Open Access Article
Open Access ArticleA stable LnMOF as a highly efficient and selective luminescent sensor for detecting malachite green in water and real samples†
Xue-Ting Wangac,
Wei Weiac,
Kai Zhang ac and
Shao-Wu Du
ac and
Shao-Wu Du *b
*b
aState Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou, Fujian 350002, China
bProvincial Key Laboratory of Functional Marine Sensing Materials, Minjiang University, Fuzhou, Fujian 350108, China. E-mail: swdu@fjirsm.ac.cn
cUniversity of Chinese Academy of Sciences, Beijing, 100049, P. R. China
First published on 7th February 2020
Abstract
A series of isostructural 3D lanthanide metal–organic frameworks (LnMOFs), with the formula n(H3O)[Ln(L)(H2O)]n·nH2O (Ln = Gd 1, Eu 2 and Tb 3, H4L = 3,5,3′,5′-oxytetrabenzoic acid), have been successfully synthesized by solvothermal reactions. Single-crystal X-ray diffraction analysis reveals that 1–3 are constructed from wave-like Ln–carboxylate chains which are further connected by the ligands to form 3D channel-type frameworks. Further experiments suggest that 3 is thermally stable up to 322 °C and exhibits outstanding chemical stability in aqueous solutions with the pH ranging from 3 to 11. Significantly, 3 can be utilized for the first time to detect malachite green (a synthetic antibiotic to cure saprolegniasis) in aqueous media even in the presence of other interfering antibiotics, with a high sensitivity (Ksv = 8.33 × 104 M−1), low detection limit (DL = 0.25 μM) and good recyclability. On a more practical note, we found that the luminescence intensity of 3 showed almost no response to pH changes (pH 3–11), allowing steady sensing in real samples such as river water, simulated human serum and urine with satisfactory recoveries and RSD.
Introduction
Antibiotics are effective drugs to protect human health, and are also routinely used as feed additives for preventing bacterial infections and promoting the growth of animals in aquaculture.1 Recently, on site and real time detection of antibiotic residues in the environment has become an urgent need due to their toxicity and broad range of applications.2 Despite their superb value, abuse and overuse of antibiotics have left their residues in food, soil, and river and drinking water.3 Long-term intake of antibiotic contaminated foods and water can eventually cause serious damage to human beings through food chains. Once been ingested, antibiotics are difficult to degrade and excrete. They are prone to accumulate in human bodies, resulting in health risks.4 Among several classes of antibiotics (Table S1†), malachite green (MG), a type of synthetic triphenylmethane antibiotics, has become one of the most poisonous organic pollutants. MG has been banned in the European Union, and has been considered illegal in the United States, China and most other countries.5 However, it still can be detected in food products, water, river, human bodies, and even in human urine. Although various analytical techniques such as liquid chromatography and HPLC-MS/MS are known for detection of antibiotics,6 these methods seem to be either too expensive or time-consuming. Hence, it is an urgent issue to develop an effective and simple technique for detecting antibiotics.In recent years, design and synthesis of luminescent sensor based on lanthanide metal–organic frameworks (LnMOFs) have provoked enormous attention because lanthanide complexes usually have high luminescence efficiency, large Stokes shifts, narrow band emission and prolonged luminescence lifetimes.7 LnMOFs with high luminescence intensity can be successfully synthesized by careful selection of lanthanide ions and suitable antenna ligands.8 Due to electron (charge) or energy transfer processes,9 the addition of analytes usually cause changes of luminescence intensity and subsequently result in the emission color change of LuMOFs under UV lamp, by which one can easily determine the existence and also the concentration of analyte(s). Taking advantages of their obvious color changes, LnMOFs are capable of sensing ions and organic pollutants.10 Until now, many LnMOFs have been used to detect antibiotics, but none of them has been used to identify MG.11 In addition, most sensing experiments for detection of antibiotics in real samples are carried out in river water and serum,12 and there have been only two reports on sensing of antibiotics by LnMOF in urine solution.13
In this study, we used the 3,5,3′,5′-oxytetrabenzoic acid (H4L) as a bridging ligand to construct a series of isostructural LnMOFs, namely n(H3O)[Ln(L)(H2O)]n·nH2O (Ln = 1 Gd, 2 Eu, 3 Tb). Compound 3 has excellent stability in acidic and basic aqueous solutions. Luminescence studies showed that 2 and 3 exhibited relative high quantum efficiencies and long lifetimes, and more interestingly, the luminescence of 3 was inert to pH changes. We found that 3 had an excellent sensitivity and recyclability in detecting MG in aqueous solution, even in the presence of other interfering antibiotics. What is more, it also displayed effective detection of MG in water samples collected from Minjing River in Fuzhou, as well as in simulated human serum and urine samples based on the luminescence quenching effect.
