Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An efficient synthetic strategy for ligand-free upconversion nanoparticles†
Chunning
Sun
 *a,
Jan Ron Justin
Simke
b and
Michael
Gradzielski
*a,
Jan Ron Justin
Simke
b and
Michael
Gradzielski
 *a
*a
aStranski-Laboratorium für Physikalische und Theoretische Chemie, Institut für Chemie, Technische Universität Berlin, Strasse des 17. Juni 124, 10623 Berlin, Germany. E-mail: sunchunning@gmail.com; michael.gradzielski@tu-berlin.de
bZentraleinrichtung Elektronenmikroskopie (ZELMI), Technische Universität Berlin, Strasse des 17. Juni 135, 10623 Berlin, Germany
First published on 12th August 2020
Abstract
High-quality lanthanide-doped upconversion nanoparticles are generally synthesized by employing long-chain oleic acid as the ligand in the synthetic process, rendering them hydrophobic, and preferentially dispersed in nonpolar solvents. Thus, postsynthetic surface modifications are required prior to practical applications. Herein, we have developed a facile approach to remove the surface ligand from oleate-stabilized upconversion nanoparticles using short-chain acids as stripping agents by a simple vortexing method on a time scale of 10 seconds. This method allows for the fast and efficient hydrophobic-to-hydrophilic transition in either biphasic solvent systems or single hydrophobic media without any noticeable detrimental effects on size, shape, and phase of crystals during the ligand removal process. Furthermore, the obtained ligand-free upconversion nanoparticles can be readily transferred to the aqueous solution and further modified with water-soluble capping molecules by sequential surface functionalization. As an example, by coupling polymer functionalized upconversion nanoparticles with organic dyes, we are able to construct nanoprobes for selective fluorescence sensing for Cu2+.
Introduction
Lanthanide-doped upconversion nanoparticles are capable of absorbing two or more low energy photons sequentially via metastable and long-lived intermediate energy states. By this process, electrons are promoted to higher energy levels, which leads to higher-energy light emissions, ranging from the visible to the ultraviolet region, and for this property, upconversion nanoparticles have attracted considerable attention in the past decades.1 In stark contrast to semiconductor quantum dots and organic dyes, upconversion nanoparticles feature excellent chemical and optical properties, including large anti-Stokes shifts, long fluorescence lifetimes, sharp multicolour emissions, low cytotoxicity, as well as high chemical- and photo-stability.2 Based on these outstanding merits, upconversion nanoparticles are regarded as promising luminescent materials for applications in photodynamic therapy,3–6 bioimaging,7–9 sensing,10–13 anti-counterfeiting,14,15 and other optical fields.16High-quality upconversion nanoparticles with precisely tuned size, composition, shape, and well-designed architecture are generally synthesized by applying the long-chain oleic acid (OA) as the ligand in the synthetic process.17–22 Sterically stabilized oleate-capped upconversion nanoparticles are prone to disperse in nonpolar and hydrophobic solvents, while most applications of upconversion materials require the particles to be readily dispersed in hydrophilic media. To address this problem, postsynthetic surface modifications are essential for rendering upconversion nanoparticles dispersible in the aqueous solution or polar organic media prior to subsequent practical applications. Heretofore, surface post-modifications of oleate-capped upconversion nanoparticles including ligand exchange,23–28 oxidation,29–31 and removal,32,33 amphiphilic polymer deposition,34–38 and silica coating39,40 have been applied. However, these existing methods involve either laborious procedures or time-consuming reactions. A generalized, fast, and efficient strategy for the hydrophobic-to-hydrophilic transition of upconversion nanoparticles is largely missing.
