Open Access Article
Open Access ArticleLactic acid bacteria exhibit similar antioxidant capacities in Caenorhabditis elegans- and Campylobacter jejuni-infected mice†
Xing Jina,
Yufeng Hea,
Zhenmin Liu*b,
Yonghua Zhou*c,
Xiaohua Chend,
Gang Wang *aefg,
Zhihong Sunh,
Jianxin Zhaoaeg,
Hao Zhang
*aefg,
Zhihong Sunh,
Jianxin Zhaoaeg,
Hao Zhang aegij and
Wei Chen
aegij and
Wei Chen aeik
aeik
aState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China. E-mail: wanggang@jiangnan.edu.cn; Fax: +86-510-85912155; Tel: +86-510-85912155
bState Key Laboratory of Dairy Biotechnology, Shanghai Engineering Research Center of Dairy Biotechnology, Dairy Research Institute, Bright Dairy & Food Co., Ltd., Shanghai 200436, P. R. China. E-mail: liuzhenmin@brightdairy.com
cKey Laboratory of National Health and Family Planning Commission on Parasitic Disease Control and Prevention, Jiangsu Provincial Key Laboratory on Parasite and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi 214064, P. R. China. E-mail: zhouyonghua@jipd.com
dCollege of Life Sciences and Environment, Hengyang Normal University, Hengyang 421008, P. R. China
eSchool of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China
fInternational Joint Research Laboratory for Probiotics, Jiangnan University, Wuxi 214122, P. R. China
g(Yangzhou) Institute of Food Biotechnology, Jiangnan University, Yangzhou 225004, P. R. China
hKey Laboratory of Dairy Biotechnology and Engineering, Education Ministry of P. R. China, Inner Mongolia Agricultural University, Inner Mongolia 010018, P. R. China
iNational Engineering Research Center for Functional Food, Jiangnan University, Wuxi 214122, P. R. China
jWuxi Translational Medicine Research Center and Jiangsu Translational Medicine Research Institute Wuxi Branch, Wuxi 214122, P. R. China
kBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University (BTBU), Beijing 100048, P. R. China
First published on 21st January 2020
Abstract
Caenorhabditis elegans (C. elegans) is increasingly used as a model organism to screen probiotics for ageing research. In this study, a C. elegans lifespan analysis model exposed to H2O2 and juglone (for chronic and acute assays) was used to preselect lactic acid bacteria (LAB) with resistance to oxidative stress. The mechanism of oxidative stress protection was analyzed. Seven out of ten LAB strains screened for namely 427, X13, 9-5, 422, Z5, G14 and H29M-8M demonstrated higher levels of protection to C. elegans, ranging from 70% to 85% survival rate, in comparison to the inactive strains 408, 13-7 and 430 (35% to 45% survival rate). The survival rates of the seven oxidative stress tolerant strains were correlated to increase in catalase (CAT), superoxide dismutase (SOD) and malondialdehyde (MDA) levels and decrease in reactive oxygen species (ROS) expression in C. elegans during H2O2 and juglone exposure. In addition, the transcription of mitogen-activated protein kinase (MAPK) and Nrf2-Keap1-ARE pathway related genes was elevated in the oxidative stress protection of LAB isolates. Finally, a significant correlation was found between the ability of these LAB strains to protect C. elegans from H2O2 or juglone exposure and their antioxidative ability in Campylobacter jejuni-infected mice. Thus the results indicated that the oxidative stress-based lifespan model of C. elegans is useful for screening of LAB with antioxidant efficacy in pathogen-infected mammals.
Introduction
Oxidative stress does great harm to the health of organisms, including humans. For example, the etiology of Parkinson's disease, heart failure, cancer, atherosclerosis and Alzheimer's disease is linked to damage due to oxidative stress.1–6 Oxidative stress is attributed to excessive accumulation of reactive oxygen species (ROS) derived from endogenous or exogenous sources. In response to oxidative stress induced damage, organisms may activate production of antioxidant enzymes or phase II metabolising enzymes, such as superoxide dismutase (SOD), haem oxygenase-1 (HO-1), catalase (CAT) and glutathione-S-transferases (GSTs),7 via antioxidant responsive elements (AREs) in the promoter regions of these enzymes' genes. In addition, nuclear factor erythroid-2-related factor 2 (Nrf2), which belongs to the basic leucine zipper transcription factor (NF-E2) family, has been verified to play a vital antioxidant role in adjusting gene expression by binding to ARE to activate the Nrf2-ARE signalling pathway.8 In addition, the evolutionarily conserved endocrine insulin/IGF-1-like signalling (IIS) pathway and mitogen-activated protein kinases signalling pathway (MAPK) form part of the host's immune defence against oxidative stress.Lactic acid bacteria (LAB) are generally recognised as safe and widely used in food industry. Their health-promoting properties, such as anti-carcinogenic activity, inhibition of foodborne pathogens, anti-mutagenic and hypocholesterolaemic activity, lead to their increased uses.9,10
Although many methods exist for quantifying the antioxidant capacities of natural LAB,11,12 there is few in vivo methods for the rapid large-scale screening of LAB antioxidant for their ability to prolong the life of organisms. It has been reported that Lactobacillus paracasei Fn 032, Lactobacillus rhamnosus GG and Lactobacillus spp. can scavenge hydroxyl radicals in colon fermentation-mimetic systems.13 In addition, orally administered Lactococcus lactis and Lactobacillus plantarum were shown to protect against TNBS-induced colitis by generating SOD in mice and rats.14
However, all animal models have the disadvantages of high cost, long cycle, cumbersome operation and insignificant immune response indicators, and do not permit rapid large-scale screening for LAB capable of effectively protection against oxidative stress. In these respects, the uses of less complex test organisms may offer an attractive and practical solution.15
The small, free-living (non-parasitic) soil nematode (nematode) Caenorhabditis elegans (C. elegans) is widely applied in ecotoxicological studies as an in vivo model, due to its short generation time, diminutive form and defined genetic background. C. elegans has been used as a model for investigation of oxidative injury and its influence on ageing.16 Nematodes have also been shown to be a suitable model for screening of anti-Salmonella infection microorganisms and antibacterial compounds.17,18 Moreover, there is a correlation between the longevity of C. elegans and the activity of insulin-like growth factor and diabetes in mammals.19 Few studies have used C. elegans as a model for screening LAB for their ability to protect against oxidative damage and thereby enhance longevity. Also, no study has reported on the relationship between the antioxidant properties of LAB and their ability to alleviate pathogen infection-generated oxidative damage in animals.
In this study, a C. elegans lifespan model was used to investigate the biological response of nematodes to oxidative stress, enabling rapid evaluation of the mitigation effects of LAB. A Toxoplasma gondii (T. gondii)-induced acute ileitis model that abrogated the resistance of mice to Campylobacter jejuni infection was also used to validate the relationship between the antioxidation properties of LAB in nematodes and its mitigation of oxidative damage in Campylobacter jejuni-infected mice.
Materials and methods
C. elegans, LAB and Campylobacter jejuni
The N2 wild-type strain of C. elegans was used in this study, cultivated and maintained at 20 °C. S medium, M9 buffer and nematode growth medium (NGM) were used for culturing the nematodes and conducting the experimental lifespan studies.Escherichia coli (E. coli) OP50 was provided as feed for the nematodes. The E. coli was grown at 37 °C for 24 h in Luria–Bertani medium to a concentration of 108 colony-forming units (CFU) per mL.
