrared fluorescent protein for in vivo imaging, Nat. Biotechnol., 2011, 29, 757–761 CrossRef CAS PubMed.
- X. Shu, A. Royant, M. Z. Lin, T. A. Aguilera, V. Lev-Ram, P. A. Steinbach and R. Y. Tsien, Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome, Science, 2009, 324, 804–807 CrossRef PubMed.
- K. Konig, Multiphoton microscopy in life sciences, J. Microsc., 2000, 200, 83–104 CrossRef CAS PubMed.
- R. Weissleder, A clearer vision for in vivo imaging, Nat. Biotechnol., 2001, 19, 316–317 CrossRef CAS PubMed.
- A. M. Smith, M. C. Mancini and S. Nie, Second window for in vivo imaging, Nat. Nanotechnol., 2009, 4, 710–711 CrossRef CAS PubMed.
- C. L. Amiot, S. Xu, S. Liang, L. Pan and J. X. Zhao, Near-infrared fluorescent materials for sensing of biological targets, Sensors, 2008, 8, 3082–3105 CrossRef CAS PubMed.
- G. Hong, A. L. Antaris and H. Dai, Near-infrared fluorophores for biomedical imaging, Nat. Biomed. Eng., 2017, 1, 0010 CrossRef CAS.
- K. A. Rumyantsev, K. K. Turoverov and V. V. Verkhusha, Near-infrared bioluminescent proteins for two-color multimodal imaging, Sci. Rep., 2016, 6, 36588 CrossRef CAS PubMed.
- Y. Tsukasaki, M. Morimatsu, G. Nishimura, T. Sakata, H. Yasuda, A. Komatsuzaki, T. M. Watanabe and T. Jin, Synthesis and optical properties of emission-tunable PbS/CdS core-shell quantum dots for in vivo fluorescence imaging in the second near-infrared window, RSC Adv., 2014, 4, 41164–41171 RSC.
- M. M. Karasev, O. V. Stepanenko, K. A. Rumyantsev, K. K. Turoverov and V. V. Verkhusha, Near-infrared fluorescent proteins and their applications, Biochemistry, 2019, 84, S32–S50 CAS.
- D. M. Shcherbakova and V. V. Verkhusha, Near-infrared fluorescent proteins for multicolor in vivo imaging, Nat. Methods, 2013, 10, 751–754 CrossRef CAS PubMed.
- D. Yu, W. C. Gustafson, C. Han, C. Lafaye, M. Noirclerc-Savoye, W.-P. Ge, D. A. Thayer, H. Huang, T. B. Kornberg, A. Royant, L. Y. Jan, Y. N. Jan, W. A. Weiss and X. Shu, An improved monomeric infrared fluorescent protein for neuronal and tumor brain imaging, Nat. Commun., 2014, 5, 3626 CrossRef CAS PubMed.
- D. M. Shcherbakova, M. Baloban, A. V. Emelyanov, M. Brenowitz, P. Guo and V. V. Verkhusha, Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging, Nat. Commun., 2016, 7, 12405 CrossRef CAS PubMed.
- D. M. Shcherbakova, O. V. Stepanenko, K. K. Turoverov and V. V. Verkhusha, Near-Infrared fluorescent proteins: multiplexing and optogenetics across scales, Trends Biotechnol., 2018, 36, 1230–1243 CrossRef CAS PubMed.
- D. M. Shcherbakova, N. C. Cammer, T. M. Huisman, V. V. Verkhusha and L. Hodgson, Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET, Nat. Chem. Biol., 2018, 14, 591–600 CrossRef CAS PubMed.
- D. Yu, M. A. Baird, J. R. Allen, E. S. Howe, M. P. Klassen, A. Reade, K. Makhijani, Y. Song, S. Liu, Z. Murthy, S.-Q. Zhang, O. D. Weiner, T. B. Kornberg, Y. N. Jan, M. W. Davidson and X. Shu, A naturally monomeric infrared fluorescent protein for protein labeling in vivo, Nat. Methods, 2015, 12, 763–765 CrossRef CAS PubMed.
- O. S. Oliinyk, A. A. Shemetov, S. Pletnev, D. M. Shcherbakova and V. V. Verkhusha, Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing, Nat. Commun., 2019, 10, 279 CrossRef PubMed.
- M. E. Auldridge, K. A. Satyshur, D. M. Anstrom and K. T. Forest, Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein, J. Biol. Chem., 2012, 287, 7000–7009 CrossRef CAS PubMed.
- K. D. Piatkevich, F. V. Subach and V. V. Verkhusha, Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome, Nat. Commun., 2013, 4, 2153 CrossRef PubMed.
- C. E. Badr and B. A. Tannous, Bioluminescence imaging: progress and applications, Trends Biotechnol., 2011, 29, 624–633 CrossRef CAS PubMed.
- R. Kojima, H. Takakura, T. Ozawa, Y. Tada, T. Nagano and Y. Urano, Rational design and development of near-infrared-emitting firefly luciferins available in vivo, Angew. Chem., Int. Ed., 2013, 52, 1175–1179 CrossRef CAS PubMed.
- S. Iwano, R. Obata, C. Miura, M. Kiyama, K. Hama, M. Nakamura, Y. Amano, S. Kojima, T. Hirano, S. Maki and H. Niwa, Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light, Tetrahedron, 2013, 69, 3847–3856 CrossRef CAS.
- A. P. Jathoul, H. Grounds, J. C. Anderson and M. A. Pule, A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging, Angew. Chem., Int. Ed., 2014, 53, 13059–13063 CrossRef CAS PubMed.