Among them, the ligand removal strategy is considered as a versatile approach to prepare hydrophilic upconversion nanoparticles, as it provides more possibilities for the further functionalization of upconversion nanoparticles. For instance, nitrosonium tetrafluoroborate was employed to replace the oleate ligand to prepare hydrophilic nanoparticles, which were partially capped by the BF4− anion, and the quasi-ligand-free nanoparticles were further functionalized by grafting different capping molecules on the surface. However, the air-sensitive and oxidative stripping reagent limits its application.32 Another strategy was to use HCl solution (pH 4) to obtain ligand-free upconversion nanoparticles from oleate-capped upconversion nanoparticles based on an acid–base reaction. However, the stripping process occurred between solid oleate-capped upconversion nanoparticles and acidic aqueous solution in a solid–liquid system, which lowered the removal efficiency of the oleate ligand from the surface of nanoparticles and took hours of reaction time for complete ligand exfoliation.33
Here we report a fast, highly efficient, and easy-to-process approach to produce ligand-free upconversion nanoparticles by a simple vortexing method with short-chain acids as stripping agents. Biphasic and single solvent systems are introduced to remove the capping ligand from the surface of oleate-stabilized upconversion nanoparticles in just seconds time. More importantly, the resulting ligand-free upconversion nanoparticles can be readily transferred to the aqueous solution and further functionalized by water-soluble capping molecules.
Results and discussion
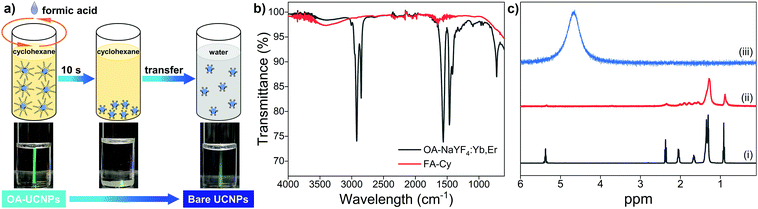
Monodisperse hexagonal oleate-capped NaYF4: 20 mol% Yb, 2 mol% Er nanoparticles (abbreviated as OA-UCNPs) are synthesized by the high-temperature coprecipitation method in octadecene using OA as the capping ligand.22 The resulting OA-UCNPs can be dispersed in nonpolar solvents (cyclohexane, chloroform, or toluene) owing to the presence of the oleate ligand on their surface. In order to realize the ligand removal from the surface of OA-UCNPs, biphasic and single solvent systems are introduced. For the ligand removal occurs in biphasic solvent systems, a wide variety of polar solvents including acetonitrile, dimethylformamide, dimethyl sulfoxide, formamide, methanol, and N-methyl-2-pyrrolidone are employed to form liquid–liquid interfaces with cyclohexane, and the reaction is simply performed by a short contact of the organic dispersion of OA-UCNPs with immiscible polar solvents containing short-chain organic acids on a vortex mixer, leading to the in-solution transfer of upconversion nanoparticles to the polar phase (Fig. S1, ESI†). In terms of single solvent systems, the ligand removal takes place when cyclohexane, chloroform, or toluene is adopted as the dispersant to stabilize OA-UCNPs. Ligand exfoliation is realized by direct addition of short-chain acids to a single solvent system, and ligand-free UCNPs are generated upon the sufficient interaction of short-chain acids with the hydrophobic solvent through vigorous shaking in a short time. More importantly, the ligand-free UCNPs obtained in the above-mentioned systems via the vortexing method can be easily transferred to the aqueous solution.Herein, a single hydrophobic solvent system, where formic acid (FA) is employed as the stripping agent and cyclohexane is used as the hydrophobic solvent for OA-UCNPs, is taken as a typical example (abbreviated as FA-Cy). Typically, 2 mL of cyclohexane solution containing OA-UCNPs (10 mg mL−1) was added to a 4 mL glass vial, followed by the addition of 0.5 mmol FA. After vigorous shaking (3000 rpm) for 10 s on a vortex mixer, ligand-free UCNPs were precipitated out from the cyclohexane. Ligand-free UCNPs are obtained by centrifugation and can be easily transferred to the aqueous solution. The ligand removal process is depicted in Fig. 1a. OA-UCNPs undergo effective collision with FA when the mixed solution is under vigorous shaking on a vortex mixer, and the acid–base reaction between FA and oleate ligand occurs instantaneously, resulting in the formation of OA via protonation. The resultant OA releases from the surface of nanoparticles and subsequently dissolves in cyclohexane solution. Moreover, the obtained ligand-free UCNPs cannot be dispersed in either cyclohexane or FA and thus are prone to precipitate out from the mixed solution. The fast acid–base reaction between FA and oleate ligand together with the spatial separation of the stripped OA and bare UCNPs enable the efficient ligand removal in the FA-Cy system. The ligand removal process is tracked by Fourier transform infrared (FT-IR), proton nuclear magnetic resonance (1H NMR), transmission electron microscopy (TEM), X-ray powder diffraction (XRD), thermogravimetric analysis (TGA), and ζ-potential measurements.