Ten strains of LAB were isolated from human and poultry faeces, and traditional fermented food. All the strains were stored in the Culture Collection of Food Microorganisms at Jiangnan University (Table 1). No human experiments were conducted. The contributors were not expected to encounter risk or discomfort in the process of gathering the faecal samples, and written informed consent to handle faecal samples for public health purposes was obtained from the contributors or, where relevant, their legal guardians. All the LAB strains were identified through 16S gene sequencing and cultivated in deMan, Rogosa and Sharpe (MRS) agar at a 2% (v/v) inoculum size at 37 °C for 18–20 h. LAB were kept in 30% glycerol at −80 °C for long-term storage. LAB were sub-cultured twice before being used in tests at 2% (v/v) inoculation.
| Strain | Species | Year of isolation | Location of isolation | Origin |
|---|---|---|---|---|
| 427 | L. plantarum | 2014 | Ledong, Hainan | Traditional pickles |
| X13 | L. crispatus | 2015 | Zhongxiang, Hubei | Healthy poultry feces |
| 9-5 | L. reuteri | 2015 | Yangzhou, Jiangsu | Healthy adult feces |
| 422 | L. fermentum | 2010 | Leshan, Sichuan | Traditional pickles |
| Z5 | L. salivarius | 2013 | Huhehaote, Neimengu | Traditional koumiss |
| G14 | L. reuteri | 2016 | Bama, Guangxi | Healthy adult feces |
| H29M-8M | P. pentosaceus | 2017 | Zhongxiang, Hubei | Healthy poultry feces |
| 408 | L. plantarum | 2014 | Lingao, Hainan | Traditional pickles |
| 13-7 | L. salivarius | 2016 | Bama, Guangxi | Healthy adult feces |
| 430 | L. plantarum | 2014 | Lingao, Hainan | Traditional pickles |
The Campylobacter jejuni strain NCTC 11168 was purchased from ATCC. A Campylobacter jejuni-selective supplement (Oxoid) and 5% sterile sheep blood (Xinrui, China) were added to Columbia blood agar base plates (Oxoid, UK) and Campylobacter jejuni strains were grown in this medium under a gas condition of (5% O2, 10% CO2, 85% N2) for 48 h at 37 °C.
Lifespan experiment of C. elegans under oxidative stress
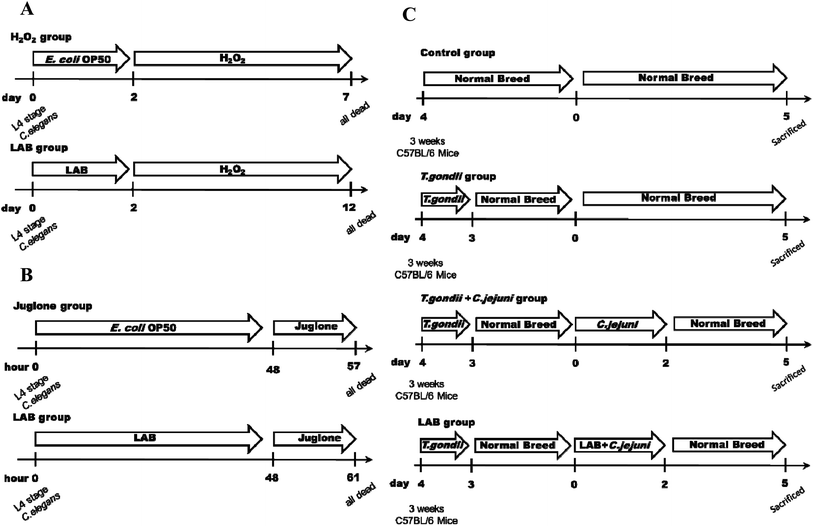
Growth and reproduction of nematodes were synchronised as previously described.20 After synchronisation, the eggs were placed on E. coli OP50 NGM plates for 72 h until they reached the L4 stage. All experiments were carried out at 20 °C and at least three independent replications were executed for every assay. To determine the appropriate chronic and acute concentrations to induce oxidative damage in C. elegans, 0.001%, 0.003% and 0.006% (v/v) H2O2 and 100, 200 and 400 μM juglone were tested. To evaluate the ability of the LAB strains to protect the nematodes against oxidative stress, the nematodes were fed respective LAB strains at a concentration of 109 CFU mL−1 for the first two days. Fifty nematodes (100 nematodes in total) were then treated with 0.003% (v/v) H2O2 or 200 μM juglone in 12-well tissue culture plates and incubated at 20 °C. Under H2O2 stress, count of live nematodes was performed every 12 h, until all of the nematodes were death. Under juglone stress, count of live nematodes was performed every hour, until all the nematodes were death. A negative control group of nematodes were fed only E. coli OP50 for the first two days, and then 50 nematodes (100 nematodes in total) were treated with H2O2 or juglone (Fig. 1A and B). The numbers of live nematodes were recorded, and the probability of their survival was calculated as described previously.Measurement of catalase (CAT), dismutase (SOD) and malondialdehyde (MDA)
After a 0.003% (v/v) H2O2 (12 h exposure) and a 200 μM juglone (30 min exposure) treatment, the nematodes were homogenised for measurement of CAT, SOD and MDA. Each group comprised 100 nematodes, and at least three independent replications were performed for each assay. The measurement of CAT, SOD and MDA was conducted with homologous kits from Nanjing Jiancheng Institute of Biotechnology (Nanjing, China) according to the supplier's instructions.Measurement of extracellular ROS
After treatment with 0.003% (v/v) H2O2 (12 h exposure) or 200 μM juglone (30 min exposure), the nematodes were washed three times with M9 buffer and were then incubated with 5 μg mL−1 of H2DCFDA for 10 min and 10 μM Dihydroethidium (DHE) for 1 hour respectively. The nematodes were then washed three times with PBS to remove the superficial H2DCFDA and DHE, and then treated with 30 mM sodium azide (NaN3) to narcotise the fluorescent stained nematodes. Nematodes were then photographed by fluorescent microscope (Nikon Eclipse Ti–S, Japan). ImageJ software recorded the relative fluorescent intensity of H2DCFDA and DHE. At least three independent replications were performed for each assay.RNA extraction, reverse transcription and quantitative real-time PCR analysis
After treatment with 0.003% (v/v) H2O2 (12 h exposure) or 200 μM juglone (30 min exposure), approximately 200 nematodes from each treatment group were assayed. The whole RNA of C. elegans and of the bacteria was extracted as previously described.21The transcription level of mitogen-activated protein kinase (MAPK) pathway genes (pmk-1 and sek-1), IIS signalling pathway genes (skn-1) and Nrf2-Keap1-ARE pathway genes (sod-3) in C. elegans was determined by quantitative polymerase chain reaction (qPCR) assay.22 The housekeeping gene GapA was used to determine and normalize the level of mRNA transcription of the C. elegans. The annealing temperature of the RT-qPCR assay was 56 °C. The delta Ct method was used to analyse the RT-qPCR data and determine the relative abundance of the target genes (in terms of fold-changes).20,23
Induction of acute ileitis for Campylobacter jejuni infection and LAB intervention in mice
Three-week-old female C57BL/6 mice obtained from Shanghai Laboratory Animal Center (Shanghai, China) were used in the tests. Six mice were housed in each cage, with a 12 h light–dark cycle and under a controlled environment (humidity, 45% ± 5%; temperature, 22 °C ± 2 °C). All of the experimental procedures (#JIPD2017030) were approved by the Animal Care and Use Committee at the Jiangsu Institute of Parasitic Diseases.To induce ileitis, the C57BL/6 mice were infected orally with 100 T. gondii cysts (ME49 strain; obtained from the Jiangsu Institute of Parasitic Diseases, Remington, Wuxi, China).24 As shown in Fig. 1C, for the four days thereafter the mice were successively (at 1 h intervals) treated with LAB (109 CFU in 0.3 mL) or PBS (0.3 mL) and were subsequently treated with Campylobacter jejuni NCTC 11168 (109 CFU in 0.3 mL) by gavage on two consecutive days.