Experimental section
Materials and instrumentations
All reagents were commercially available and used as received without any further purification. Synthetic human urine and serum samples were supplied by Jiangsu Yilesa Biotechnology Co., Ltd. Elemental analyses for C and H were carried out using an elemental Vario EL III analyser. Experimental powder X-ray diffraction (PXRD) was recorded on a Miniflex 600 diffractometer with CuKα radiation (λ = 1.5406 Å) at a scan speed of 1.6° min−1. UV/Vis spectra were recorded using a Lambda950 spectrometer. Thermogravimetric analyses (TAG) were performed on a NETZSCH STA449C instrument heated from 30 to 800 °C (heating rate of 10 °C min−1 under a constant stream of nitrogen). Infrared spectra were recorded on a PerkinElmer FTIR Spectrum One spectrometer using KBr as pellets. Emission spectra were obtained from an Edinburgh FLS fluorescence spectrophotometer and the quantum yields and luminescence lifetimes were measured on an FLS 1000 and FLS 980, respectively.Preparations of 1–3
A mixture of Ln(CH3COO)3·6H2O (Ln = Gd 1, Eu 2, Tb 3) (0.2 mmol), H4L (0.1 mmol), oxalic acid (0.1 mmol), CH3COOH and H2O (3 mL) was placed in a 25 mL Teflon-lined autoclave and heated at 140 °C for 3 days. Then, the reaction was slowly cooled down to room temperature to obtain yellow block crystals. After filtration, the product was washed with ethanol and air-dried. Yields based on H4L: 57.4% (32 mg) for 1; 56.2% (31 mg) for 2; 61.3% (34 mg) for 3. Elemental analysis (%) calcd for 1: C16H13O12Gd (554.51), C, 34.62; H, 2.34; found: C, 34.65; H, 2.41; For 2: C16H13O12Eu (549.23), C, 34.96; H, 2.37; found: C, 35.00; H, 2.12; For 3: C16H13O12Tb (556.19), C, 34.52; H, 2.34; found: C, 34.49; H, 2.00. FT-IR (KBr pellet: cm−1, Fig. S1†) for 1: 3641 (m), 3177 (m), 3086 (m), 1885 (w), 1551 (vs), 1380 (vs), 1259 (s), 1118 (s), 1017 (s), 785 (s), 714 (s), 552 (m), 492 (m), 421 (m). For 2: 3641 (m), 3167 (m), 3076 (m), 1895 (w), 1551 (vs), 1380 (vs), 1270 (s), 1118 (s), 1007 (s), 785 (s), 714 (s), 552 (m), 502 (m), 421 (m). For 3: 3641 (m), 3177 (m), 3076 (m), 1895 (w), 1562 (vs), 1380 (vs), 1270 (s), 1118 (s), 1017 (s), 785 (s), 714 (s), 552 (m), 512 (m), 431 (m).Crystal structure determination
Single-crystal X-ray diffraction data were collected on a Bruker D8-venture diffractometer equipped with a graphite-monochromated MoKα radiation (λ = 0.71073 Å) at room temperature. The structures were solved by the direct method using Olex2 with the ShelXS14 structure solution program and refined by the least squares minimisation with the ShelXL refinement package.15 Metal atoms in each compound were located from the E-maps. All the nonhydrogen atoms were refined anisotropically. The hydrogen atoms were generated geometrically and refined isotropically. Hydrogen atoms of the water molecules coordinated to the Ln3+ ions were restrained at fixed positions. Crystallographic details and results of the structure refinements are compiled in Table S2.† Selected bond lengths and angles are listed in Table S3.† The X-ray data for 1–3 have been deposited at the Cambridge Crystallographic Data Centre with the reference numbers of CCDC 1942126–1942128.†Results and discussion