The successful removal of the oleate ligand is confirmed by FT-IR (Fig. 1b). The transmission bands of OA-UCNPs at 2921 and 2851 cm−1 are attributed to asymmetric and symmetric stretching vibrations of methylene (–CH2–) groups in the long alkyl chain. A weak peak at 3006 cm−1, assigning to the ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H stretching vibration, can be clearly observed in the spectrum. Moreover, two peaks centred at 1560 and 1464 cm−1 can be assigned to the asymmetric and symmetric stretching vibrations of the carboxylate group. These characteristic peaks prove the presence of oleate ligand on the untreated OA-UCNPs. However, they disappear in the FA-Cy sample, indicating the success in the ligand exfoliation. In addition, the broad band centred around 3500 cm−1, assigning to the solvated water molecules, is consistent with the hydrophilic nature of ligand-free UCNPs. To exclude disturbances of the lanthanide dopants on the 1H NMR signal, undoped NaYF4 nanoparticles are applied instead for the 1H NMR study (Fig. 1c). Compared with the 1H NMR spectrum of pure OA in CDCl3, the undoped oleate-capped NaYF4 sample reveals the characteristic bands of OA with broadened signals, originating from the inhomogeneous chemical environment and restricted rotational freedom of the oleate ligand anchored on the surface of nanoparticles. These signals vanish after the acid treatment and dispersing the sample in D2O, thereby corroborating the efficient ligand removal from the surface of OA-UCNPs.
C–H stretching vibration, can be clearly observed in the spectrum. Moreover, two peaks centred at 1560 and 1464 cm−1 can be assigned to the asymmetric and symmetric stretching vibrations of the carboxylate group. These characteristic peaks prove the presence of oleate ligand on the untreated OA-UCNPs. However, they disappear in the FA-Cy sample, indicating the success in the ligand exfoliation. In addition, the broad band centred around 3500 cm−1, assigning to the solvated water molecules, is consistent with the hydrophilic nature of ligand-free UCNPs. To exclude disturbances of the lanthanide dopants on the 1H NMR signal, undoped NaYF4 nanoparticles are applied instead for the 1H NMR study (Fig. 1c). Compared with the 1H NMR spectrum of pure OA in CDCl3, the undoped oleate-capped NaYF4 sample reveals the characteristic bands of OA with broadened signals, originating from the inhomogeneous chemical environment and restricted rotational freedom of the oleate ligand anchored on the surface of nanoparticles. These signals vanish after the acid treatment and dispersing the sample in D2O, thereby corroborating the efficient ligand removal from the surface of OA-UCNPs.