Determination of antioxidant index of liver in mice
Mouse liver (0.1 g) was ground with saline (0.9 mL), and resulting homogenate was centrifuged at 8000 rpm for 6 min at 4 °C. The liquid supernatant was extracted and its biochemical indices were assayed. Total antioxidant capacity (TOC) and glutathione (GSH) were measured by assay kits from Nanjing Jiancheng Institute of Biotechnology (Nanjing, China), according to the supplier's protocols.Statistical analysis
Statistical analysis was performed with GraphPad Prism 5.0 and Origin 9.0, and significance analysis was performed with SPSS Statistics 20.0. The data were expressed as means ± standard deviations (SDs). Kaplan–Meier survival analysis was used to assess the survival rate of C. elegans. The groups were compared using a two-tailed Student's t-test, and a two-sided p-value of less than 0.05 was considered to indicate statistical significance. Mean values with different superscript letters over the bars are significantly different (i.e., p < 0.05).Results
LAB intervention showed different protective effects on C. elegans against oxidative stress
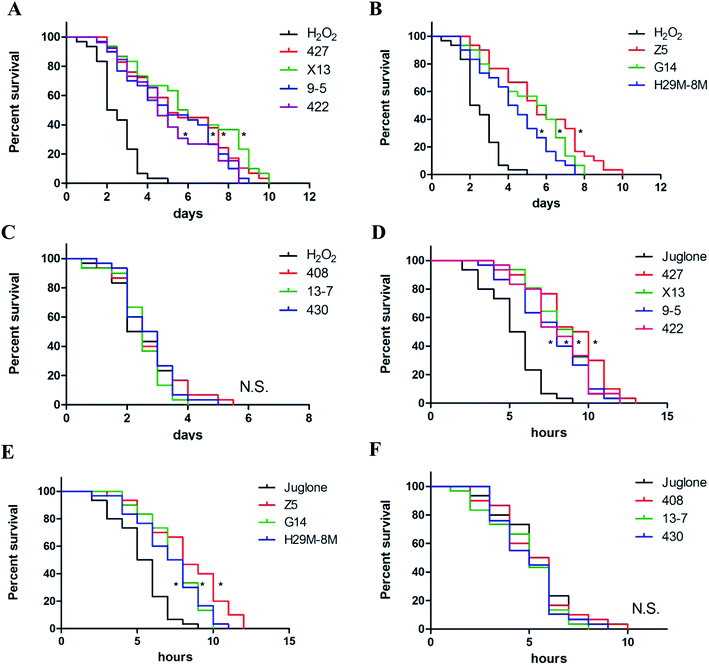
Ten LAB strains were assessed to determine their capacity to protect C. elegans against oxidative damage from exposure to H2O2 or juglone. Several different concentrations of H2O2 (0.001%, 0.003% and 0.006% (v/v)) and juglone (100, 200 and 400 μM) were used to determine the optimal concentration for inducing chronic and acute oxidative damage (Fig. S1†). Consistent with many previous studies, 0.003% (v/v) H2O2 and 200 μM juglone were chosen as the oxidant concentrations used in this study. As shown in Fig. 2A–C and Table 2, treatment with only H2O2 killed ∼50% of the nematodes within ∼3 days of their exposure, whereas 72.86–85.37% of the nematodes pre-treated with 427, X13, 9-5, 422, Z5, G14 or H29M-8M strains of LAB before exposure to H2O2 were still alive at this time. The time till death of all nematodes in the H2O2-exposed groups that were pre-treated with the LAB strains was more than twice as long as that for nematodes in the H2O2-treated group that were not pre-treated with the LAB strains. These LAB strains were also investigated for their ability to protect nematodes exposed to juglone for 6 h. The average lifespan of C. elegans exposed to juglone and treated only with E. coli OP50 was half of those pre-treated with the LAB strains (Fig. 2D–F and Table 3). Correspondingly, only 20–30% of the nematodes treated with 427, X13, 9-5, 422, Z5, G14 or H29M-8M prior to exposure to juglone were dead after 6 h, attending a longer average lifespan of ∼4 h than that of nematodes exposed to juglone and OP50 (i.e., lifespan of ∼10 h for LAB-treated nematodes versus ∼6 h for OP50-treated nematodes). However, LAB strains 408, 13-7 and 430 did not confer resistance to oxidative stress from H2O2 or juglone.| Groupsb | Survival (%) | DT50c (day) | P |
|---|---|---|---|
| a Summary of two or more separate experiments. Survival of worms on the last day (day 3) of the assays was estimated with the Kaplan–Meier survival analysis.b H2O2: treatment with E. coli OP50 in the first 2 days and then under H2O2 exposure when C. elegans at L4 stage. LAB: treatment with LAB in the first 2 days and then under H2O2 exposure when C. elegans at L4 stage. Day 0 marked the point at which the nematodes were first exposed to H2O2.c DT50, the time at which half of the worms were dead. | |||
| H2O2 | 45.39 | 2.07 | — |
| 427 | 80.48 | 5.11 | <0.01 |
| X13 | 82.67 | 5.79 | <0.01 |
| 9-5 | 78.56 | 5.14 | <0.01 |
| 422 | 81.03 | 4.89 | <0.01 |
