Open Access Article
Open Access ArticleNanonewton forces between Staphylococcus aureus surface protein IsdB and vitronectin†
Marion
Mathelié-Guinlet
 a,
Felipe
Viela
a,
Felipe
Viela
 a,
Giampiero
Pietrocola
a,
Giampiero
Pietrocola
 b,
Pietro
Speziale
b,
Pietro
Speziale
 *b and
Yves F.
Dufrêne
*b and
Yves F.
Dufrêne
 *ac
*ac
aLouvain Institute of Biomolecular Science and Technology, UCLouvain, Croix du Sud, 4-5, bte L7.07.07, B-1348 Louvain-la-Neuve, Belgium. E-mail: yves.dufrene@uclouvain.be
bDepartment of Molecular Medicine, Unit of Biochemistry, University of Pavia, Viale Taramelli 3/b, 27100 Pavia, Italy. E-mail: pietro.speziale@unipv.it
cWalloon Excellence in Life Sciences and Biotechnology (WELBIO), B-1300 Wavre, Belgium
First published on 30th October 2020
Abstract
Single-molecule experiments have recently revealed that the interaction between staphylococcal surface proteins and their ligands can be extremely strong, equivalent to the strength of covalent bonds. Here, we report on the unusually high binding strength between Staphylococcus aureus iron-regulated surface determinant B (IsdB) and vitronectin (Vn), an essential human blood protein known to interact with bacterial pathogens. The IsdB–Vn interaction is dramatically strengthened by mechanical tension, with forces up to 2000 pN at a loading rate of 105 pN s−1. In line with this, flow experiments show that IsdB-mediated bacterial adhesion to Vn is enhanced by fluid shear stress. The stress-dependent binding of IsdB to Vn is likely to play a role in promoting bacterial adhesion to human cells under fluid shear stress conditions.
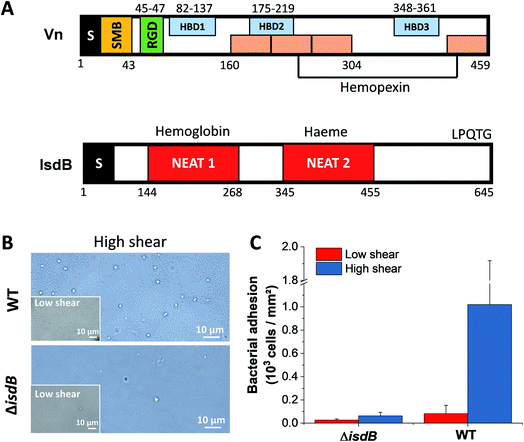
Staphylococcus aureus, a human commensal of the microbiota frequently colonizing the respiratory tract and the skin,1,2 is also an opportunistic pathogen causing a range of illnesses from minor skin and respiratory infections to life-threatening diseases such as endocarditis and sepsis.2–4 In the past decades, due to its key role in nosocomial infections and to the emergence of antibiotic-resistant strains, S. aureus has become a worldwide clinical problem.2,5S. aureus can invade a variety of mammalian cells, from endothelial6,7 and epithelial cells8–10 to osteoblasts,11,12 thereby enabling it to evade host immune defenses. Attachment of bacterial pathogens to host cells and biomaterials is the first step leading to infection. Key players in cell adhesion are bacterial adhesins which specifically bind to host surfaces and proteins from the extracellular matrix (ECM).13 Among these, vitronectin (Vn) plays major roles in bacterial pathogenesis by supporting complement escape to the immune system, adhesion and cellular invasion.14,15 Vn is a ∼75 kDa (sequence ∼ 450 residues) multifunctional glycoprotein found as folded monomers in the plasma and as multimers in the ECM.16 It comprises a somatomedin-B domain, followed by an RGD sequence, four hemopexin-like repeats and three heparin-binding domains (Fig. 1A).16,17
S. aureus can bind Vn with high-to-moderate affinity (10−10 to 10−8 M) depending on the bacterial recognition sites involved.18–20 Many microbial pathogens, including S. aureus were found to bind human Vn via the same C-terminal heparin-binding domain, leaving the complement regulatory region of Vn accessible and subsequently allowing versatile host immune responses.21,22 Of note, similarly to Moraxella catarrhalis and Haemophilus influenzae14 both multimeric and monomeric forms of Vn have the capability to interact with S. aureus. But, multimeric forms of Vn bind to S. pneumoniae and S. aureus more efficiently than monomeric forms, likely due to better exposition of bacterial binding-sites in such conformations.23–26 Furthermore, Vn also enhances staphylococcal adhesion to biomaterials like catheters.27