- T. Kuchimaru, S. Iwano, M. Kiyama, S. Mitsumata, T. Kadonosono, H. Niwa, S. Maki and S. Kizaka-Kondoh, A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging, Nat. Commun., 2016, 7, 11856 CrossRef CAS PubMed.
- J. C. Anderson, H. Grounds, A. P. Jathoul, J. A. H. Murray, S. J. Pacman and L. Tisi, Convergent synthesis and optical properties of near-infrared emitting bioluminescent infra-luciferins, RSC Adv., 2017, 7, 3975–3982 RSC.
- R. Saito, T. Kuchimaru, S. Higashi, S. W. Lu, M. Kiyama, S. Iwano, R. Obata, T. Hirano, S. Kizaka-Kondoh and S. A. Maki, Synthesis and luminescence properties of near-infrared N-heterocyclic luciferin analogues for in vivo optical imaging, Bull. Chem. Soc. Jpn., 2019, 92, 608–618 CrossRef CAS.
- M. Kiyama, S. Iwano, S. Otsuka, S. W. Lu, R. Obata, A. Miyawaki, T. Hirano and S. A. Maki, Quantum yield improvement of red-light-emitting firefly luciferin analogues for in vivo bioluminescence imaging, Tetrahedron, 2018, 74, 652–660 CrossRef CAS.
- S. Iwano, M. Sugiyama, H. Hama, A. Watakabe, N. Hasegawa, T. Kuchimaru, K. Z. Tanaka, M. Takahashi, Y. Ishida, J. Hata, S. Shimozono, K. Namiki, T. Fukano, M. Kiyama, H. Okano, S. Kizaka-Kondoh, T. J. McHugh, T. Yamamori, H. Hioki, S. Maki and A. Miyawaki, Single-cell bioluminescence imaging of deep tissue in freely moving animals, Science, 2018, 359, 935–939 CrossRef CAS PubMed.
- H. W. Yeh, T. Wu, M. Chen and H. W. Ai, Identification of factors complicating bioluminescence imaging, Biochemistry, 2019, 58, 1689–1697 CrossRef CAS PubMed.
- C. Wu, K. Mino, H. Akimoto, M. Kawabata, K. Nakamura, M. Ozaki and Y. Ohmiya, in vivo far-red luminescence imaging of a biomarker based on BRET from Cypridina bioluminescence to an organic dye, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15599–15603 CrossRef CAS PubMed.
- K. Saito, Y. F. Chang, K. Horikawa, N. Hatsugai, Y. Higuchi, M. Hashida, Y. Yoshida, T. Matsuda, Y. Arai and T. Nagai, Luminescent proteins for high-speed single-cell and whole-body imaging, Nat. Commun., 2012, 3, 1262 CrossRef PubMed.
- F. Weihs and H. Dacres, Red-shifted bioluminescence resonance energy transfer: improved tools and materials for analytical in vivo approaches., Trends Anal. Chem., 2019, 116, 61–73 CrossRef CAS.
- O. V. Stepanenko, G. S. Bublikov, O. V. Stepanenko, D. M. Shcherbakova, V. V. Verkhusha, K. K. Turoverov and I. M. Kuznetsova, A knot in the protein structure – probing the near-infrared fluorescent protein iRFP designed from a bacterial phytochrome, FEBS J., 2014, 281, 2284–2298 CrossRef CAS PubMed.
- M. Isomura, K. Yamada, K. Noguchi and A. Nishizono, Near-infrared fluorescent protein iRFP720 is optimal for in vivo fluorescence imaging of rabies virus infection, J. Gen. Virol., 2017, 98, 2689–2698 CrossRef PubMed.
- K. E. Luker, P. Pata, I. I. Shemiakina, A. Preverzeva, A. C. Stacer, D. S. Shcherbo, V. Z. Pletnev, M. Skolnaja, K. A. Lukyanov, G. D. Luker, I. Pata and D. M. Chudakov, Comparative study reveals better far-red fluorescent protein for whole body imaging, Sci. Rep., 2015, 5, 10332 CrossRef CAS PubMed.
- M. E. Matlashov, D. M. Shcherbakova, J. Alvelid, M. Baloban, F. Pennacchietti, A. A. Shemetov, I. Testa and V. V. Verkhusha, A set of monomeric near-infrared fluorescent proteins for multicolor imaging across scales, Nat. Commun., 2020, 11, 1–12 Search PubMed.
- T. Lamparter, N. Michael, F. Mittmann and B. Esteban, Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11628–11633 CrossRef CAS PubMed.
- B. Karniol and R. D. Vierstra, The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 2807–2812 CrossRef CAS PubMed.
- X. X. Zhou and M. Z. Lin, Photoswitchable fluorescent proteins: ten years of colorful chemistry and exciting applications, Curr. Opin. Chem. Biol., 2013, 17, 682–690 CrossRef CAS PubMed.
- K. D. Piatkevich, H. J. Suk, S. B. Kodandaramaiah, F. Yoshida, E. M. DeGennaro, M. Drobizhev, T. E. Hughes, R. Desimone, E. S. Boyden and V. V. Verkhusha, Near-Infrared fluorescent proteins Engineered from Bacterial Phytochromes in Neuroimaging, Biophys. J., 2017, 113, 2299–2309 CrossRef CAS PubMed.
- K. Wang, W. Sun, C. T. Richie, B. K. Harvey, E. Betzig and N. Ji, Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue, Nat. Commun., 2015, 6, 1–6 Search PubMed.