| Z5 | 85.37 | 5.71 | <0.01 |
| G14 | 82.18 | 5.96 | <0.01 |
| H29M-8M | 72.86 | 4.54 | <0.01 |
| 408 | 40.21 | 2.63 | — |
| 13-7 | 35.66 | 2.21 | — |
| 430 | 45.85 | 3.07 | — |
| Groupsb | Survival (%) | DT50c (hour) | P |
|---|---|---|---|
| a Summary of two or more separate experiments. Survival of worms on the last hour (hour 6) of the assays was estimated with the Kaplan–Meier survival analysis.b Juglone: treatment with E. coli OP50 in the first 2 days and then under juglone exposure when C. elegans at L4 stage. LAB: treatment with LAB in the first 2 days and then under juglone exposure when C. elegans at L4 stage. Hour 0 marked the point at which the nematodes were first exposed to juglone.c DT50, the time at which half of the worms were dead. | |||
| Juglone | 48.25 | 6.05 | — |
| 427 | 79.32 | 10.23 | <0.01 |
| X13 | 80.14 | 9.15 | <0.01 |
| 9-5 | 70.49 | 8.67 | <0.01 |
| 422 | 79.86 | 8.41 | <0.01 |
| Z5 | 72.14 | 8.97 | <0.01 |
| G14 | 80.09 | 7.87 | <0.01 |
| H29M-8M | 70.32 | 7.09 | <0.01 |
| 408 | 45.73 | 5.78 | — |
| 13-7 | 45.79 | 5.94 | — |
| 430 | 39.99 | 6.08 | — |
The increase in levels of antioxidant enzymes in C. elegans under LAB treatment and oxidative stress
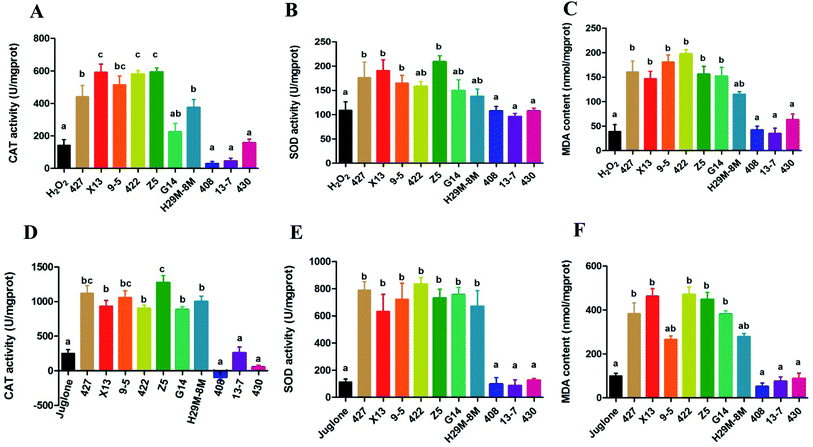
The data in Fig. 3A–C indicated production of CAT, SOD and MDA in the control and LAB pre-treated C. elegans groups exposed to H2O2 for 12 h. Most of the LAB pre-treated groups were quite strongly protected, exhibiting significant increases in the levels of CAT, SOD and MDA (P < 0.05) and a prolonged lifespan compared with the H2O2 group not pre-treated with LAB. The oxidative damage in nematodes pre-treated with E. coli OP50 and exposed to juglone was more extensive than that in nematodes pre-treated with E. coli OP50 and exposed to H2O2 (Fig. 3A–C).The overall trend in antioxidant levels for groups pre-treated with 427, X13, 9-5, 422, Z5, G14 or H29M-8M and exposed to juglone was consistent with the H2O2 exposure experiments. However, as shown in Fig. 3D–F, the actual levels of CAT, SOD, and MDA in these LAB pre-treated, H2O2-exposed groups were greater than those of the LAB pre-treated, juglone-exposed groups. However, LAB strains 408, 13-7 and 430 conveyed no resistance to H2O2- or juglone-mediated oxidative stress, and also showed no significant effect on levels of CAT, SOD, and MDA.
The decrease of ROS expression in C. elegans under LAB-treatment during H2O2 or juglone exposure
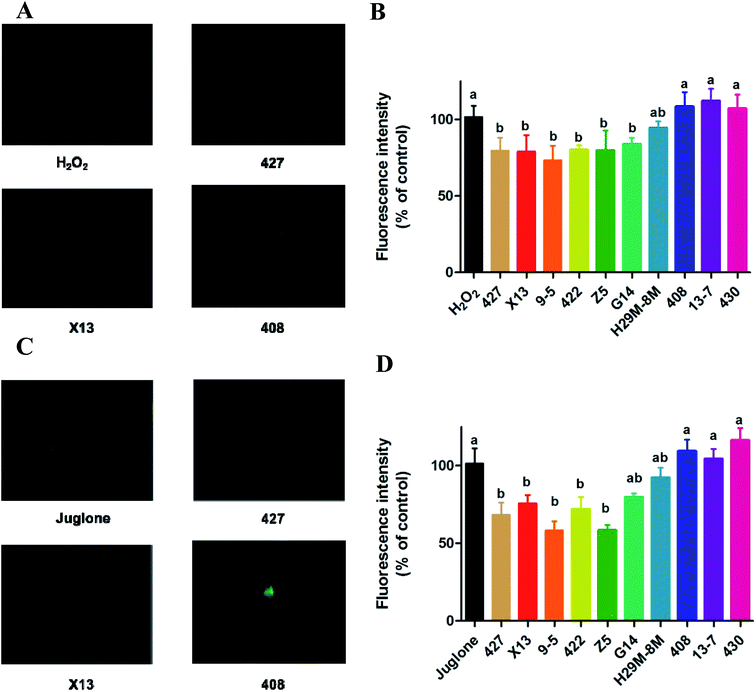
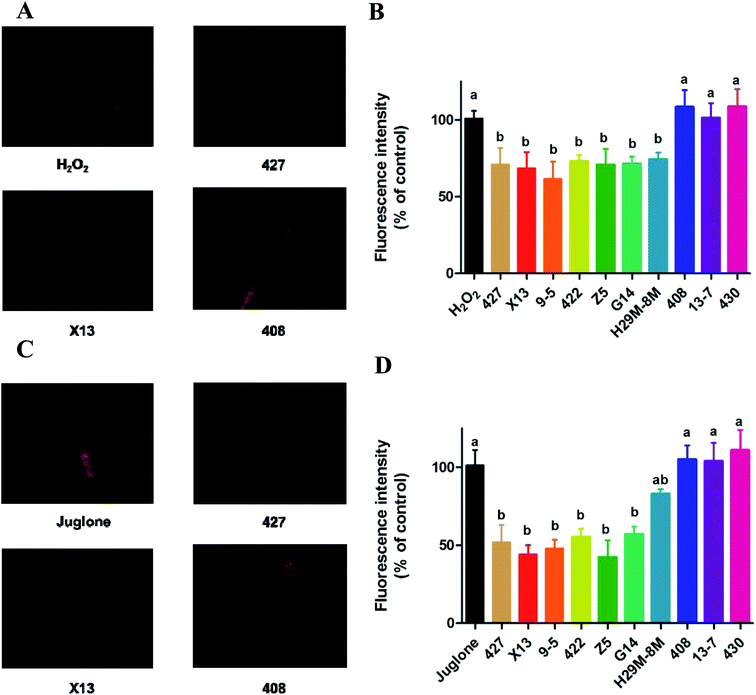
The generation of reactive oxygen species (ROS) in N2 nematodes exposing to H2O2 or juglone was assayed and shown in Fig. 4 and 5. The results showed that nematodes pre-treated with LAB were protected against H2O2 stress, having a prolonged lifespan and lower ROS levels than the non LAB pre-treated, H2O2-exposed group (Fig. 4A, B and 5A, B). This suggested that LAB pre-treatment resulted in a decrease in the ROS levels in nematodes exposed to H2O2 or juglone.The fluorescence microscope photographs showed that the nematodes exposed to juglone had more extensive ROS damage than the H2O2 control group. Fig. 4C, D and 5C, D showed that the fluorescence intensity of nematodes pre-treated with 427, X13, 9-5, 422 or Z5 strains of LAB and exposed to juglone were significantly less than that of the non-pre-treated, juglone-exposed group, demonstrating that the LAB pre-treatment could reduce the ROS levels in nematodes. The results for nematodes pre-treated with LAB before H2O2 or juglone exposure were mostly consistent with the lifespan and enzyme levels of nematodes under oxidative stress.