Staphylococci express iron-regulated surface determinant (Isd) proteins that are upregulated under iron-restricted conditions.28,29 These proteins, whose main task is to bind hemoglobin and extract heme to provide the cell with a source of iron, contain one to three near iron transporter (NEAT) motifs (Fig. 1A). These domains consist of ∼120 amino acids with a conserved eight-stranded beta-sandwich fold that mediates the binding to hemoproteins or heme through an hydrophobic pocket.30 In addition, Isd proteins are involved in adhesion, colonization and pathogenesis.31 For instance, IsdA and IsdB were shown to promote adhesion to squamous epithelial cells and platelets respectively, and their subsequent invasion.32,33 IsdC from Staphylococcus lugdunensis increases adhesion to abiotic surfaces but also in between cells, subsequently favoring biofilm formation.34 Zapotoczna et al. suggested that S. aureus IsdB interacts with specific integrins that bind ligands with an RGD motif, improving further cell invasion.35 Vn is one such ligand which has distinct RGD sequences and bacterial-binding domains.14 It has therefore been suggested that IsdB stabilizes Vn-binding integrins, allowing more vitronectin to bind.35
IsdB has been shown to act as a receptor for Vn (multimeric and monomeric conformation), under iron starvation conditions under which those adhesins are expressed at the surface of S. aureus.36 Adherence to and subsequent invasion of mammalian cells were then promoted by the bridge Vn can form between IsdB and αvβ3 integrin on the host cell. Despite the potential role of Vn-binding by Isd proteins in S aureus pathogenesis, the molecular details of such interaction and the adhesins involved have not yet been elucidated. Here we demonstrate that IsdB and Vn form molecular complexes with very high mechanical stability, which is achieved under high physical stress.
Results
IsdB mediates strong bacterial adhesion to Vn-coated surfaces
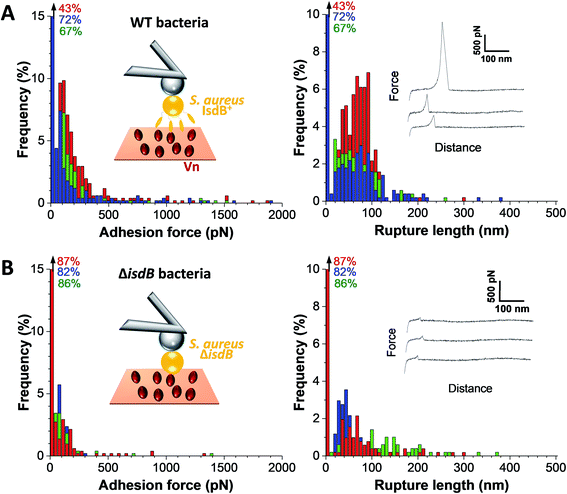
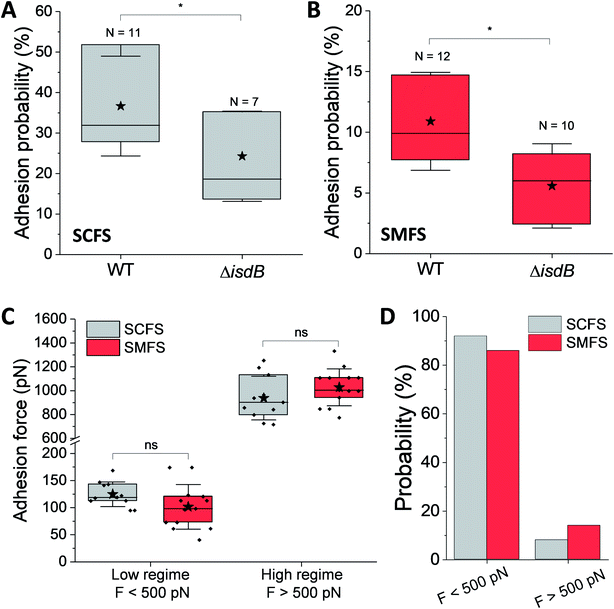
We first tested the ability of S. aureus SH1000 to bind to Vn using a macroscopic adhesion assay, where bacterial suspensions were flowed over Vn-coated substrates mounted into a microparallel flow chamber. As S. aureus can encounter Vn in the bloodstream, allowing for a better colonization of endothelial cells, bacterial adhesion was first studied under flow at low and high shear stress, 12 and 120 s−1 respectively lying in the moderate range of physiological values which can broadly vary depending on blood vessels37 (Fig. 1B and C). Adhering bacteria were quantified by direct observation by optical microscopy. While WT bacteria poorly adhered at low shear rate, adhesion was largely increased at high shear rate, corresponding to normal venous shear rates. Mutant cells lacking IsdB poorly adhered whatever the flow conditions. This shows that IsdB supports S aureus adhesion to Vn surfaces under fluid flow, in a shear stress dependent manner.This prompted us to study the molecular interactions