- K. D. Piatkevich, E. E. Jung, C. Straub, C. Linghu, D. Park, H. J. Suk, D. R. Hochbaum, D. Goodwin, E. Pnevmatikakis, N. Pak, T. Kawashima, C. T. Yang, J. L. Rhoades, O. Shemesh, S. Asano, Y. G. Yoon, L. Freifeld, J. L. Saulnier, C. Riegler, F. Engert, T. Hughes, M. Drobizhev, B. Szabo, M. B. Ahrens, S. W. Flavell, B. L. Sabatini and E. S. Boyden, A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters article, Nat. Chem. Biol., 2018, 14, 352–360 CrossRef CAS PubMed.
- Y. Qian, K. D. Piatkevich, B. Mc Larney, A. S. Abdelfattah, S. Mehta, M. H. Murdock, S. Gottschalk, R. S. Molina, W. Zhang, Y. Chen, J. Wu, M. Drobizhev, T. E. Hughes, J. Zhang, E. R. Schreiter, S. Shoham, D. Razansky, E. S. Boyden and R. E. Campbell, A genetically encoded near-infrared fluorescent calcium ion indicator, Nat. Methods, 2019, 16, 171–174 CrossRef CAS PubMed.
- Y. Qian, D. M. O. Cosio, K. D. Piatkevich, S. Aufmkolk, W.-C. Su, O. T. Celiker, A. Schohl, M. H. Murdock, A. Aggarwal, Y.-F. Chang, P. W. Wiseman, E. S. Ruthazer, E. S. Boyden and R. E. Campbell, Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging, bioRxiv, 2020, 2020.04.08.032433.
- M. T. N. Tran, J. Tanaka, M. Hamada, Y. Sugiyama, S. Sakaguchi, M. Nakamura, S. Takahashi and Y. Miwa, In vivo image analysis using iRFP transgenic mice, Exp. Anim., 2014, 63, 311–319 CrossRef CAS PubMed.
- A. K. Hock, E. C. Cheung, T. J. Humpton, T. Monteverde, V. Paulus-Hock, P. Lee, E. McGhee, A. Scopelliti, D. J. Murphy, D. Strathdee, K. Blyth and K. H. Vousden, Development of an inducible mouse model of iRFP713 to track recombinase activity and tumour development in vivo, Sci. Rep., 2017, 7, 1–9 CrossRef CAS PubMed.
- C. T. Richie, L. R. Whitaker, K. W. Whitaker, J. Necarsulmer, H. A. Baldwin, Y. Zhang, L. Fortuno, J. J. Hinkle, P. Koivula, M. J. Henderson, W. Sun, K. Wang, J. C. Smith, J. Pickel, N. Ji, B. T. Hope and B. K. Harvey, Near-infrared fluorescent protein iRFP713 as a reporter protein for optogenetic vectors, a transgenic Cre-reporter rat, and other neuronal studies, J. Neurosci. Methods, 2017, 284, 1–14 CrossRef CAS PubMed.
- A. Fukuda, N. Honda, Y. Fujioka, S. Sekiguchi, Y. Mizuno, F. Miwa, Y. Sugiyama, K. Hayashi, K. Nishimura and K. Hisatake, Non-invasive in vivo imaging of UCP1 expression in live mice via near-infrared fluorescent protein iRFP720, PLoS One, 2019, 14, 1–16 Search PubMed.
- B. Gu, E. Posfai and J. Rossant, Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos, Nat. Biotechnol., 2018, 36, 632–637 CrossRef CAS PubMed.
- K. McDole, L. Guignard, F. Amat, A. Berger, G. Malandain, L. A. Royer, S. C. Turaga, K. Branson and P. J. Keller, In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level, Cell, 2018, 175, 859–876 CrossRef CAS.
- A. Rehemtulla, L. D. Stegman, S. J. Cardozo, S. Gupta, D. E. Hall, C. H. Contag and B. D. Ross, Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging, Neoplasia, 2000, 2, 491–495 CrossRef CAS PubMed.
- B. A. Rabinovich, Y. Ye, T. Etto, J. Q. Chen, H. I. Levitsky, W. W. Overwijk, L. J. N. Cooper, J. Gelovani and P. Hwu, Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14342–14346 CrossRef CAS PubMed.
- J. B. Kim, K. Urban, E. Cochran, S. Lee, A. Ang, B. Rice, A. Bata, K. Campbell, R. Coffee, A. Gorodinsky, Z. Lu, H. Zhou, T. K. Kishimoto and P. Lassota, Non-invasive detection of a small number of bioluminescent cancer cells in vivo, PLoS One, 2010, 5, e9364 CrossRef PubMed.
- Y. Liang, P. Walczak and J. W. Bulte, Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells, J. Biomed. Opt., 2012, 17, 016004 CrossRef PubMed.
- Y. Nakajima, T. Yamazaki, S. Nishii, T. Noguchi, H. Hoshino, K. Niwa, V. R. Viviani and Y. Ohmiya, Enhanced beetle luciferase for high-resolution bioluminescence imaging, PLoS One, 2010, 5, e10011 CrossRef PubMed.
- M. Chang, K. P. Anttonen, S. L. G. Cirillo, K. P. Francis and J. D. Cirillo, Real-time bioluminescence imaging of mixed mycobacterial infections, PLoS One, 2014, 9, e108341 CrossRef.
- V. Villalobos, S. Naik, M. Bruinsma, R. S. Dothager, M. H. Pan, M. Samrakandi, B. Moss, A. Elhammali and D. Piwnica-Worms, Dual-color click beetle luciferase heteroprotein fragment complementation assays, Chem. Biol., 2010, 17, 1018–1029 CrossRef CAS.