LAB strains upregulated immune gene transcription to protect C. elegans against oxidative stress
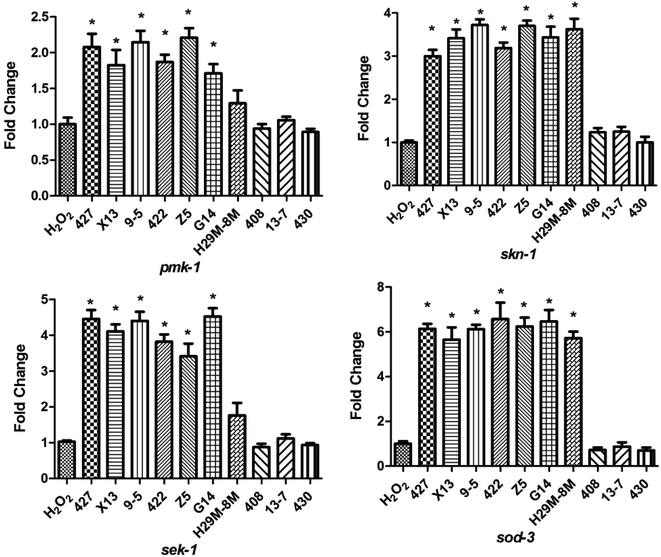
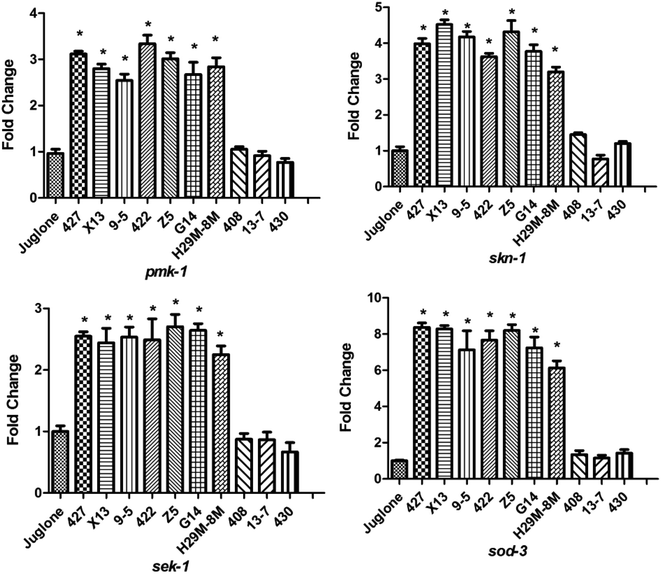
To determine the mechanism by which the ten LAB strains varied in their protection conferred on the nematodes (high or low) against oxidative stress, the transcription levels of the four immunity genes (pmk-1, sek-1, skn-1 and sod-3) of the nematodes were assayed after H2O2 (after 12 h) and juglone (after 30 min) treatment. As shown in Fig. 6, when nematodes were treated with LAB strains conferring high levels of protection (such as 427, X13, Z5), the transcription levels of the immunity genes were significantly elevated compared with those of nematodes exposed to H2O2 after treatment with E. coli OP50. As shown, the transcription levels of pmk-1 and sek-1 (MAPK-signalling pathway genes) were doubled and quadrupled, respectively. At the same time, the transcription levels of skn-1 (an IIS signalling pathway gene) and sod-3 (Nrf2-Keap1-ARE) were also increased. These may have prolonged the lifespan of the nematodes under H2O2 exposure. The transcript levels of skn-1 in the LAB pre-treated group was approximately six times the non-pre-treated, H2O2-exposed group.The increase in levels of antioxidant gene-transcripts in groups treated with effective LAB strains before H2O2 exposure was greater than that in groups treated with non-LAB strains treated before juglone exposure (Fig. 7). For example, the transcription levels of the MAPK-signalling pathway genes pmk-1 and sek-1 in the LAB-427, 9-5 or Z5 pre-treated groups under H2O2 exposure were two to three times higher than those in the group treated with E. coli OP50 before juglone exposure. At the same time, pre-treatment with 427, 9-5 or G14 significantly increased the transcription levels of the IIS signalling pathway gene skn-1 (four to five times) and the Nrf2-Keap1-ARE-signalling pathway gene sod-3 (eight times).
In contrast, when nematodes were treated with LAB strains 408, 13-7 or 430, which conferred only a low level of antioxidant protection, the transcription levels of their antioxidant genes were almost identical to those of nematodes treated with E. coli OP50 alone, before juglone exposure. To avoid the contaminations from bacterial RNA in altering the yield of RNA and subsequenty cDNA, the primers had been verified by different cDNA templates (Fig. S2†). No cross-reactivity was found in the results. In addition, whether the surface of C. elegans were cleaned did not significantly affect the changes in the level of transcription of the nematodes immune gene (Fig S3 and S4†). These data indicated that the ability of LAB to protect nematodes against oxidative stress was correlated with their influence on the levels of transcription of immune (i.e., antioxidant) genes. The role of LAB in regulating the activity of immune genes was thus shown to be linked with their role in preventing death of C. elegans exposed to H2O2 or juglone.
LAB with anti-oxidative activity in C. elegans also improved liver antioxidative parameters in mice infected with Campylobacter jejuni
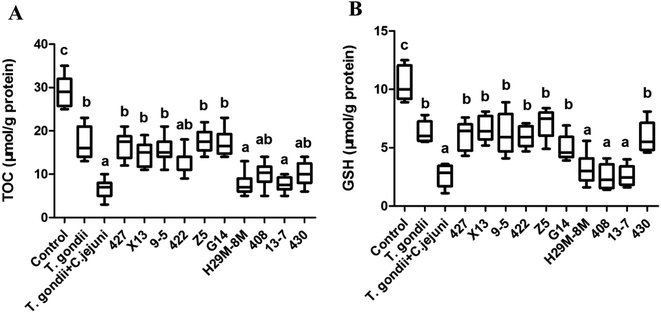
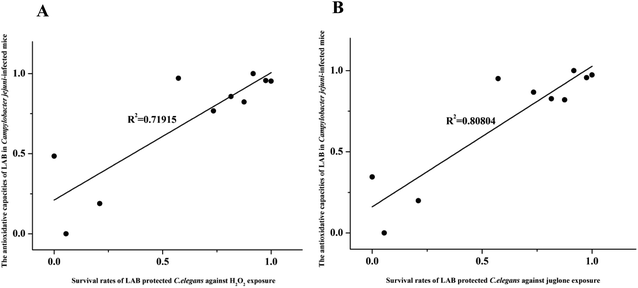
LAB strains differing in their ability to protect C. elegans against oxidative stress were further investigated to determine their ability to alleviate oxidative stress in Campylobacter jejuni-infected mice with T. gondii-induced acute ileitis. Seven days after T. gondii and Campylobacter jejuni infection, the mice showed acute ileitis and likely to die imminently. As shown in Fig. 8, the T. gondii + Campylobacter jejuni group had significantly reduced levels of TOC and GSH compared with the control group and the T. gondii group (P < 0.05). In contrast, groups pre-treated with LAB strains 427, X13, 9-5, 422, Z5 or G14 did not exhibit these changes in TOC and GSH levels (Fig. 8). In addition, the TOC and GSH activity in these LAB intervention groups were the same as that in the T. gondii group. However, LAB strains 408 and 13-7 had no effect on these activities. Obviously, the antioxidant activity of LAB strains that had ability to prolong the lifespan of nematodes subject to H2O2 or juglone exposure was higher than the antioxidant activities of the ineffective strains.Next, correlation analysis was conducted to examine the relationship between the antioxidative ability of the ten LAB strains in Campylobacter jejuni-infected mice and the survival rate of nematodes subject to H2O2 or juglone exposure after treatment with one of these ten LAB strains. In this analysis, GSH, one of the primary non-enzymatic antioxidants, was considered to represent the antioxidative capacity of LAB in Campylobacter jejuni-infected mice. The coefficients of determination (R2) values for these correlations were 0.71915 and 0.80804 for H2O2 and juglone, respectively, which indicated that the antioxidative ability of LAB strains in C. elegans under H2O2 or juglone exposure was significantly correlated to the antioxidative ability of LAB strains in Campylobacter jejuni-infected mice (Fig. 9).