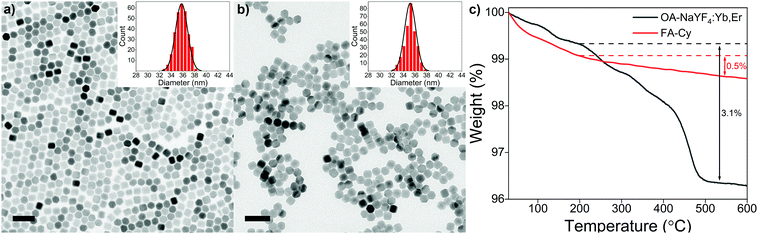
The morphology and structure of as-synthesized and acid-treated OA-UCNPs are characterized by TEM and XRD. TEM images present OA-UCNPs and FA-treated nanoparticles with a highly uniform hexagonal morphology (Fig. 2a and b). A particle analysis (at least 300 particles), performed from many such TEM images obtained from different regions of samples, demonstrates monodisperse oleate-capped and oleate-free UCNPs with an average diameter of ca. 35 nm, validating the maintained particle size of both original and acid-treated nanoparticles. TEM images and size distributions of acid-treated samples in other single and biphasic solvent systems show basically identical results (Fig. S2, ESI†), confirming the generality of this approach. Moreover, small-sized OA-UCNPs (ca. 20 nm) can be applied to the fast ligand removal through the here described method as well (Fig. S3, ESI†). XRD patterns of both samples show well-defined diffraction peaks, which can be indexed to the pure hexagonal phase of NaYF4, signifying the maintained high crystallinity after acid treatment (Fig. S4, ESI†).
TGA is conducted to evaluate the amount of ligand on both as-synthesized and acid-treated OA-UCNPs (Fig. 2c). The curves show a much lower total weight loss above 200 °C in the FA-Cy sample (∼0.5%) compared with the OA-UCNPs sample (∼3.1%). Together with an entirely different weight loss behaviour between the FA-Cy and OA-UCNPs sample, both features further prove the successful ligand removal after the acid treatment. Moreover, upon treatment with FA, bare UCNPs are easily dissolved in water, and the ζ-potential of obtained bare UCNPs is determined to be 36.3 mV (pH ∼ 5.5), owing to the uncoordinated metal cations induced by the acid treatment. This indicates the conversion of hydrophobic OA-UCNPs into stable hydrophilic colloids, and similar behaviours are observed in other acid-treated OA-UCNPs systems (Fig. S5, ESI†).
In the light of the above-described FT-IR, 1H NMR, TEM, XRD, TGA, and ζ-potential results, it can be concluded that the oleate ligand is removed by the treatment with short-chain acids in both single and biphasic solvent systems by means of the vortexing method on short time scales. At the same time, the acid treatment process has no noticeable adverse effects on size, shape, and phase of the resulting nanoparticles.
To investigate the optical property of UCNPs before and after ligand removal, upconversion luminescence (UCL) spectra are carried out on OA-UCNPs and ligand-free UCNPs dispersed in hydrophobic solvents and water, respectively. As presented in Fig. S6 (ESI†), UCNPs generate green and red UCL emissions when exposed to a 980 nm continuous-wave (CW) laser, originating from 2H11/2 → 4I15/2, 4S3/2 → 4I15/2, and 4F9/2 → 4I15/2 transitions of the Er3+ ion. UCL intensities of OA-UCNPs show no significate differences in green and red regions when dissolved in different hydrophobic solvents, whereas UCL intensities of bare UCNPs dispersed in water reduce by almost a factor of 10 compared with those of OA-UCNPs dissolved in hydrophobic media, where it is mainly attributed to the presence of high energy OH-vibrations in water.41,42 Compared with previous methods,32,33 ligand-free UCNPs prepared by the here proposed method show similar results in the morphology and UCL (Fig. S7, ESI†). However, the here developed method is much faster and reaction conditions are milder. In addition, various stripping agents and solvents can be employed for the ligand removal of OA-UCNPs. More importantly, the proposed method can be applied to HCl-sensitive materials, like NaLaF4 (Fig. S8, ESI†).43
The treatment of OA-UCNPs with FA makes the ligand-free UCNPs dispersible in water and positively charged bare UCNPs generate, favouring the attachment of water-soluble capping molecules by sequential surface functionalization, such as poly(acrylic acid) (PAA), polyvinylpyrrolidone (PVP), and polyethylenimine (PEI).44 Different water-soluble capping molecules modified UCNPs are characterized by TEM, FT-IR, TGA, and UCL measurements (Fig. S9, ESI†). FT-IR and TGA measurements prove the presence of corresponding molecules capped on the surface of the modified bare UCNPs, and TEM images exhibit the identical size and morphology after the functionalization. Moreover, there are no apparent differences in the position and intensity of UCL among PAA-, PVP-, and PEI-modified UCNPs.