between IsdB and Vn using AFM. Force–distance curves were recorded by performing approach–retract cycles between single bacteria immobilized on an AFM colloidal probe and a defined area (5 μm × 5 μm) of Vn-coated substrates (single-cell force spectroscopy, SCFS, Fig. 2, for more cells see ESI Fig. 1†). From those pixel-by-pixel force–distance maps, adhesion events were determined. For three representative cells, many force curves (39%, mean from 3 cells; Fig. 2A) featured adhesion events with small forces centered at 155 ± 96 pN (mean ± s.d.; n = 531 adhesive curves; 3 cells). Of note, a few adhesion events (∼10%) of 1026 ± 450 pN (mean ± s.d.; n = 67 adhesive curves; 3 cells) were also observed. Mutant cells lacking IsdB poorly adhered to Vn, with lower forces (Fig. 2B): the adhesion probability decreased significantly from 37% to 24% (mean from 11 and 7 cells for WT and ΔisdB respectively; Fig. 4A) – supporting the specificity of the forces measured with the WT. Nonetheless the residual adhesion observed with mutant cells might reflect interactions with other Vn-receptors or unspecific attachment. It is worth mentioning that such a decrease cannot be explained by either Vn being ripped from the substrate or IsdB/Vn being removed from their corresponding surface because of the EDC/NHS chemistry used and the constant binding probability over time respectively.
Bacterial-Vn interactions ruptured at 73 ± 41 nm (mean ± s.d.; n = 531 adhesive curves; 3 cells). Vn molecules were immobilized on the substrates through multiple sites, meaning they should not substantially contribute to the measured extensions. Mature IsdB is made of ∼600 residues; assuming that each amino acid contributes 0.36 nm, the fully unfolded protein should be ∼215 nm. This suggests that bonds rupture before complete unfolding of IsdB, thus suggesting the protein is mechanically stable.
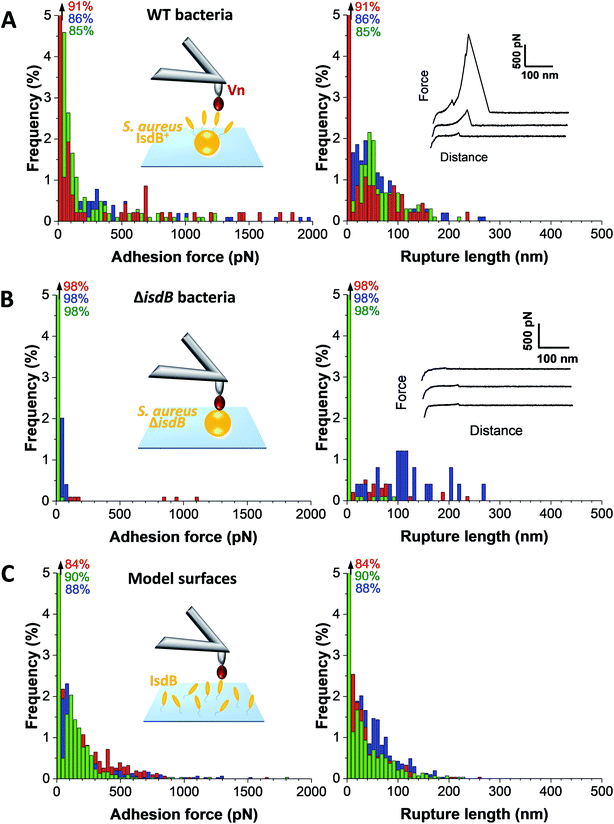
Strength and dynamics of the IsdB–Vn interaction
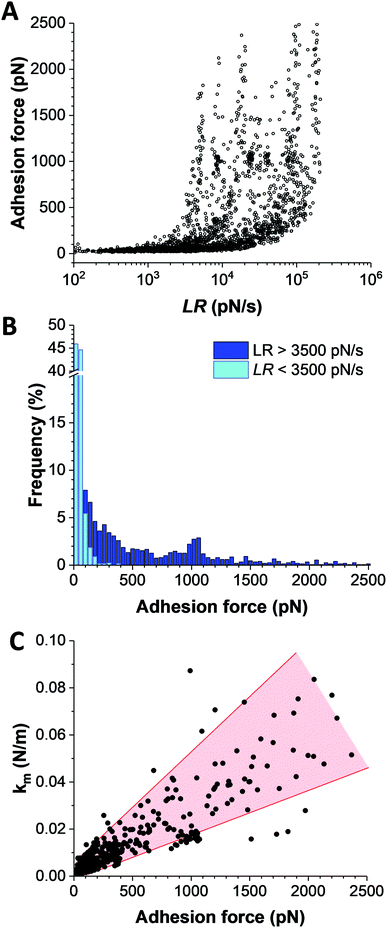
The strength of single IsdB–Vn bonds was then studied by pulling on single IsdB adhesins on top of the living bacteria immobilized on a polystyrene dish with AFM tip functionalized with Vn, in a similar pixel-by-pixel force–distance maps (single-molecule force spectroscopy, SMFS, Fig. 3, for more cells see ESI Fig. 2†). Consistent with SCFS experiments, weak forces of 137 ± 117 pN (mean ± s.d.; n = 271 adhesive curves; 3 cells) were observed for representative WT cells but also much larger ones, ranging from 500 pN to 2000 pN (∼25% of adhesive events) (Fig. 3A). Similar force distributions were obtained both in SCFS and SMFS (Fig. 4C and D): (i) in the low force regime, mean forces of 125 ± 23 pN (probability of 92%, n = 11 cells) and 101 ± 41 pN (probability of 86%, n = 13 cells) were respectively reported while (ii) in the high force regime, mean forces of 870 ± 117 pN (probability of 8%, n = 11 cells) and 1028 ± 155 pN (probability of 17%, n = 13 cells) were respectively observed. This, along with the single-peak nature of adhesion events (see retraction profiles in Fig. 2 and 3) and the absence of clear multiples of a single adhesion force unit, support the idea that we are probing single bonds rather than multiple bonds rupturing in parallel. Again, binding was specific as most adhesion events were abrogated with the mutant lacking IsdB (Fig. 3B), for which the adhesion probability dropped