Discussion
The model organism C. elegans has been used in research on the ageing of animals and humans to examine the relationships between senescence and the organism.25 The ageing process is often accompanied by oxidative stress,26 and an organism will generally clear away ROS and any deleterious infectious organisms to protect itself from oxidative stress. In this study, chronic and acute assays were designed to investigate the antioxidant properties of LAB in C. elegans through measurement of the responses of nematodes to H2O2 and juglone.27 The nematode lifespan was used as a model for oxidative stress to screen for candidate LAB strains for their protection against oxidative damage in host organisms. The findings indicated that LAB strains protected against oxidative stress not only by reducing levels of ROS in nematodes, but also by activating the nematodes' antioxidant genes. Also, the antioxidative ability of LAB in nematodes could reflect the antioxidative ability of LAB strains in Campylobacter jejuni-infected mice. This offered a new method of screening for LAB with antioxidant efficacy in pathogen-infected hosts.ROS are generated by normal oxygen-consuming metabolic processes and are typically related to oxidative stress.28 ROS comprise oxygen ions and peroxides, and these species can be both endogenously and exogenously generated. The detection of ROS levels in vivo can intuitively reflect the extent of oxidative stress in hosts. It has been reported that L. fermentum CECT5716, L. coryniformis CECT5711 and L. gasseri CECT5714 verifiably reduce ROS activity and the expression of NADPH oxidase, considered a major generator of ROS, in spontaneously hypertensive rats.29–32 Patel et al. confirmed that L. acidophilus intervention reduced ROS expression in calf thymus macrophages to enhance the immune response.33 In this study, the LAB pre-treatment groups that had the strongest protective effect against H2O2- or juglone-mediated oxidative stress in nematodes also significantly reduced the levels of ROS (P < 0.005) compared with the groups treated only with E. coli OP50. An alleviation of overall free radical status, a decrease in both the oxidation of lipids and carbonylation of proteins and an increase in antioxidant defences are known to correlate with diminished levels of ROS.34 Therefore, the analyses showed that the protection against oxidative stress conferred to C. elegans by the active LAB strains that prolonged nematode lifespan also correlated with the effect of LAB on ROS levels in a mouse disease model.
The antioxidant enzyme systems of organisms are usually studied under oxidative stress. In mitochondria, superoxide is one of the primary generators of ROS. Concurrently, the catalytic action of SOD decomposes the superoxide into H2O2 and H2O, and thus SOD is a central manager of ROS levels.35 Kullisaar et al. verified L. fermentum had the ability to stimulate the expression of Mn-SOD to counter oxidative damage.36 The ability of LAB to affect local delivery of SOD offers a new approach in nullification of enteric diseases typified by the production of ROS. To this end, it has been reported that L. casei BL23 could induce SOD expression in hosts with Crohn's disease, which explained the ability of the L. casei BL23-treated group to recover faster from initial weight loss, milder intestinal inflammation and increased intestinal enzymatic activities than the placebo group.14,28 CAT takes part in cellular antioxidant defence by decomposing H2O2 to prevent the production of hydroxyl radicals by the Fenton reaction.37 It was reported that L. casei could restrain or reduce the damage of intestinal pathologies ascribed to oxidative stress.38 MDA is a representative product in the process of lipid peroxidation. Its concentration reflects the level of free radicals in an organism, and it is also a sensitive indicator of the metabolism of free radicals in an organism. The level of MDA indirectly reflects the attack of free radicals on cells, and the activity of SOD indirectly reflects the removal of oxygen free radicals in hosts. In this study, treatment of C. elegans with active strains of LAB increased the expression of CAT, SOD and MDA in nematodes exposed to either H2O2 or juglone confirmed the antioxidant enzyme system was one of the mechanisms by which certain LAB strains protected the nematodes against oxidative stress.
With the growth and reproduction of LAB in a host's intestine, a host's immune system is regulated through pivotal LAB-mediated signalling pathways.39 Thus, the intervention of LAB may trigger a host's immune response to oxidative stress damage. Three main antioxidant signalling pathways mediated by LAB were examined in this study: the mitogen-activated protein kinase (MAPK) signalling pathway, the IIS signalling pathway and the Nrf2-Keap1-ARE signalling pathway.28,40 It has been found that the metabolite of Lactobacillus GG can induce heat shock protein (Hsp) 25 and Hsp72 to partially activate the MAPK signalling pathway in mouse colon cells.41 It has also been reported that Lactobacillus rhamnosus GG can generate soluble proteins p40 and p75, and that these signalling molecules have the ability to reduce H2O2-induced epithelial barrier disruption via a MAPK-dependent mechanism.42 In turn, data from this study showed that the activity of LAB strains, which demonstrated a protective effect against oxidative stress in C. elegans was correlated with the upregulation of the MAPK pathway genes pmk-1 and sek-1.
The antioxidant capacity of these LAB in nematodes was also reflected in their activation of the IIS signalling pathway gene skn-1, where the IIS pathway is an evolutionarily conserved endocrine signalling pathway that regulates food storage and growth.43 The skn-1 gene codes for SKN-1, which is a target of the IIS pathway. The SKN-1 is a transcription factor that orchestrates the oxidative stress response in adult C. elegans, independent of DAF-16 activity, by activating the expression of genes encoding for enzymes including CATs, SODs, and some GSHs.44–46 Overexpression of SKN-1 in intestinal cells increases lifespan and oxidative stress resistance.40 The antioxidant LAB strains that conferred protection for C. elegans against oxidative stress in this study, significantly enhanced the transcription of skn-1.
Another widely researched biological system, the Nrf2-Keap1-ARE pathway, regulates and controls the transformation of exogenous stimuli into eukaryotic transcriptional responses. The activation of Nrf2 has been demonstrated to upregulate genes, which are involved in ROS detoxification, thus protecting against oxidative damage or electrophilic environmental stressors.47,48 Nrf2 levels were observably increased in the livers of high cholesterol diet mice fed with Lactobacillus plantarum.49 It has also been shown that Lactobacillus plantarum FC225 has antioxidant and hypolipidemic effects and that Nrf2 expression is markedly promoted in mice fed a high-fat diet.50 Somewhat consistent with these observations, we observed that the transcription of sod-3, a Nrf2-Keap1-ARE pathway gene, was significantly enhanced by those LAB strains that greatly enhanced the lifespan of nematodes exposed to H2O2 or juglone. Moreover, in juglone acute assays, the upregulation of sod-3 was more distinctly correlated with LAB treatment. All of the signalling pathways mentioned above are highly conserved and represented by numerous homologous genes in mammals. They are also known to play a pivotal role in growth promotion, acclimatisation, and resistance to oxidative stress in C. elegans.