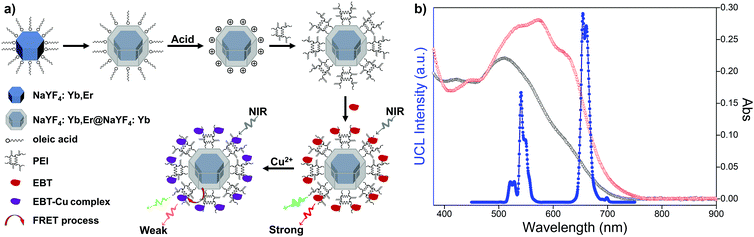
Owing to the unique property of upconversion nanoparticles, they have been widely applied in optical fields. To explore the sensing application, upconversion-based fluorescence resonance energy transfer (FRET) nanoprobes for the detection of Cu2+ were prepared by coupling PEI-modified bare upconversion nanoparticles with the Eriochrome Black T (EBT) dye. The synthetic process is depicted in Fig. 3a. OA-UCNPs core nanoparticles and core–shell structured oleate-capped NaYF4: 20 mol% Yb, 2 mol% Er@NaYF4: 20 mol% Yb nanoparticles (abbreviated as OA-csUCNPs) were first synthesized according to the previous work.45 Ligand-free csUCNPs were obtained by the here developed vortexing method, and PEI-functionalized csUCNPs (abbreviated as PEI-csUCNPs) were obtained by the attachment of PEI molecules, followed by assembling of the EBT dye on the surface of PEI-csUCNPs to prepare nanocomposites (denoted as csUCNPs/EBT). Upon the addition of Cu2+ at pH 3, the spectral overlap between the red UCL emission from csUCNPs and the absorbance of the EBT-Cu adduct increases (Fig. 3b), indicating the possibility of enhanced FRET process between UCNPs and the metal–dye complex.
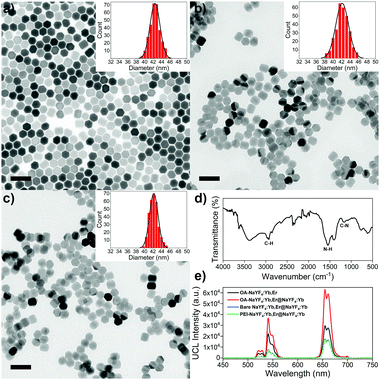
TEM image reveals that OA-csUCNPs have a relatively uniform size, and the mean diameter grows to 43 nm after the shell deposition on the core OA-UCNPs (Fig. 4a). XRD results show that both the core-only and core–shell structured upconversion nanoparticles are the hexagonal phase (Fig. S4, ESI†). The UCL intensity of the core–shell nanoparticles is enhanced by a factor of two compared with that of the core-only OA-UCNPs (Fig. 4e). As a result of the ligand exfoliation by FA, bare csUCNPs can be easily dispersed in water and further functionalized by PEI, which is confirmed by the FT-IR spectrum (Fig. 4d). The size and shape remain unchanged after the ligand removal and further surface modification (Fig. 4b and c), albeit with a loss of green and red UCL intensity in the aqueous environment (Fig. 4e). The EBT dye can be easily attached to the surface of PEI-csUCNPs through electrostatic interactions, and the amount of the EBT is calculated to be about 187 μM from detailed spectrophotometric titration spectra (Fig. S10, ESI†).