from 11% (mean on 12 WT cells) to 6% (mean on 10 ΔisdB cells) (Fig. 4B). The rupture lengths were 63 ± 44 nm, which is smaller than the length of unfolded adhesins (∼210 nm), again confirming the rupture before complete unfolding. Finally, we also probed recombinant IsdB proteins, showing that, overall, they feature similar binding force properties for Vn (Fig. 3C). However, on closer examination we found that strong forces > 500 pN were hardly observed. This suggests that when adhesins are randomly attached on a surface they cannot engage into strong bonds, unlike when they are natively oriented on living bacteria.Bacterial adhesion is strongly influenced by physical stress like fluid flow.38,39 To sustain these mechanical forces, S. aureus express a series of surface adhesions that firmly bind to host cells and ECM proteins.40 SdrG, ClfA, and ClfB have been shown to bind their ligands through ultrastrong forces, activated by mechanical tension.41–43 To test whether this also happens with IsdB, the adhesion force between Vn tips and IsdB cells was measured at different retraction speeds (from 0.5 μm s−1 to 10 μm s−1, see ESI Fig. 3†), in turn different loading rates (LRs), estimated from the force vs. time curves (Fig. 5A). As exemplified for one representative cell, (i) no significant dependency was found between adhesion force and rupture length, at a defined retraction speed, and (ii) the rupture length distributions (and average values) obtained when probing the cell at different speeds were not significantly different (ESI Fig. 3†). We are thus confident that the same adhesion events are probed along the loading rate variations. Then, unbinding forces were analyzed over discrete ranges of LRs (Fig. 5B, see also ESI Fig. 4†). While adhesion forces showed a spread distribution neither described by the classical Bell Evans44 or Friddle-Noy-De Yoreo45 models, these measurements revealed that only weak forces of 51 ± 45 pN were observed at low LRs (<3500 pN s−1), while strong forces of 506 ± 538 pN dominated at high LRs (>3500 pN s−1). It is worth mentioning that, at the highest LRs probed here, the measured forces might actually be underestimated by ∼10% due to the cantilever dynamics, meaning that the actual “true” rupture forces might even be higher at the highest LRs.46 Though it is difficult to uncorrelate the mechanical tension exerted on the bond from the actual measurement of the rupture force, it is clear that the probability of observing strong rupture forces increases with the LR, while the population of lower forces becomes depleted (ESI Fig. 4†). Such shift towards higher forces, both in terms of probability and strength suggests a strengthening of the IsdB–Vn bond with tensile force. This behavior is fully consistent with the higher adhesion of IsdB bacteria to Vn-coated surfaces when the shear rate is increased (Fig. 1C). Finally, we assessed the spring constant of the molecular complex (km) using the slope of the linear portion of the force vs. tip position curves (Fig. 5C). Compared to weak forces, strong forces > 500 pN were associated with higher molecular stiffness (km = 4.2 ± 3.9 pN nm−1vs. km = 35.3 ± 22.3 pN nm−1 respectively; mean ± s.d. over 6 cells). Altogether our data suggest that the shift towards strong forces results from a change in the conformational state of IsdB, from a weak- to a strong-binding state, as observed for catch bonds.