Structure description
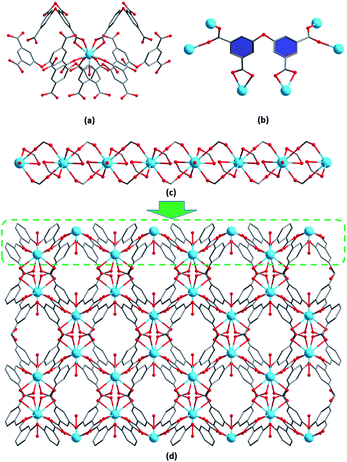
Single-crystal X-ray diffraction analysis reveals that 1–3 are isostructural and crystallize in the orthorhombic Pnna space group. Hence, we only take 3 as an example to describe its structure in detail. The asymmetric unit of 3 contains a half of crystallographically independent Tb3+ ion, a half of L4− ligand, a half of coordinated water molecule, a half of H3O+ cation and a half of lattice water molecule. As shown in Fig. 1a, the nine-coordinated environment of Tb3+ is fulfilled by eight carboxylate oxygen atoms derived from the L4− ligand and one oxygen atom from a coordinated water molecule, leading to a distorted tricapped trigonal-prismatic geometry. The four carboxylate groups of the H4L ligand are all deprotonated and adopt (κ1-κ1)-μ2 and (κ1-κ1)-μ1 binding modes to connect six Tb3+ ions (Fig. 1b). The neighbouring Tb3+ ions are alternately bridged by two bridging carboxylate groups, generating a wave-like chain along the b direction (Fig. 1c). These chains are further linked by chelating carboxylate groups to form a 3D porous framework structure with oxygen-rich channels viewed along the c direction (Fig. 1d). The protonated water cations and lattice water molecules are accommodated in the channels. There exit hydrogen bonds between the lattice water molecules, H3O+ cations and the carboxylate oxygen atoms (O7⋯O1 = 2.8008 Å, ∠O7–H7A⋯O1 = 163.136°; O8⋯O7 = 2.7446 Å, ∠O8–H8A⋯O7 = 151.267°, symmetry code A: −x + 1, y − 1/2, z + 1/2) (Fig. S2†).Stability investigation
Powder X-ray diffraction (PXRD) experiments have been carried out to examine the phase purity of the crystalline products (Fig. S3†). The high purity of 1–3 was confirmed by the experimental PXRD patterns of as-synthesized samples, which match well with their simulated spectra based on the single-crystal X-ray data.As MOF stability is essential for practical applications, the stability test of 1–3 was performed. Compounds 1–3 have similar TGA curves which display three main steps of weight loss. The first weight loss of 3.5% at 135 °C can be attributed to the loss of lattice water molecules (calcd. 3.2%) (Fig. S4†). Upon further heating, the weight loss of 6.6% at 218 °C is due to the removal of coordinated water molecules (calcd. 6.7%). The third weight loss of 9.7% at 322 °C corresponds to the loss of protonated water cations (calcd. 9.9%). Further increasing the temperature caused the decomposition of framework. In addition, the stability of 3 in aqueous solutions with different pH values was also investigated. The PXRD pattern of 3 did not change after being soaked in water with pH ranging from 3 to 11 for 24 h, indicating of excellent chemical stability of 3 in aqueous solutions over a wide pH range (Fig. 2).