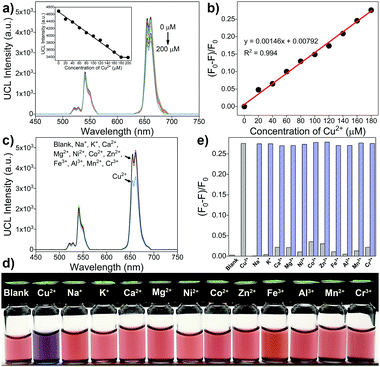
To examine the sensing ability of the upconversion-based FRET nanoprobes, different concentrations of Cu2+ are added to the Britton Robinson buffer (BRB) solution (100 mM, pH 3) containing the csUCNPs/EBT nanocomposites. A significant decrease in the UCL intensity at 654 nm proportional to the Cu2+ concentration could be observed, owing to the redshift absorption of the EBT–Cu complex and thus leading to an enhanced FRET process from the csUCNPs to the metal–dye complex (Fig. 5a and b). The detection limit for Cu2+ is approximately 9.4 μM according to the 3σ rule. Moreover, no apparent lessening of the UCL intensity and colour change is observed upon the addition of other metal ions (Fig. 5c–e), suggesting an excellent selectivity of the nanoprobes for Cu2+ sensing without interference by the presence of other metal ions. Therefore, the csUCNPs/EBT nanocomposites can be served as sensitive and selective nanoprobes for Cu2+ ions.
Conclusions
In summary, by introducing biphasic and single solvent systems, we have developed an efficient synthetic strategy for transferring hydrophobic OA-UCNPs to ligand-free ones using short-chain acids as stripping agents by a simple vortexing method. A wide range of polar solvents can be applied to form biphasic interfaces with cyclohexane, facilitating in-solution ligand exfoliation and allowing the transfer of bare UCNPs from the hydrophobic phase to the polar phase. Moreover, short-chain acids can be added directly as well to remove the surface ligand when cyclohexane, chloroform, or toluene is adopted as the dispersant for OA-UCNPs. The here described method is simply performed at ambient conditions on the second time scales without affecting the size, shape, and phase of the crystals. Additionally, the here developed method can be applied to HCl-sensitive materials as well. More importantly, ligand-free UCNPs are readily functionalized by sequential surface functionalization with water-soluble capping molecules. By coupling PEI-csUCNPs with EBT dye, we are able to construct FRET nanoprobes, providing the ability for selective Cu2+ sensing. We believe that this facile approach can be applied to other oleate-stabilized nanoparticles, allowing for the hydrophobic-to-hydrophilic transition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
C. Sun acknowledges the financial support from the China Scholarship Council (CSC, No. 201404910463) and TU Berlin. Furthermore, we are grateful to Ina Speckmann and Christina Eichenauer for the XRD and TGA measurements, respectively.Notes and references
- Y. Wang, K. Zheng, S. Song, D. Fan, H. Zhang and X. Liu, Chem. Soc. Rev., 2018, 47, 6473–6485 RSC.
- J. Zhou, Q. Liu, W. Feng, Y. Sun and F. Li, Chem. Rev., 2015, 115, 395–465 CrossRef CAS PubMed.
- Y. Shao, B. Liu, Z. Di, G. Zhang, L.-D. Sun, L. Li and C.-H. Yan, J. Am. Chem. Soc., 2020, 142, 3939–3946 CrossRef CAS PubMed.
- L. He, M. Brasino, C. Mao, S. Cho, W. Park, A. P. Goodwin and J. N. Cha, Small, 2017, 13, 1700504 CrossRef PubMed.
- L. Francés-Soriano, M. A. Zakharko, M. González-Béjar, P. A. Panchenko, V. Herranz-Pérez, D. Pritmov, M. A. Grin, A. Mironov, J. M. García-Verdugo, O. A. Fedorova and J. Pérez-Prieto, Chem. Mater., 2018, 30, 3677–3682 CrossRef.
- M. Tang, X. Zhu, Y. Zhang, Z. Zhang, Z. Zhang, Q. Mei, J. Zhang, M. Wu, J. Liu and Y. Zhang, ACS Nano, 2019, 13, 10405–10418 CrossRef CAS PubMed.
- J. Liu, R. Zhang, C. Shang, Y. Zhang, Y. Feng, L. Pan, B. Xu, T. Hyeon, W. Bu, J. Shi and J. Du, J. Am. Chem. Soc., 2020, 142, 7858–7867 CrossRef CAS PubMed.