Discussion
The structure of IsdB is unique in that it contains two NEAT motifs, structurally solved, that bind to hemoglobin or haem.16 While IsdA which has one such motif binds multiple ligands – haem, fibrinogen, fibronectin, cytokeratin 10, and loricrin –,47 the ligand-binding activities of IsdB are much less understood. IsdB promotes adhesion to human cells as well as platelets,35 but whether this involves specific ligand interactions is currently unknown. Recently, IsdB has been identified as a specific, high affinity Vn-binding protein and both NEAT motifs of the protein individually bind Vn with an affinity in the nanomolar range.36 Furthermore, IsdB binding to Vn was specifically inhibited by heparin and high concentrations of NaCl. Biological implications of such interaction involve the interference of IsdB-bound Vn with the assembly and deposition of the terminal complement complex on the bacterial surface and adherence to and invasion of epithelial and endothelial cells by S. aureus. We have shown here that IsdB and Vn form molecular complexes that are extremely stable, eventually reaching high strength in the nN regime. We expect this interaction to be of biological importance as Vn plays multiple functions in bacterial pathogenesis, including mediating complement escape, bacterial adherence, and cellular invasion.14The probability of forming strong bonds increases with the rate of applied force, strong IsdB–Vn adhesion being only observed at high tensile load. In line with this, macroscopic adhesion assay demonstrate higher bacterial adhesion levels at increased shear stress. As previously hypothesized in the case of the prototypical strong SdrG–Fg bonds,48 such force-enhanced activity is reminiscent of a catch bond behavior, i.e. the counterintuitive capacity of a bond to be longer-lived under high external stress than under low stress, as opposed to the increasing shorter-lived slip bonds. The transition from weak to strong interaction correlates with an increase in molecular stiffness. For the mechanically stable multidomain cellulosome protein complex,49 steered molecular dynamics and single-molecule experiments revealed that strong forces (600–750 pN) can be achieved if the complex directs force along pathways nonparallel to the pulling direction. This supports a model whereby force induces conformational changes in the adhesin, from a weak-binding folded state, to a strong-binding extended state. We speculate that IsdB might sense mechanical forces acting on the cell surface and induce intracellular responses.
Vn is present in the ECM and in soluble form in biological fluids and shows similarity, in terms of adhesion properties, with fibrinogen (Fg) and fibronectin (Fn). It is thus interesting that our finding of a strong interaction triggered by mechanical force is qualitatively similar to what has been recently described for the some staphylococcal adhesins binding either Fg or Fn through DLL41–43,48 and β-zipper50,51 mechanisms. The strong DLL interaction results from an intricate hydrogen bond network between the ligand peptide backbone and the N2N3 subdomains.48 Under load, the screw-like arrangement of hydrogen bonds maintains the peptide in a perfect shear geometry. In the β-zipper interaction, the well-studied FnBPA–Fn interactions involve the binding of Fn-binding repeats (FnBRs) to Fn-FI modules, subsequently forming additional β-strands along triple peptide β-sheets in the Fn molecule.51
What is the molecular basis of the IsdB–Vn interaction? Hallströmet et al. found that several pathogens recognize Vn, via the same C-terminal heparin-binding domain (amino acids 352–374).21 This allowed microbes to utilize the complement inhibitory function of Vn, as the N-terminal functional region of vitronectin was not shielded. This contributes to evasion of direct lysis by complement, which is an important step in pathogenesis. We therefore speculate that both NEAT domains of IsdB bind the C-terminal site of Vn, as already proposed by Pietrocola et al.36
That ligand-binding is strengthened by mechanical force highlights the importance of physical stress in activating S. aureus–hosts interactions.52 In the human body, the pathogen can be exposed to variable amounts of Vn as well as different shear stress conditions. Notably, Vn is abundantly present in periodontal, lung and kidney tissues while being less available in skin and gastrointestinal tissues. The stress-dependent IsdB–Vn adhesion may thus help the bacteria to efficiently colonize host cells through weak or strong interactions depending on the physical stress they experience.