- M. Yasunaga, Y. Nakajima and Y. Ohmiya, Dual-color bioluminescence imaging assay using green- and red-emitting beetle luciferases at subcellular resolution, Anal. Bioanal. Chem., 2014, 406, 5735–5742 CrossRef CAS PubMed.
- M. Cronin, A. R. Akin, S. A. Collins, J. Meganck, J. B. Kim, C. K. Baban, S. A. Joyce, G. M. van Dam, N. Zhang, D. van Sinderen, G. C. O’Sullivan, N. Kasahara, C. G. Gahan, K. P. Francis and M. Tangney, High resolution in vivo bioluminescent imaging for the study of bacterial tumor targeting, PLoS One, 2012, 7, e30940 CrossRef CAS PubMed.
- Y. Guo, R. I. Ramos, J. S. Cho, N. P. Donegan, A. L. Cheung and L. S. Miller, In vivo bioluminescence imaging to evaluate systemic and topical antibiotics against community-acquired methicillin-resistant Staphylococcus aureus-infected skin wounds in mice, Antimicrob. Agents Chemother., 2013, 57, 855–863 CrossRef CAS PubMed.
- T. Xu, S. Ripp, G. S. Sayler and D. M. Close, Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells, PLoS One, 2014, 9, e96347 CrossRef PubMed.
- D. M. Close, S. S. Patterson, S. Ripp, S. J. Baek, J. Sanseverino and G. S. Sayler, Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line, PLoS One, 2010, 5, e12441 CrossRef PubMed.
- A. M. Loening, T. D. Fenn, A. M. Wu and S. S. Gambhir, Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output, Protein Eng., Des. Sel., 2006, 19, 391–400 CrossRef CAS PubMed.
- A. M. Loening, A. M. Wu and S. S. Gambhir, Red-shifted Renilla reniformis luciferase variants for imaging in living subjects, Nat. Methods, 2007, 4, 641–643 CrossRef CAS PubMed.
- W. W. Lorenz, M. J. Cormier, D. J. Okane, D. Hua, A. A. Escher and A. A. Szalay, Expression of the Renilla reniformis luciferase gene in mammalian cells, J. Biolumin. Chemilumin., 1996, 11, 31–37 CrossRef CAS PubMed.
- B. A. Tannous, D. E. Kim, J. L. Fernandez, R. Weissleder and X. O. Breakefield, Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo, Mol. Ther., 2005, 11, 435–443 CrossRef CAS PubMed.
- S. B. Kim, H. Suzuki, M. Sato and H. Tao, Superluminescent variants of marine luciferases for bioassays, Anal. Chem., 2011, 83, 8732–8740 CrossRef CAS PubMed.
- S. V. Markova, S. Golz, L. A. Frank, B. Kalthof and E. S. Vysotski, Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa – a novel secreted bioluminescent reporter enzyme, J. Biol. Chem., 2004, 279, 3212–3217 CrossRef CAS PubMed.
- S. V. Markova, M. D. Larionova, L. P. Burakova and E. S. Vysotski, The smallest natural high-active luciferase: cloning and characterization of novel 16.5 kDa luciferase from copepod Metridia longa, Biochem. Biophs. Res. Commun., 2015, 457, 77–82 CrossRef CAS PubMed.
- S. E. Webb and A. L. Miller, Aequorin-based genetic approaches to visualize Ca2+ signaling in developing animal systems, Biochim. Biophys. Acta, 2012, 1820, 1160–1168 CrossRef CAS.
- E. M. Thompson, P. Adenot, F. I. Tsuji and J. P. Renard, Real-time imaging of transcriptional activity in live mouse preimplantation embryos using a secreted luciferase, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 1317–1321 CrossRef CAS.
- O. Shimomura, F. H. Johnson and T. Masugi, Cypridina bioluminescence: light-emitting oxyluciferin–luciferase complex, Science, 1969, 164, 1299–1300 CrossRef CAS PubMed.
- M. P. Hall, J. Unch, B. F. Binkowski, M. P. Valley, B. L. Butler, M. G. Wood, P. Otto, K. Zimmerman, G. Vidugiris, T. Machleidt, M. B. Robers, H. A. Benink, C. T. Eggers, M. R. Slater, P. L. Meisenheimer, D. H. Klaubert, F. Fan, L. P. Encell and K. V. Wood, Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate, ACS Chem. Biol., 2012, 7, 1848–1857 CrossRef CAS PubMed.
- A. A. Homaei, A. B. Mymandi, R. Sariri, E. Kamrani, R. Stevanato, S. M. Etezad and K. Khajeh, Purification and characterization of a novel thermostable luciferase from Benthosema pterotum, J. Photochem. Photobiol., B, 2013, 125, 131–136 CrossRef CAS PubMed.
- N. R. Conley, A. Dragulescu-Andrasi, J. Rao and W. E. Moerner, A selenium analogue of firefly D-luciferin with red-shifted bioluminescence emission, Angew. Chem., Int. Ed., 2012, 51, 3350–3353 CrossRef CAS PubMed.
- A. Shakhmin, M. P. Hall, T. Machleidt, J. R. Walker, K. V. Wood and T. A. Kirkland, Coelenterazine analogues emit red-shifted bioluminescence with NanoLuc, Org. Biomol. Chem., 2017, 15, 8559–8567 RSC.
- M. P. Hall, C. C. Woodroofe, M. G. Wood, I. Que, M. Van't Root, Y. Ridwan, C. Shi, T. A. Kirkland, L. P. Encell, K. V. Wood, C. Löwik and L. Mezzanotte, Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging, Nat. Commun., 2018, 9, 132 CrossRef PubMed.