- V. Kilin, G. Campargue, I. Fureraj, S. Sakong, T. Sabri, F. Riporto, A. Vieren, Y. Mugnier, C. Mas, D. Staedler, J. Michael Collins, L. Bonacina, A. Vogel, J. A. Capobianco and J. Wolf, ACS Nano, 2020, 14, 4087–4095 CrossRef CAS PubMed.
- X. Song, S. Li, H. Guo, W. You, X. Shang, R. Li, D. Tu, W. Zheng, Z. Chen, H. Yang and X. Chen, Angew. Chem., Int. Ed., 2019, 58, 18981–18986 CrossRef CAS PubMed.
- X. Liu, X. Li, X. Qin, X. Xie, L. Huang and X. Liu, Adv. Mater., 2017, 29, 1702315 CrossRef PubMed.
- E. Hemmer, M. Quintanilla, F. Légaré and F. Vetrone, Chem. Mater., 2015, 6, 235–244 CrossRef.
- L. Marciniak, K. Prorok and A. Bednarkiewicz, J. Mater. Chem. C, 2017, 5, 7890–7897 RSC.
- J. Zhao, J. Gao, W. Xue, Z. Di, H. Xing, Y. Lu and L. Li, J. Am. Chem. Soc., 2017, 140, 578–581 CrossRef PubMed.
- W. Ren, G. Lin, C. Clarke, J. Zhou and D. Jin, Adv. Mater., 2020, 32, 1901430 CrossRef CAS PubMed.
- T. Sun, B. Xu, B. Chen, X. Chen, M. Li, P. Shi and F. Wang, Nanoscale, 2017, 9, 2701–2705 RSC.
- B. Zhou, B. Shi, D. Jin and X. Liu, Nat. Nanotechnol., 2015, 10, 924–936 CrossRef CAS PubMed.
- G. S. Yi and G. M. Chow, Adv. Funct. Mater., 2006, 16, 2324–2329 CrossRef CAS.
- J.-C. Boyer, F. Vetrone, L. A. Cuccia and J. A. Capobianco, J. Am. Chem. Soc., 2006, 128, 7444–7445 CrossRef CAS PubMed.
- H.-X. Mai, Y.-W. Zhang, R. Si, Z.-G. Yan, L.-D. Sun, L.-P. You and C.-H. Yan, J. Am. Chem. Soc., 2006, 128, 6426–6436 CrossRef CAS PubMed.
- N. J. J. Johnson, A. Korinek, C. Dong and F. C. J. M. van Veggel, J. Am. Chem. Soc., 2012, 134, 11068–11071 CrossRef CAS PubMed.
- X. Wang, J. Zhuang, Q. Peng and Y. Li, Nature, 2005, 437, 121–124 CrossRef CAS PubMed.
- Z. Li and Y. Zhang, Nanotechnology, 2008, 19, 345606 CrossRef PubMed.
- J.-C. Boyer, M.-P. Manseau, J. I. Murray and F. C. J. M. van Veggel, Langmuir, 2010, 26, 1157–1164 CrossRef CAS PubMed.
- N. J. J. Johnson, N. M. Sangeetha, J.-C. Boyer and F. C. J. M. van Veggel, Nanoscale, 2010, 2, 771–777 RSC.
- N. Bogdan, F. Vetrone, R. Roy and J. A. Capobianco, J. Mater. Chem., 2010, 20, 7543–7550 RSC.
- G.-S. Yi and G.-M. Chow, Chem. Mater., 2007, 19, 341–343 CrossRef CAS.
- M. Nyk, R. Kumar, T. Y. Ohulchanskyy, E. J. Bergey and P. N. Prasad, Nano Lett., 2008, 8, 3834–3838 CrossRef CAS PubMed.
- W. Ren, S. Wen, S. A. Tawfik, Q. P. Su, G. Lin, L. A. Ju, M. J. Ford, H. Ghodke, A. van Oijen and D. Jin, Chem. Sci., 2018, 9, 4352–4358 RSC.