Experimental procedures
Bacterial strains and growth conditions
S. aureus SH1000 (both WT and ΔisdB cells) were first grown in Brain Heart Infusion (BHI) broth overnight, at 37 °C and under shaking at 200 rpm, to reach their stationary phase. They were then diluted 100× and grown in RPMI overnight (37 °C, 200 rpm) to create iron-restricted conditions. For AFM experiments, cells were harvested by centrifugation at 3000 × g for 5 minutes and washed twice with PBS. Construction of S. aureus isdB deletion mutant was performed as reported previously.35Isolation of Vn from human plasma
Human Vn was purified from human plasma on a heparin- Sepharose Hi-TrapTM column (GE Healthcare) following the protocol reported by Pietrocola et al.36Functionalization of substrates and cantilevers with Vn
Gold-coated glass coverslips and cantilevers (OMCL-TR400PB-1, Olympus Ltd., Tokyo, Japan; nominal spring constant ∼ 0.02 N m−1) were immersed overnight in an ethanol solution containing 1 mM of 10% 16-mercaptododecahexanoic acid/90% 1-mercapto-1-undecanol (Sigma), rinsed with ethanol and dried with N2. Substrates and cantilevers were then immersed for 30 min into a solution containing 10 mg mL−1N-hydroxysuccinimide (NHS) and 25 mg mL−1 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (Sigma) and rinsed with Ultrapure water (ELGA LabWater). Finally, they were incubated with 0.1 mg mL−1 of Vn for 1 h, rinsed further with PBS buffer, and then immediately used without dewetting.Bacterial adhesion to vitronectin-coated surfaces
For flow experiments, S. aureus bacterial suspensions were flowed over vitronectin-coated surfaces for 2 min. using a fluidic chamber and a peristaltic pump (Miniplus, Gilson).53 Two different flow rates were tested, 2 and 20 mL min−1, corresponding to shear rates of 12 and 120 s−1 respectively that match the ones encountered in veins.37 Loosely attached bacteria were removed by flowing PBS during 2 min using the corresponding flow rates. Adhering bacteria were imaged using an inverted microscope (Leica DM16000) and counted using the ImageJ image analysis software (NIH Image).Single-cell force spectroscopy
Colloidal probes were prepared as described earlier.54 Briefly, single silica microsphere (6.1 μm diameter, Bangs laboratories) were attached with a thin layer of UV-curable glue (NOA 63, Norland Edmund Optics) on triangular shaped tip-less cantilevers (NP-O10, Bruker). Those newly modified cantilevers were then immersed for 1 h in Tris-buffered saline (TBS; Tris, 50 mM; NaCl, 150 mM; pH 8.5) containing 4 mg mL−1 dopamine hydrochloride (Sigma-Aldrich), rinsed in TBS, and directly used to catch single cells. The spring constant was finally determined by the thermal noise method and gave k ∼ 0.08 N m−1. For single-cell experiments, 100 μL of a suspension of 1 × 106 cells were dropped on a glass Petri dish, next to a Vn-coated substrate, the whole being immersed in PBS. The colloidal probe was brought into contact with a bacterium and retracted to catch the cell through electrostatic interactions with polydopamine. The cell probe was then positioned over Vn-substrates without dewetting. Cell probes were used to measure interaction forces on Vn-surfaces at room temperature by recording multiple forces curves (16 × 16) on different spots, a maximum applied force of 250 pN, and approach and retraction speeds of 1 μm s−1 and a contact time of 500 ms. Data were analyzed with the data processing software from JPK Instruments (Berlin, Germany). Adhesion force and rupture distance histograms were obtained by calculating the adhesion force and rupture distance of the last peak for each curve.Single-molecule force spectroscopy
For SMFS experiments, cantilevers (k ∼ 0.02 N m−1) were prepared as described above and bacteria were immobilized on polystyrene substrates. SMFS measurements were performed at room temperature in PBS buffer with a NanoWizard IV atomic force microscope (JPK Instruments). Adhesion maps were obtained by recording 32 × 32 force–distance curves on areas of 500 by 500 nm2 with an applied force of 250 pN, a constant approach and retraction speed of 1 μm s−1 and a contact time of 500 ms. SMFS data were processed and analyzed the same was as for SCFS experiments.Author contributions
MMG, FV, GP, PS and YFD designed the experiments, analysed the data and wrote the article. MMG and FV collected the data.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Work at the Université catholique de Louvain was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 693630), the FNRS-WELBIO (grant n°WELBIO-CR-2015A-05), the National Fund for Scientific Research (FNRS), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y. F. D. is Research Director at the FNRS.References
- A. L. Byrd, Y. Belkaid and J. A. Segre, Nat. Rev. Microbiol., 2018, 16, 143–155 CrossRef CAS.
- C. P. Parlet, M. M. Brown and A. R. Horswill, Trends Microbiol., 2019, 27, 497–507 CrossRef CAS.
- D. C. Angus and T. van der Poll, N. Engl. J. Med., 2013, 369, 840–851 CrossRef CAS.