- P. Iglesias and J. A. Costoya, A novel BRET-based genetically encoded biosensor for functional imaging of hypoxia, Biosens. Bioelectron., 2009, 24, 3126–3130 CrossRef CAS PubMed.
- A. Takai, M. Nakano, K. Saito, R. Haruno, T. M. Watanabe, T. Ohyanagi, T. Jin, Y. Okada and T. Nagai, Expanded palette of Nano-lanterns for real-time multicolor luminescence imaging, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4352–4356 CrossRef CAS PubMed.
- B. R. Branchini, D. M. Ablamsky and J. C. Rosenberg, Chemically modified firefly luciferase is an efficient source of near-infrared light, Bioconjugate Chem., 2010, 21, 2023–2030 CrossRef CAS PubMed.
- M. Abe, R. Nishihara, Y. Ikeda, T. Nakajima, M. Sato, N. Iwasawa, S. Nishiyama, R. Paulmurugan, D. Citterio, S. B. Kim and K. Suzuki, Near-infrared bioluminescence imaging with a through-bond energy transfer caseette, ChemBioChem, 2019, 20, 1919–1923 CrossRef CAS PubMed.
- L. Xiong, A. J. Shuhendler and J. Rao, Self-luminescing BRET-FRET near-infrared dots for in vivo lymph-node mapping and tumor imaging, Nat. Commun., 2012, 3, 1193 CrossRef PubMed.
- T. Kuchimaru, T. Suka, K. Hirota, T. Kadonosono and S. Kizaka-Kondoh, A novel injectable BRET-based in vivo imaging probe for detecting the activity of hypoxia-inducible factor regulated by the ubiquitin-proteasome system, Sci. Rep., 2016, 6, 34311 CrossRef CAS PubMed.
- R. Liu, J. Tang, Y. Xu and Z. Dai, Bioluminescence Imaging of inflammation in vivo based on bioluminescence and fluorescence resonance energy transfer using nanobubble ultrasound contrast agent, ACS Nano, 2019, 13, 5124–5132 CrossRef CAS PubMed.
- X. Xu, H. An, D. Zhang, H. Tao, Y. Dou, X. Li, J. Huang and J. Zhang, A self-illuminating nanoparticle for inflammation imaging and cancer therapy, Sci. Adv., 2019, 5, eaat2953 CrossRef PubMed.
- M. K. So, C. Xu, A. M. Loening, S. S. Gambhir and J. Rao, Self-illuminating quantum dot conjugates for in vivo imaging, Nat. Biotechnol., 2006, 24, 339–343 CrossRef CAS PubMed.
- M. K. So, A. M. Loening, S. S. Gambhir and J. Rao, Creating self-illuminating quantum dot conjugates, Nat. Protoc., 2006, 1, 1160–1164 CrossRef CAS.
- Y. Zhang, M. So, A. M. Loening, H. Yao, S. S. Gambhir and J. Rao, HaloTag protein-mediated site-specific conjugation of bioluminescent proteins to quantum dots, Angew. Chem., Int. Ed., 2006, 45, 4936–4940 CrossRef CAS PubMed.
- H. Yao, Y. Zhang, F. Xiao, Z. Xia and J. Rao, Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases, Angew. Chem., Int. Ed., 2007, 46, 4346–4349 CrossRef CAS PubMed.
- Y. Xing, M. K. So, A. L. Koh, R. Sinclair and J. Rao, Improved QD-BRET conjugates for detection and imaging, Biochem. Biophys. Res. Commun., 2008, 372, 388–394 CrossRef CAS.
- K. A. Cissell, S. Campbell and S. K. Deo, Rapid, single-step nucleic acid detection, Anal. Bioanal. Chem., 2008, 391, 2577–2581 CrossRef CAS.
- Z. Xia, Y. Xing, M. K. So, A. L. Koh, R. Sinclair and J. Rao, Multiplex detection of protease activity with quantum dot nanosensors prepared by intein-mediated specific bioconjugation, Anal. Chem., 2008, 80, 8649–8655 CrossRef CAS PubMed.
- J. Du, C. Yu, D. Pan, J. Li, W. Chen, M. Yan, T. Segura and Y. Lu, Quantum-dot-decorated robust transductable bioluminescent nanocapsules, J. Am. Chem. Soc., 2010, 132, 12780–12781 CrossRef CAS PubMed.
- N. Ma, A. F. Marshall and J. Rao, Near-infrared light emitting luciferase via biomineralization, J. Am. Chem. Soc., 2010, 132, 6884–6885 CrossRef CAS PubMed.
- C. Wu, K. Kawasaki, S. Ohgiya and Y. Ohmiya, Chemical studies on the BRET system between the bioluminescence of Cypridina and quantum dots, Photochem. Photobiol. Sci., 2011, 10, 1531–1534 RSC.
- M. Kumar, D. Zhang, D. Broyles and S. K. Deo, A rapid, sensitive, and selective bioluminescence resonance energy transfer (BRET)-based nucleic acid sensing system, Biosens. Bioelectron., 2011, 30, 133–139 CrossRef CAS PubMed.
- G. A. Quinones, S. C. Miller, S. Bhattacharyya, D. Sobek and J. P. Stephan, Ultrasensitive detection of cellular protein interactions using bioluminescence resonance energy transfer quantum dot-based nanoprobes, J. Cell. Biochem., 2012, 113, 2397–2405 CrossRef CAS PubMed.