- H. Hu, M. Yu, F. Li, Z. Chen, X. Gao, L. Xiong and C. Huang, Chem. Mater., 2008, 20, 7003–7009 CrossRef CAS.
- H.-P. Zhou, C.-H. Xu, W. Sun and C.-H. Yan, Adv. Funct. Mater., 2009, 19, 3892–3900 CrossRef CAS.
- Z. Chen, H. Chen, H. Hu, M. Yu, F. Li, Q. Zhang, Z. Zhou, T. Yi and C. Huang, J. Am. Chem. Soc., 2008, 130, 3023–3029 CrossRef CAS PubMed.
- A. Dong, X. Ye, J. Chen, Y. Kang, T. Gordon, J. M. Kikkawa and C. B. Murray, J. Am. Chem. Soc., 2011, 133, 998–1006 CrossRef CAS PubMed.
- N. Bogdan, F. Vetrone, G. A. Ozin and J. A. Capobianco, Nano Lett., 2011, 11, 835–840 CrossRef CAS PubMed.
- C. Wang, H. Tao, L. Cheng and Z. Liu, Biomaterials, 2011, 32, 6145–6154 CrossRef CAS PubMed.
- S. Cui, H. Chen, H. Zhu, J. Tian, X. Chi, Z. Qian, S. Achilefu and Y. Gu, J. Mater. Chem., 2012, 22, 4861–4873 RSC.
- M. Deng, N. Tu, F. Bai and L. Wang, Chem. Mater., 2012, 24, 2592–2597 CrossRef CAS.
- S. H. Nam, Y. M. Bae, Y. I. Park, J. H. Kim, H. M. Kim, J. S. Choi, K. T. Lee, T. Hyeon and Y. D. Suh, Angew. Chem., Int. Ed., 2011, 50, 6093–6097 CrossRef CAS PubMed.
- L.-L. Li, R. Zhang, L. Yin, K. Zheng, W. Qin, P. R. Selvin and Y. Lu, Angew. Chem., Int. Ed., 2012, 51, 6121–6125 CrossRef CAS PubMed.
- R. Abdul Jalil and Y. Zhang, Biomaterials, 2008, 29, 4122–4128 CrossRef CAS PubMed.
- J.-N. Liu, W.-B. Bu and J.-L. Shi, Acc. Chem. Res., 2015, 48, 1797–1805 CrossRef CAS PubMed.
- R. Arppe, I. Hyppanen, N. Perala, R. Peltomaa, M. Kaiser, C. Wurth, S. Christ, U. Resch-Genger, M. Schaferling and T. Soukka, Nanoscale, 2015, 7, 11746–11757 RSC.
- F. T. Rabouw, P. T. Prins, P. Villanueva-Delgado, M. Castelijns, R. G. Geitenbeek and A. Meijerink, ACS Nano, 2018, 12, 4812–4823 CrossRef CAS PubMed.
- B. Chen, B. Ren and F. Wang, Chem. Mater., 2019, 31, 9497–9503 CrossRef CAS.
- W. Kong, T. Sun, B. Chen, X. Chen, F. Ai, X. Zhu, M. Li, W. Zhang, G. Zhu and F. Wang, Inorg. Chem., 2017, 56, 872–877 CrossRef CAS PubMed.
- F. Wang, R. Deng, J. Wang, Q. Wang, Y. Han, H. Zhu, X. Chen and X. Liu, Nat. Mater., 2011, 10, 968–973 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, ligand density calculation, schematic illustration of ligand removal process in a biphasic solvent system, TEM images and ζ-potential of ligand-free UCNPs prepared in different systems, XRD results of oleate and oleate-free UCNPs and csUCNPs, characterization of polymer-modified UCNPs including TEM, FT-IR, TGA and UCL spectra, UCL spectra of oleate-capped and bare UCNPs, spectrophotometric titration spectra. See DOI: 10.1039/d0ma00411a |
| This journal is © The Royal Society of Chemistry 2020 |