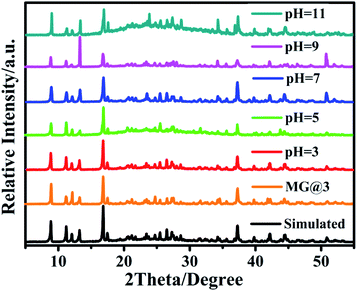 | ||
| Fig. 2 XRD patterns of 3 after immersion in aqueous solutions with pH ranging from 3 to 11 and in a MG solution. | ||
Luminescent properties
The solid-state luminescence of H4L and 1–3 was recorded. Because the lowest energy level of Gd3+ is too high to receive energy from the ligand, the characteristic emission of Gd3+ at 311 nm cannot be observed in the luminescent spectrum of 1. Instead, it shows a broad emission band centered at 500 nm upon excitation at 335 nm, which can be attributed to the π*–π transition of the ligand (Fig. S5†).16 Compounds 2 and 3 emit red and green light respectively under UV light irradiation, which is visible to the naked eyes. Upon excitation at 330 nm, the emission spectrum of 2 consists of four peaks at 594, 614, 652 and 700 nm, which are due to the 5D0 → 7FJ (J = 1, 2, 3, 4) transitions of Eu3+ (Fig. S6†). When excited at 330 nm, the emission spectrum of 3 exhibits four bands at 490, 544, 584, and 615 nm assigned to 5D4 → 7FJ (J = 6, 5, 4, 3) transitions of Tb3+ (Fig. S7†). The luminescence decay times and quantum yields of 2 and 3 were also measured. As shown in Fig. S8,† the decay times are 517.9 μs for 2 and 763.7 μs for 3. The quantum yields of 2 and 3 are 10.98 and 19.10%, respectively.To evaluate the luminescence stability of 3, the emission spectra of 3 in aqueous solutions at different pH values were recorded. The luminescence intensity remains almost constant when pH varies from 3 to 11 (Fig. 3a). This pH-independent luminescence property is not commonly observed in the Tb3+ complexes. The advantages of high luminescence efficiency and stability make 3 a promising candidate for application in luminescence sensing of antibiotics in aqueous solutions, as well as in some real samples that may have different pH values. Antibiotics, widely used to treat human and animal diseases, are among one of the major water pollutants. Herein, the luminescence sensing properties of 3 toward antibiotics in aqueous solutions were investigated. Compound 3 (5 mg) was dispersed in 3 mL of aqueous solution containing respectively 0.01 mol L−1 of ciprofloxacin (CPFX), lincomycin hydrochloride (LIN), penicillin (PCL), azithromycin (AZM), roxithromycin (ROX), potassium antimony tartrate (PAT), and malachite green (MG). Then the variation in emission intensity was monitored. The changes in luminescence intensity are displayed in Fig. 3b and S9.† As we can see, all of these antibiotics decrease or increase in varying degrees the luminescence intensities of 3 at 544 nm. Since the luminescence was nearly completely quenched by MG, 3 could be served as a probe to detect MG.
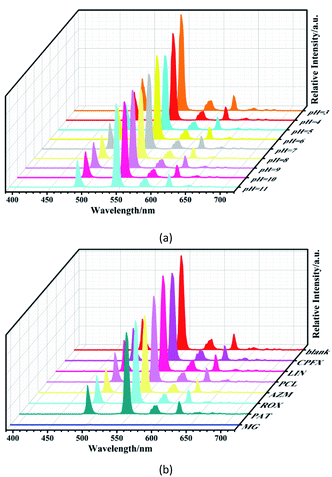 | ||
| Fig. 3 (a) Emission spectra of 3 dispersed in aqueous solutions with different pH values; (b) emission spectra of 3 dispersed in aqueous solutions with different antibiotics. | ||
Luminescence sensing of antibiotics
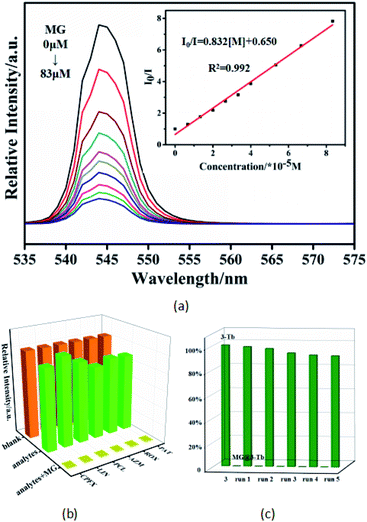
The luminescence sensing ability of 3 for MG was further tested. The titration experiment was performed using different concentrations of MG. As shown in Fig. 4a, the luminescence intensity of 3 decreases gradually when the concentration of MG increases from 0 to 83 μM. This quenching effect can be quantitatively analyzed using the Stern–Volmer equation:| (I0/I) = 1 + Ksv[M] |
Possible mechanism for luminescence quenching