- Q. Wu and M. Chu, Self-illuminating quantum dots for highly sensitive in vivo real-time luminescent mapping of sentinel lymph nodes, Int. J. Nanomed., 2012, 7, 3433–3443 CAS.
- R. Alam, D. M. Fontaine, B. R. Branchini and M. M. Maye, Designing quantum rods for optimized energy transfer with firefly luciferase enzymes, Nano Lett., 2012, 12, 3251–3256 CrossRef CAS PubMed.
- M. Hasegawa, Y. Tsukasaki, T. Ohyanagi and T. Jin, Bioluminescence resonance energy transfer coupled near-infrared quantum dots using GST-tagged luciferase for in vivo imaging, Chem. Commun., 2013, 49, 228–230 RSC.
- R. Alam, J. Zylstra, D. M. Fontaine, B. R. Branchini and M. M. Maye, Novel multistep BRET-FRET energy transfer using nanoconjugates of firefly proteins, quantum dots, and red fluorescent proteins, Nanoscale, 2013, 5, 5303–5306 RSC.
- R. Alam, L. M. Karam, T. L. Doane, J. Zylstra, D. M. Fontaine, B. R. Branchini and M. M. Maye, Near infrared bioluminescence resonance energy transfer from firefly luciferase-quantum dot bionanoconjugates, Nanotechnology, 2014, 25, 495606 CrossRef PubMed.
- A. Samanta, S. A. Walper, K. Susumu, C. L. Dwyer and I. L. Medintz, An enzymatically-sensitized sequential and concentric energy transfer relay self-assembled around semiconductor quantum dots, Nanoscale, 2015, 7, 7603–7614 RSC.
- A. Kamkaew, H. Sun, C. G. England, L. Cheng, Z. Liu and W. Cai, Quantum dot–NanoLuc bioluminescence resonance energy transfer enables tumor imaging and lymph node mapping in vivo, Chem. Commun., 2016, 52, 6997–7000 RSC.
- R. Alam, L. M. Karam, T. L. Doane, K. Coopersmith, D. M. Fontaine, B. R. Branchini and M. M. Maye, Probing bioluminescence resonance energy transfer in quantum rod–luciferase nanoconjugates, ACS Nano, 2016, 10, 1969–1977 CrossRef CAS PubMed.
- S. Tsuboi and T. Jin, Bioluminescence resonance energy transfer (BRET)-coupled annexin V-functionalized quantum dots for near-infrared optical detection of apoptotic cells, ChemBioChem, 2017, 18, 2231–2235 CrossRef CAS PubMed.
- S. Tsuboi and T. Jin, Recombinant protein (luciferase-IgG binding domain) conjugated quantum dots for BRET-coupled near-infrared imaging of epidermal growth factor receptors, Bioconjugate Chem., 2018, 29, 1466–1474 CrossRef CAS PubMed.
- Z. Xia and J. Rao, Biosensing and imaging based on bioluminescence resonance energy transfer, Curr. Opin. Biotechnol., 2009, 20, 37–44 CrossRef CAS PubMed.
- S. Tsuboi and T. Jin, BRET based dual-colour (visible/near-infrared) molecular imaging using a quantum dot/EGFP–luciferase conjugate, RSC Adv., 2019, 9, 34964–34971 RSC.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy. Springer, New York, 3rd edn, 2006 Search PubMed.
- A. Dragulescu-Andrasi, C. T. Chan, A. De, T. F. Massoud and S. S. Gambhir, Bioluminescence resonance energy transfer (BRET) imaging of protein–protein interactions within deep tissues of living subjects, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12060–12065 CrossRef CAS.
- N. Kosaka, M. Mitsunaga, S. Bhattacharyya, S. C. Miller, P. L. Choyke and H. Kobayashi, Self-illuminating in vivo lymphatic imaging using a bioluminescence resonance energy transfer quantum dot nano-particle, Contrast Media Mol. Imaging, 2011, 6, 55–59 CrossRef CAS PubMed.
- F. Ding, Y. Zhan, X. Lu and Y. Sun, Recent advances in near-infrared II fluorophores for multifunctional biomedical imaging, Chem. Sci., 2018, 9, 4370–4380 RSC.
- G. Hong, S. Diao, J. Chang, A. L. Antaris, C. Chen, B. Zhang, S. Zhao, D. N. Atochin, P. L. Huang, K. I. Andreasson, C. J. Kuo and H. Dai, Through-skull fluorescence imaging of the brain in a new near-infrared window, Nat. Photon., 2014, 8, 723–730 CrossRef CAS.
- G. Hong, J. C. Lee, J. T. Robinson, U. Raaz, L. Xie, N. F. Huang, J. P. Cooke and H. Dai, Multifunctional in vivo vascular imaging using near-infrared II fluorescence, Nat. Med., 2012, 18, 1841–1846 CrossRef CAS.
- K. Welsher, S. P. Sherlock and H. Dai, Deep-tissue anatomical imaging of mice using carbon nanotube nanoprobes in the second near-infrared window, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 8943–8948 CrossRef CAS PubMed.
- K. Welsher, Z. Liu, S. P. Sherlock, J. T. Robinson, Z. Chen, D. Daranciang and H. Dai, A route to brightly fluorescent carbon nanotubes for near-infrared imaging in mice, Nat. Nanotechnol., 2009, 4, 773–780 CrossRef CAS PubMed.
- K. Welsher, Z. Liu Z, D. Daranciang and H. Dai, Selective probing and imaging of cells with single walled carbon nanotubes as near-infrared fluorescent molecules, Nano Lett., 2008, 8, 586–590 CrossRef CAS PubMed.