Due to the limits of the growth temperature and microbiota of C. elegans compared with mammals, the reliability of our assay that correlated the antioxidative efficiency of certain LAB strains in C. elegans with the ability of these strains to also protect against oxidative stress in mammals should be confirmed. Infection with Campylobacter, a genus of pathogenic microorganisms, often generates symptoms such as bloody diarrhoea, abdominal pain and fever, and LAB such as L. gasseri SBT2055 and L. salivarius NRRL B-30514 have been shown to prevent and protect against Campylobacter infections.51 However, our study was the first to correlate the antioxidant properties of LAB in C. elegans with the ability of LABs to alleviate oxidative damage in animals caused by Campylobacter jejuni infection. Mice co-infected with T. gondii and Campylobacter jejuni exhibited an obvious increase in Campylobacter jejuni infection and more serious oxidative stress damage, which indicated that the mice's resistance to Campylobacter jejuni colonisation was eliminated and the immune system was destroyed by T. gondii infection.24,52
GSH is the main non-enzymatic cellular antioxidant, and synergises with selenium-dependent glutathione peroxidase to effectively remove peroxynitrite and hydroxyl radicals and hydrogen peroxides. GSH is also responsible for reducing H2O2, in a process catalysed by glutathione peroxidases (GSH-PX). Lactobacillus fermentum53,54 and Clostridium butyricum55 have both been reported to stimulate high levels of GSH production, leading to suppression of hepatic oxidative stress in mice. TOC is a comprehensive index to measure the functional status of antioxidant system, and is typically understood to reflect the ability of antioxidant enzyme and non-enzymatic systems in response to external stimulation and control the state of free radical metabolism. Treating a Type 2 diabetes model with Bifidobacterium lactis Bb12 and Lactobacillus acidophilus La5 improved levels of GSH and TOC compared with a placebo.56 Human experiments in healthy older persons showed that the administration of Lactobacillus fermentum ME-3 significantly increased the TOC.57 In this study, the TOC and GSH levels were found to correlate with the activity of LAB strains against oxidative stress in C. elegans and the ability to mitigate oxidative damage in Campylobacter jejuni-infected mice. The correlation analysis showed the lifespan model of C. elegans against oxidative stress can be applied to screen LAB for antioxidant efficacy in pathogen-infected mammals.
Conclusion
The C. elegans model caused by H2O2 or juglone to oxidative damage and Campylobacter jejuni-infected mice model were used to screen LAB with antioxidant effect in vivo. The results indicated the lifespan model of C. elegans against oxidative stress can to some extent be applied to screen LAB for antioxidant efficacy in pathogen-infected mammals.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Zhihong Sun for his help on measurement of extracellular ROS with Dihydroethidium. This work was supported by the National Natural Science Foundation of China (No. 31671839), the National Natural Science Foundation of China Key Program (No. 31530056), the National Natural Science Foundation of China (No. 31601444, No. 31301407), the Fundamental Research Funds for the Central Universities (JUSRP51501), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the national first-class discipline program of Food Science and Technology (JUFSTR20180102), the Program of Collaborative Innovation Centre of Food Safety and Quality Control in Jiangsu Province and the project/research supported by Scientific Research Fund of Hunan Provincial Education Department (15B034).References
- E. Junn, M. M. Mouradian and V. Dias, Journal of Parkinson's Disease, 2013, 3, 461–491 Search PubMed.
- Y. Zhao and B. Zhao, Oxid. Med. Cell. Longevity, 2013, 2013, 1–11 Search PubMed.
- H. Tsutsui, S. Kinugawa and S. Matsushima, Am. J. Physiol.: Heart Circ. Physiol., 2011, 301, 2181–2190 CrossRef PubMed.
- P. Marchio, S. Guerra-Ojeda, J. M. Vila, M. Aldasoro, V. M. Victor and M. D. Mauricio, Oxid. Med. Cell. Longevity, 2019, 1–33, DOI:10.1155/2019/8563845.
- D. Harrison, K. K. Griendling, U. Landmesser, B. Hornig and H. Drexler, Am. J. Cardiol., 2003, 91, 7–11 CrossRef.
- B. Halliwell, Biochem. J., 2007, 401, 1–11 CrossRef CAS PubMed.
- A. Comandini, V. Marzano, G. Curradi, G. Federici, A. Urbani and C. Saltini, Pulm. Pharmacol. Ther., 2010, 23, 482–492 CrossRef CAS PubMed.
- W. Li, Y. Guo, C. Zhang, R. Wu, A. Y. Yang, J. Gaspar and A. N. Kong, Chem. Res. Toxicol., 2016, 29, 2071–2095 Search PubMed.
- A. Bah, H. Albano, J. B. Barbosa, I. Fhoula, Y. Gharbi, A. Najjari, A. Boudabous, P. Teixeira and H. I. Ouzari, BioMed Res. Int., 2019, 1–11, DOI:10.1155/2019/6937837.
- C. M. Yong Zhang, J. Zhao, H. Xu and Q. Hou, Oncotarget, 2017, 8, 24719–24727 Search PubMed.
- M.-Y. Lin and C.-L. Yen, J. Agric. Food Chem., 1999, 47, 3661–3664 CrossRef CAS PubMed.
- M.-Y. Lin and C.-L. Yen, J. Agric. Food Chem., 1999, 47, 1460–1466 CrossRef CAS PubMed.
- J. Sun, X. L. Hu, G. W. Le and Y. H. Shi, Lett. Appl. Microbiol., 2010, 50, 264–269 CrossRef CAS PubMed.
- J. G. LeBlanc, S. del Carmen, A. Miyoshi, V. Azevedo, F. Sesma, P. Langella, L. G. Bermudez-Humaran, L. Watterlot, G. Perdigon and A. de Moreno de LeBlanc, J. Biotechnol., 2011, 151, 287–293 CrossRef CAS PubMed.
- J. Richmon, ILAR J., 2002, 43, 63–68 CrossRef PubMed.
- P. L. LARSEN, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8905–8909 CrossRef CAS PubMed.
- T. Ikeda, C. Yasui, K. Hoshino, K. Arikawa and Y. Nishikawa, Appl. Environ. Microbiol., 2007, 73, 6404–6409 CrossRef CAS PubMed.
- Y. M. Zhou, L. Shao, J. A. Li, L. Z. Han, W. J. Cai, C. B. Zhu and D. J. Chen, Biosci., Biotechnol., Biochem., 2011, 75, 1746–1751 CrossRef CAS PubMed.
- C. Kenyon, Philos. Trans. R. Soc. London, Ser. B, 2011, 366, 9–16 CrossRef CAS PubMed.
- M. Zhou, J. Zhu, H. Yu, X. Yin, P. M. Sabour, L. Zhao, W. Chen and J. Gong, J. Appl. Microbiol., 2014, 117, 217–226 CrossRef CAS PubMed.
- M. Zhou, H. Yu, X. Yin, P. M. Sabour, W. Chen and J. Gong, PLoS One, 2014, 9, 1–11 Search PubMed.
- X. Yin, J. R. Chambers, R. Wheatcroft, R. P. Johnson, J. Zhu, B. Liu and C. L. Gyles, Appl. Environ. Microbiol., 2009, 75, 4975–4983 CrossRef CAS PubMed.