To further pinpoint how MG affect the luminescence intensity of 3, the possible mechanism for the luminescence quenching by MG was investigated. There are, in general, three main mechanisms for the luminescence quenching of LnMOFs by analytes, which include collapse of framework structures and charge or energy transfer. The PXRD pattern of 3 after sensing was almost the same as that of the pristine sample, excluding the possibility of structural disruption of the framework (Fig. S10†). Another possible mechanism involves electron transfer from the conduction band (CB) of the sensor to the LUMO of the analyte, instead to the ground state of the sensor, and the lower the LUMO energy of the analyte, the easier the electron can transfer.18 As listed in Table S4,† the order of LUMO energy of antibiotics is MG < 3 < other antibiotics, implying that the charge transfer from ligand to MG would be one of the causes of luminescence quenching. Meanwhile, the inner filter effect (IFE), which occurs when the absorption band of the analyte has a certain degree of overlap with the excitation or emission spectrum of the sensor, is also responsible for luminescence quenching.19 The greater the overlap, the more effective IFE will be generated. As shown in Fig. S11,† different from other antibiotics, there is a large overlap between the excitation spectrum of 3 and the absorption spectrum of MG. As a result, MG can restrain light absorption of 3 and influence the energy transfer from the ligand to the Tb3+ ion.20 Therefore, the luminescence quenching in this system may be contributed to the combination effect of charge transfer and IFE effect.Analytical application in real samples
To explore the practical application of 3 for detecting MG in real samples, the luminescence response of the sensor to different concentrations of MG in river water was measured by the standard addition method. The river water samples were taken from Minjiang River in Fuzhou. Five rounds of testing were conducted at each concentration. As shown in Table 1, the measured concentrations of MG by luminescence probe of 3 are close to the actual values, with satisfactory recoveries of 99.24–107% and acceptable relative standard deviations (RSD). Similarly, we also measured the concentrations of MG in simulated human serum and urine. The results demonstrate that 3 has the potential for quantitative detecting MG at the level of micromole in the actual samples of river water, serum and urine.| Samples | Added (μM) | Founded (μM) | RSD(%) | Recovery (%) |
|---|---|---|---|---|
| River water | 6.999 | 7.000 | 8.083 | 100.0 |
| 9.341 | 10.00 | 10.93 | 107.0 | |
| 30.23 | 30.00 | 8.066 | 99.24 | |
| Serum | 5.399 | 7.000 | 3.863 | 107.9 |
| 10.56 | 10.00 | 11.00 | 105.6 | |
| 30.98 | 30.00 | 4.145 | 103.3 | |
| Urine | 7.260 | 7.000 | 2.754 | 103.7 |
| 10.18 | 10.00 | 3.110 | 101.8 | |
| 32.30 | 30.00 | 7.589 | 107.6 |
Conclusions
In conclusion, three isomorphic 3D LnMOFs (Ln = Gd, Eu and Tb) were successfully synthesized via a solvothermal reaction of lanthanide ions with multidentate 3,5,3′,5′-oxytetrabenzoic acid ligand. The Tb3+ compound exhibits high chemical stability in aqueous solutions at low and high pH values. Furthermore, the luminescence intensity of 3 is not affected by pH, allowing it to be used as a luminescence sensor for detecting specific organic pollutants under more extreme conditions. To the best of our knowledge, compound 3 represents the first example of LnMOFs used in sensing MG in aqueous solutions even in the presence of other interfering antibiotics. It displays a strong luminescence response to MG with excellent sensing performance in terms of high selectivity and sensitivity, together with good recyclability. This sensing method has been further applied for the determination of MG in real samples of river water, as well as simulated human serum and urine. Owing to their outstanding chemical and luminescence stability, these materials might have great potential for use in sensing environmental contaminants under harsh conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21972060 and 21571175), and the State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences.References
- (a) S. J. Culp and F. A. Beland, J. Am. Coll. Toxicol., 1996, 15, 219 CrossRef; (b) K. Kümmerer, Chemosphere, 2009, 75, 417 CrossRef PubMed.