- Z. Liu, W. Cai, L. He, N. Nakayama, K. Chen, X. Sun, X. Chen and H. Dai, in vivo biodistribution and highly efficient tumor targeting of carbon nanotubes in mice, Nat. Nanotechnol., 2007, 2, 47–52 CrossRef CAS PubMed.
- T. Jin and Y. Imamura, Applications of highly bright PbS quantum dots for non-invasive near-infrared fluorescence imaging in the second optical window, ECS J. Solid State Sci. Technol., 2016, 5, R3138–R3145 CrossRef CAS.
- S. Tsuboi, S. Yamada, Y. Nakane, T. Sakata, H. Yasuda and T. Jin, Water-soluble near-infrared fluorophores emitting over 1000 nm and their applications to in vivo imaging in the second optical window (1000–1400 nm), ECS J. Solid State Sci. Technol., 2018, 7, R3093–R3101 CrossRef CAS.
- H. Wan, J. Yue, S. Zhu, T. Uno, X. Zhang, Q. Yang, K. Yu, G. Hong, J. Wang, L. Li, Z. Ma, H. Gao, Y. Zhong, J. Su, A. L. Antaris, Y. Xia, J. Luo, Y. Liang and H. Dai, A bright organic NIR-II nanofluorophore for three-dimensional imaging into biological tissues, Nat. Commun., 2018, 9, 1171 CrossRef.
- S. Zhu, S. Herraiz, J. Yue, M. Zhang, H. Wan, Q. Yang, Z. Ma, Y. Wang, J. He, A. L. Antaris, Y. Zhong, S. Diao, Y. Feng, Y. Zhou, K. Yu, G. Hong, Y. Liang, A. J. Hsueh and H. Dai, 3D NIR-II molecular imaging distinguishes targeted organs with high-performance NIR-II bioconjugates, Adv. Mater., 2018, 30, 1705799 CrossRef PubMed.
- V. Zubkovs, A. Antonucci, N. Schuergers, B. Lambert, A. Latin, R. Ceccarelli, A. Santinelli, A. Rogov, D. Ciepielewski and A. A. Boghossian, Spinning-disc confocal microscopy in the second near-infrared window (NIR-II), Sci. Rep., 2018, 8, 13770 CrossRef PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Imaging intracellular fluorescent proteins at nanometer resolution, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- M. Bates, S. A. Jones and X. Zhuang, Stochastic optical reconstruction microscopy (STORM): A method for superresolution fluorescence imaging, Cold Spring Harb. Protoc., 2013, 6, 498–520 Search PubMed.
- M. Zhang, H. Chang, Y. Zhang, J. Yu, L. Wu, W. Ji, J. Chen, B. Liu, J. Lu, Y. Liu, J. Zhang, P. Xu and T. Xu, Rational design of true monomeric and bright photoactivatable fluorescent proteins, Nat. Methods, 2012, 9, 727–729 CrossRef CAS.
- H. Shroff, C. G. Galbraith, J. A. Galbraith, H. White, J. Gillette, S. Olenych, M. W. Davidson and E. Betzig, Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20308–20313 CrossRef CAS PubMed.
- D. M. Shcherbakova, P. Sengupta, J. Lippincott-Schwartz and V. V. Verkhusha, Photocontrollable fluorescent proteins for superresolution imaging, Annu. Rev. Biophys., 2014, 43, 303–329 CrossRef CAS PubMed.
- M. J. Rust, M. Bates and X. Zhuang, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat. Methods, 2006, 3, 793–796 CrossRef CAS PubMed.
- S. Van De Linde, A. Loschberger, T. Klein, M. Heidbreder, S. Wolter, M. Heilemann and M. Sauer, Direct stochastic optical reconstruction microscopy with standard fluorescent probes, Nat. Protoc., 2011, 6, 991–1009 CrossRef CAS PubMed.
- T. A. Klar, S. Jakobs, M. Dyba, A. Egner and S. W. Hell, Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8206–8210 CrossRef CAS PubMed.
- R. E. Thompson, D. R. Larson and W. W. Webb, Precise nanometer localization analysis for individual fluorescent probes, Biophys. J., 2002, 82, 2775–2783 CrossRef CAS.
- T. Grotjohann, I. Testa, M. Reuss, T. Brakemann, C. Eggeling, S. W. Hell and S. Jakobs, rsEGFP2 enables fast RESOLFT nanoscopy of living cells, eLife, 2012, 1, e00248 CrossRef PubMed.
- A. Chmyrov, J. Keller, T. Grotjohann, M. Ratz, E. d'Este, S. Jakobs, C. Eggeling and S. W. Hell, Nanoscopy with more than 100
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ‘doughnuts’, Nat. Methods, 2013, 10, 737–740 CrossRef CAS PubMed.
000 ‘doughnuts’, Nat. Methods, 2013, 10, 737–740 CrossRef CAS PubMed. - T. Grotjohann, I. Testa, M. Leutenegger, H. Bock, N. T. Urban, F. Lavoie-Cardinal, K. L. Willing, C. Eggeling, S. Jakobs and S. W. Hell, Diffraction-unlimited all-optical imaging and writing with a photochromic GFP, Nature, 2011, 478, 204–208 CrossRef CAS PubMed.
- M. Andresen, A. C. Stiel, J. Fölling, D. Wenzel, A. Schönle, A. Egner, C. Eggelin, S. W. Hell and S. Jakobs, Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy, Nat. Biotechnol., 2008, 26, 1035–1040 CrossRef CAS PubMed.