- J. Zhu, X. Yin, H. Yu, L. Zhao, P. Sabour and J. Gong, Infect. Immun., 2011, 79, 1688–1695 CrossRef CAS PubMed.
- L. M. Haag, A. Fischer, B. Otto, R. Plickert, A. A. Kuhl, U. B. Gobel, S. Bereswill and M. M. Heimesaat, PLoS One, 2012, 7, 1–13 Search PubMed.
- H. Nakagawa, T. Shiozaki, E. Kobatake, T. Hosoya, T. Moriya, F. Sakai, H. Taru and T. Miyazaki, Aging Cell, 2016, 15, 227–236 CrossRef CAS PubMed.
- T. L. a. T. Finkel, Exp. Cell Res., 2008, 314, 1918–1922 CrossRef PubMed.
- J. L. Rodríguez-Chávez, E. Coballase-Urrutia, D. Ortega-Cuéllarc and G. Delgado-Lamas, RSC Adv., 2016, 6, 12032–12041 RSC.
- B. Halliwell, Ann. Rheum. Dis., 1995, 54, 505–510 CrossRef CAS PubMed.
- H.-L. H. Chih-Chung Lin, R.-H. Shih, P.-L. Chi, S.-Ei Cheng, J.-C. Chen and C.-M. Yang, Cell Commun. Signaling, 2012, 10, 1–15 CrossRef PubMed.
- M. Gomez-Guzman, M. Toral, M. Romero, R. Jimenez, P. Galindo, M. Sanchez, M. J. Zarzuelo, M. Olivares, J. Galvez and J. Duarte, Mol. Nutr. Food Res., 2015, 59, 2326–2336 CrossRef CAS PubMed.
- I. T. Lee and C. M. Yang, Mediators Inflammation, 2013, 2013, 1–13 Search PubMed.
- I. Rahman, S. K. Biswas and A. Kode, Eur. J. Pharmacol., 2006, 533, 222–239 CrossRef CAS PubMed.
- B. Patel, P. Kumar, R. Banerjee, M. Basu, A. Pal, M. Samanta and S. Das, Mol. Immunol., 2016, 75, 69–83 CrossRef CAS PubMed.
- V. J. Koller, B. Marian, R. Stidl, A. Nersesyan, H. Winter, T. Simic, G. Sontag and S. Knasmuller, Food Chem. Toxicol., 2008, 46, 1221–1229 CrossRef CAS PubMed.
- Y. Wang, Y. Wu, Y. Wang, H. Xu, X. Mei, D. Yu, Y. Wang and W. Li, Nutrients, 2017, 9, 1–15 CAS.
- M. Z. Tiiu Kullisaar, M. Mikelsaar, T. Vihalemm, H. Annuk, C. Kairane and A. Kilk, Int. J. Food Microbiol., 2002, 72, 215–224 CrossRef.
- Y. S. Ho, Y. Xiong, W. Ma, A. Spector and D. S. Ho, J. Biol. Chem., 2004, 279, 32804–32812 CrossRef CAS PubMed.
- A. de Moreno de LeBlanc, J. G. LeBlanc, G. Perdigon, A. Miyoshi, P. Langella, V. Azevedo and F. Sesma, J. Med. Microbiol., 2008, 57, 100–105 CrossRef CAS PubMed.
- J. E. Irazoqui, J. M. Urbach and F. M. Ausubel, Nat. Rev. Immunol., 2010, 10, 47–58 CrossRef CAS PubMed.
- Z. P. a. F. J. S. Katherine and I. Zhou, Katherine Mansf. Stud., 2011, 3, 1–21 CrossRef.
- Y. Tao, K. A. Drabik, T. S. Waypa, M. W. Musch, J. C. Alverdy, O. Schneewind, E. B. Chang and E. O. Petrof, Am. J. Physiol.: Cell Physiol., 2006, 290, 1018–1030 CrossRef PubMed.
- A. Seth, F. Yan, D. B. Polk and R. K. Rao, Am. J. Physiol.: Gastrointest. Liver Physiol., 2008, 294, 1–25 CrossRef PubMed.
- S. Broughton and L. Partridge, Biochem. J., 2009, 418, 1–12 CrossRef CAS PubMed.
- J. H. An and T. K. Blackwell, Genes Dev., 2003, 17, 1882–1893 CrossRef CAS PubMed.
- K. V. Jae, H. An, M. Lucke, H. Inoue, N. Hisamoto, K. Matsumoto and T. Keith Blackwell, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16275–16280 CrossRef PubMed.
- M. H. Jennifer, M. A. Tullet, J. H. An, J. Baker, Ji Y. Hwang, S. Liu, R. P. Oliveira, R. Baumeister and T. Keith Blackwell, Cell, 2008, 132, 1025–1038 CrossRef PubMed.
- R. M. Jones, C. Desai, T. M. Darby, L. Luo, A. A. Wolfarth, C. D. Scharer, C. S. Ardita, A. R. Reedy, E. S. Keebaugh and A. S. Neish, Cell Rep., 2015, 12, 1217–1225 CrossRef CAS PubMed.
- M. Kobayashi, L. Li, N. Iwamoto, Y. Nakajima-Takagi, H. Kaneko, Y. Nakayama, M. Eguchi, Y. Wada, Y. Kumagai and M. Yamamoto, Mol. Cell. Biol., 2009, 29, 493–502 CrossRef CAS PubMed.
- L. X. Wang, K. Liu, D. W. Gao and J. K. Hao, World J. Gastroenterol., 2013, 19, 3150–3156 CrossRef CAS PubMed.
- D. Gao, Z. Gao and G. Zhu, Food Funct., 2013, 4, 982–989 RSC.
- K. Nishiyama, Y. Seto, K. Yoshioka, T. Kakuda, S. Takai, Y. Yamamoto and T. Mukai, PLoS One, 2014, 9, 1–9 Search PubMed.
- G. Wang, Y. He, X. Jin, Y. Zhou, X. Chen, J. Zhao, H. Zhang and W. Chen, Front. Microbiol., 2018, 9, 1–13 CrossRef PubMed.
- T. Kullisaar, E. Songisepp, M. Aunapuu, K. Kilk, A. Arend, M. Mikelsaar, A. Rehema and M. Zilmer, Appl. Biochem. Microbiol., 2010, 46, 481–486 CrossRef CAS.
- M. Z. Tiiu Kullisaar, M. Mikelsaar, T. Vihalemm, H. Annuk, C. Kairane and A. Kilk, Int. J. Food Microbiol., 2002, 72, 215–224 CrossRef.
- H. Endo, M. Niioka, N. Kobayashi, M. Tanaka and T. Watanabe, PLoS One, 2013, 8, 1–19 Search PubMed.
- J. M.-N. P. D. Hanie, S. Ejtahed MSc., A. Homayouni-Rad PhD, M. Niafar MD., PhD, M. Asghari-Jafarabadi PhD and V. Mofid MSc., Nutrition, 2012, 28, 539–543 CrossRef PubMed.
- E. Songisepp, J. Kals, T. Kullisaar, R. Mandar, P. Hutt, M. Zilmer and M. Mikelsaar, Nutr. J., 2005, 4, 1–10 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra06105c |
| This journal is © The Royal Society of Chemistry 2020 |