- L. Thomer, O. Schneewind and D. Missiakas, Annu. Rev. Pathol.: Mech. Dis., 2016, 11, 343–364 CrossRef CAS.
- P. Moreillon and Y.-A. Que, Lancet, 2004, 363, 139–149 CrossRef.
- S. K. Ogawa, E. R. Yurberg, V. B. Hatcher, M. A. Levitt and F. D. Lowy, Infect. Immun., 1985, 50, 218–224 CrossRef CAS.
- B. E. Menzies and I. Kourteva, Infect. Immun., 1998, 66, 5994–5998 CrossRef CAS.
- K. W. Bayles, C. A. Wesson, L. E. Liou, L. K. Fox, G. A. Bohach and W. R. Trumble, Infect. Immun., 1998, 66, 336–342 CrossRef CAS.
- K. Dziewanowska, J. M. Patti, C. F. Deobald, K. W. Bayles, W. R. Trumble and G. A. Bohach, Infect. Immun., 1999, 67, 4673–4678 CrossRef CAS.
- B. Sinha, P. P. François, O. Nüße, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann and K.-H. Krause, Cell. Microbiol., 1999, 1, 101–117 CrossRef CAS.
- J. K. Ellington, S. S. Reilly, W. K. Ramp, M. S. Smeltzer, J. F. Kellam and M. C. Hudson, Microb. Pathog., 1999, 26, 317–323 CrossRef CAS.
- M. Jevon, C. Guo, B. Ma, N. Mordan, S. P. Nair, M. Harris, B. Henderson, G. Bentley and S. Meghji, Infect. Immun., 1999, 67, 2677–2681 CrossRef CAS.
- T. J. Foster, Trends Microbiol., 2019, 27, 927–941 CrossRef CAS.
- B. Singh, Y.-C. Su and K. Riesbeck, Mol. Microbiol., 2010, 78, 545–560 CrossRef CAS.
- B. Singh, Y. Su and K. Riesbeck, Open Life Sci., 2011, 6(6), 973–980 CAS.
- I. Schvartz, D. Seger and S. Shaltiel, Int. J. Biochem. Cell Biol., 1999, 31, 539–544 CrossRef CAS.
- K. Shin, B. C. Lechtenberg, L. M. Fujimoto, Y. Yao, S. S. Bartra, G. V. Plano and F. M. Marassi, Sci. Adv., 2019, 5, eaax5068 CrossRef CAS.
- J. I. Fuquay, D. T. Loo and D. W. Barnes, Infect. Immun., 1986, 52, 714–717 CrossRef CAS.
- G. S. Chhatwal, K. T. Preissner, G. Müller-Berghaus and H. Blobel, Infect. Immun., 1987, 55, 1878–1883 CrossRef CAS.
- O. D. Liang, M. Maccarana, J.-I. Flock, M. Paulsson, K. T. Preissner and T. Wadström, Biochim. Biophys. Acta, Mol. Basis Dis., 1993, 1225, 57–63 CrossRef CAS.
- T. Hallström, B. Singh, P. Kraiczy, S. Hammerschmidt, C. Skerka, P. F. Zipfel and K. Riesbeck, PLoS One, 2016, 11, e0147709 CrossRef.
- O. D. Liang, J.-I. Flock and T. Wadstrom, J. Biochem., 1994, 116, 457–463 CrossRef CAS.
- S. Bergmann, A. Lang, M. Rohde, V. Agarwal, C. Rennemeier, C. Grashoff, K. T. Preissner and S. Hammerschmidt, J. Cell Sci., 2009, 122, 256–267 CrossRef CAS.
- M. Mahawar and P. Joshi, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 2008, 149, 410–418 CrossRef.
- P. Zhuang, A. I. Chen and C. B. Peterson, J. Biol. Chem., 1997, 272, 6858–6867 CrossRef CAS.
- C. Sa E Cunha, N. J. Griffiths and M. Virji, PLoS Pathog., 2010, 6(5), e1000911 CrossRef.
- F. Lundberg, S. Schliamser and Å. Ljungh, J. Med. Microbiol., 1997, 46, 285–296 CrossRef CAS.
- K. P. Haley and E. P. Skaar, Microbes Infect., 2012, 14, 217–227 CrossRef CAS.
- J. Sheldon and D. Heinrichs, Front. Cell. Infect. Microbiol., 2012, 2, 41 Search PubMed.
- J. C. Grigg, G. Ukpabi, C. F. M. Gaudin and M. E. P. Murphy, J. Inorg. Biochem., 2010, 104, 341–348 CrossRef CAS.