- T. Brakemann, A. C. Stiel, G. Weber, M. Andresen, I. Testa, T. Grotjohann, M. Leutenegger, U. Plessmann, H. Urlaub, C. Eggeling, M. C. Wahl, S. W. Hell and S. Jakobs, A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching, Nat. Biotechnol., 2011, 29, 942–947 CrossRef CAS PubMed.
- R. Heintzmann, T. M. Jovin and C. Cremer, Saturated patterned excitation microscopy – a concept for optical resolution improvement, J. Opt. Soc. Am. A, 2002, 19, 1599–1609 CrossRef PubMed.
- E. Mudry, K. Belkebir, J. Girard, J. Savatier, E. Le Moal, C. Nicoletti, M. Allain and A. Sentenac, Structured illumination microscopy using unknown speckle patterns, Nat. Photonics, 2012, 6, 312–315 CrossRef CAS.
- L. Shao, P. Kner, E. H. Rego and M. G. L. Gustafsson, Super-resolution 3D microscopy of live whole cells using structured illumination, Nat. Methods, 2011, 8, 1044–1046 CrossRef CAS PubMed.
- E. H. Rego, L. Shao, J. J. Macklin, L. Winoto, G. A. Johansson, N. Kamps-Hughes, M. W. Davidson and M. G. L. Gustafsson, Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50 nm resolution, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E135–E143 CrossRef CAS PubMed.
- R. Turcotte, Y. Liang, M. Tanimoto, Q. Zhang, Z. Li, M. Koyama, E. Betzig and N. Ji, Dynamic super-resolution structured illumination imaging in the living brain, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 9586–9591 CrossRef CAS PubMed.
- A. Doi, R. Oketani, Y. Nawa, Y. Nawa and K. Fujita, High-resolution imaging in two-photon excitation microscopy using in situ estimations of the point spread function, Biomed. Opt. Express, 2018, 9, 202–213 CrossRef CAS PubMed.
- H. Fang, S. Yao, Q. C. Liu, Y. Cai, S. Geng, Y. Bai, Z. Tian, A. L. Zacharias, T. Takebe, Y. Chen, Z. Guo, W. He and J. Diao, De Novo-designed near-infrared nanoaggregates for super-resolution monitoring of lysosomes in cells, in whole organoids, and in vivo, ACS Nano, 2019, 13, 14426–14436 CrossRef CAS PubMed.
- F. Wang, H. Wan, Z. Ma, Y. Zhong, Q. Sun, Y. Tian, L. Qu, H. Du, M. Zhang, L. Li, H. Ma, J. Luo, Y. Liang, W. J. Li, G. Hong, L. Liu and H. Dai, Light-sheet microscopy in the near-infrared II window, Nat. Methods, 2019, 16, 545–552 CrossRef CAS.
- L. Gao, L. Shao, B.-C. Chen and E. Betzig, 3D live fluorescence imaging of cellular dynamics using Bessel beam plane illumination microscopy, Nat. Protoc., 2014, 9, 1083–1101 CrossRef CAS PubMed.
- B. C. Chen, W. R. Legant, K. Wang, L. Shao, D. E. Milkie, M. W. Davidson, C. Janetopoulos, X. S. Wu, J. A. Hammer 3rd, Z. Liu, B. P. English, Y. Mimori-Kiyose, D. P. Romero, A. T. Ritter, J. Lippincott-Schwartz, L. Fritz-Laylin, R. D. Mullins, D. M. Mitchell, J. N. Bembenek, A. C. Reyman, R. Böhme, S. W. Grill, J. T. Wang, G. Seydoux, U. S. Tulu, D. P. Kiehart and E. Betzig, Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution, Science, 2014, 346, 1257998 CrossRef PubMed.
- J. A. Carr, D. Franke, J. R. Caram, C. F. Perkinson, M. Saif, V. Askoxylakis, M. Datta, D. Fukumura, R. K. Jain, M. G. Bawendi and O. T. Bruns, Shorwave infrared fluorescence imaging with the clinically approved nea-infrared dye indocyanine green, Proc. Natl. Acad. Sci. U. S. A., 2018, 4465–4470 CrossRef CAS.
- R. Bhavane, Z. Starosolski, I. Stupin, K. B. Ghaghada and A. Annaparagada, NIR-II fluorescence imaging using indocyanine green nanoparticles, Sci. Rep., 2018, 8, 14455 CrossRef PubMed.
- J. Y. K. Lee, J. P. Thawani, J. Pierce, R. Zeh, M. Martinez-Lage, M. Chanin, O. Venegas, S. Nims, K. Learned, J. Keating and S. Singhal, Intraoperative Near-infrared optical imaging can localize gadolinium-enhancing gliomas during surgery, Neurosurgery, 2016, 79, 856–871 CrossRef PubMed.
- R. Zeh, S. Sheikh, L. Xia, J. Pierce, A. Newton, J. Predina, S. Cho, M. Nasrallah, S. Shinghal, J. Dorsey and J. Y. K. Lee, The second window ICG technique demonsrates a broad plateau period for near-infared fluoresence tumor contrast in glioblastoma, PLoS One, 2017, 12, e0182034 CrossRef PubMed.
- J. Y. K. Lee, J. T. Pierce, J. P. Thawani, R. Zeh, S. Nie, M. Martinez-Lage and S. Singhal, Near-infared fluoresecnce image-guided suegery for intracranical meningioma, J. Neurosurg., 2018, 128, 380–390 CAS.
| This journal is © The Royal Society of Chemistry 2020 |