- G. Pishchany, A. L. McCoy, V. J. Torres, J. C. Krause, J. E. Crowe, M. E. Fabry and E. P. Skaar, Cell Host Microbe, 2010, 8, 544–550 CAS.
- R. M. Corrigan, H. Miajlovic and T. J. Foster, BMC Microbiol., 2009, 9, 22 Search PubMed.
- H. Miajlovic, M. Zapotoczna, J. A. Geoghegan, S. W. Kerrigan, P. Speziale and T. J. Foster, Microbiology, 2010, 156, 920–928 CrossRef CAS.
- A. Missineo, A. D. Poto, J. A. Geoghegan, S. Rindi, S. Heilbronner, V. Gianotti, C. R. Arciola, T. J. Foster, P. Speziale and G. Pietrocola, Infect. Immun., 2014, 82, 2448–2459 CrossRef.
- M. Zapotoczna, Z. Jevnikar, H. Miajlovic, J. Kos and T. J. Foster, Cell. Microbiol., 2013, 15, 1026–1041 CrossRef CAS.
- G. Pietrocola, A. Pellegrini, M. J. Alfeo, L. Marchese, T. J. Foster and P. Speziale, J. Biol. Chem., 2020, 295, 10008–10022 CrossRef CAS.
- M. H. Kroll, J. D. Hellums, L. V. McIntire, A. I. Schafer and J. L. Moake, Blood, 1996, 88, 1525–1541 CrossRef CAS.
- A. Persat, C. D. Nadell, M. K. Kim, F. Ingremeau, A. Siryaporn, K. Drescher, N. S. Wingreen, B. L. Bassler, Z. Gitai and H. A. Stone, Cell, 2015, 161, 988–997 CrossRef CAS.
- Y. F. Dufrêne and A. Persat, Nat. Rev. Microbiol., 2020, 1–14 Search PubMed.
- M. Otto, FEMS Microbiol. Rev., 2014, 38, 1250–1270 CrossRef CAS.
- P. Herman, S. El-Kirat-Chatel, A. Beaussart, J. A. Geoghegan, T. J. Foster and Y. F. Dufrêne, Mol. Microbiol., 2014, 93, 356–368 CrossRef CAS.
- P. Herman-Bausier, C. Labate, A. M. Towell, S. Derclaye, J. A. Geoghegan and Y. F. Dufrêne, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5564–5569 CrossRef CAS.
- P. Vitry, C. Valotteau, C. Feuillie, S. Bernard, D. Alsteens, J. A. Geoghegan and Y. F. Dufrêne, mBio, 2017, 8, e01748 CrossRef CAS.
- E. Evans, Annu. Rev. Biophys. Biomol. Struct., 2001, 30, 105–128 CrossRef CAS.
- R. W. Friddle, A. Noy and J. J. De Yoreo, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 13573–13578 CrossRef CAS.
- M. R. Uhlig, C. A. Amo and R. Garcia, Nanoscale, 2018, 10, 17112–17116 RSC.
- S. R. Clarke, M. D. Wiltshire and S. J. Foster, Mol. Microbiol., 2004, 51, 1509–1519 CrossRef CAS.
- L. F. Milles, K. Schulten, H. E. Gaub and R. C. Bernardi, Science, 2018, 359, 1527–1533 CrossRef CAS.
- C. Schoeler, R. C. Bernardi, K. H. Malinowska, E. Durner, W. Ott, E. A. Bayer, K. Schulten, M. A. Nash and H. E. Gaub, Nano Lett., 2015, 15, 7370–7376 CrossRef CAS.
- R. J. Bingham, E. Rudino-Pinera, N. A. G. Meenan, U. Schwarz-Linek, J. P. Turkenburg, M. Hook, E. F. Garman and J. R. Potts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 12254–12258 CrossRef CAS.
- U. Schwarz-Linek, J. M. Werner, A. R. Pickford, S. Gurusiddappa, J. H. Kim and E. S. Pilka, et al. , Nature, 2003, 423(6936), 177–181 CrossRef CAS.
- J. A. Geoghegan and Y. F. Dufrêne, Trends Microbiol., 2018, 26, 645–648 CrossRef CAS.
- T. Vanzieleghem, N. Couniot, P. Herman-Bausier, D. Flandre, Y. F. Dufrêne and J. Mahillon, Langmuir, 2016, 32, 7277–7283 CrossRef CAS.
- A. Beaussart, S. El-Kirat-Chatel, R. M. A. Sullan, D. Alsteens, P. Herman, S. Derclaye and Y. F. Dufrêne, Nat. Protoc., 2014, 9, 1049–1055 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0na00636j |
| This journal is © The Royal Society of Chemistry 2020 |