- S. Xu, J.-J. Shi, B. Ding, Z.-Y. Liu, X.-G. Wang, X.-J. Zhao and E.-C. Yang, Dalton Trans., 2019, 48, 1823 RSC.
- X.-D. Zhu, K. Zhang, Y. Wang, W.-W. Long, R.-J. Sa, T.-F. Liu and J. Lü, Inorg. Chem., 2018, 57, 1060 CrossRef.
- (a) X. Fu, R. Lv, J. Su, H. Li, B. Yang, W. Gu and X. Liu, RSC Adv., 2018, 8, 4766 RSC; (b) M. Chen, N. Gan, Y. Zhou, T. Li, Q. Xu, Y. Cao and Y. Chen, Talanta, 2016, 161, 867 CrossRef.
- F. Zhang, H. Yao, T. Chu, G. Zhang, Y. Wang and Y. Yang, Chem.–Eur. J., 2017, 23, 10293 CrossRef.
- (a) R. Sahraei, A. Farmany, S. S. Mortazavi and H. Noorizadeh, Environ. Monit. Assess., 2013, 185, 5817 CrossRef; (b) V. Ashok, N. Agrawal, A. Durgbanshi, J. Esteve-Romero and D. Bose, J. AOAC Int., 2014, 97, 1387 CrossRef; (c) K. Halme, E. Lindfors and K. Peltonen, Food Addit. Contam., 2004, 21, 641 CrossRef; (d) X. Fang, S. Yang, K. Chingin, L. Zhu, X. Zhang, Z. Zhou and Z. Zhao, Int. J. Environ. Res. Public Health, 2016, 13, 814 CrossRef.
- (a) Y. Wu, G.-P. Yang, Y. Zhao, W.-P. Wu, B. Liu and Y.-Y. Wang, Dalton Trans., 2015, 44, 3271 RSC; (b) Y. Yang, F. Jiang, L. Chen, J. Pang, M. Wu, X. Wan, J. Pan, J. Qian and M. Hong, J. Mater. Chem. A, 2015, 3, 13526 RSC; (c) S. Dang, E. Ma, Z.-M. Sun and H. Zhang, J. Mater. Chem., 2012, 22, 16920 RSC; (d) T. Xia, F. Zhu, K. Jiang, Y. Cui, Y. Yang and G. Qian, Dalton Trans., 2017, 46, 7549 RSC; (e) R. An, H. Zhao, H.-M. Hu, X. Wang, M.-L. Yang and G. Xue, Inorg. Chem., 2016, 55, 871 CrossRef.
- (a) Z.-Q. Liu, Y. Zhao, P. Wang, Y.-S. Kang, M. Azam, S. l. Al-Resayes, X.-H. Liu, Q.-Y. Lu and W.-Y. Sun, Dalton Trans., 2017, 46, 9022 RSC; (b) J. Zhou, H. Li, H. Zhang, H. Li, W. Shi and P. Cheng, Adv. Mater., 2015, 27, 7072 CrossRef CAS; (c) C. Bai, B. Liu, H.-M. Hu, J.-D. Li, X. Wang and G. Xue, Acta Crystallogr., Sect. C: Struct. Chem., 2019, C75, 422 CrossRef; (d) B. Xu, Q. Chen, H.-M. Hu, R. An, X. Wang and G. Xue, Cryst. Growth Des., 2015, 15, 2318 CrossRef CAS; (e) Y.-C. Hu, C. Bai, H.-M. Hu, C.-T. Li, T.-H. Zhang and W. Liu, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2019, B75, 855 CrossRef.
- M. Zheng, H. Tan, Z. Xie, L. Zhang, X. Jing and Z. Sun, ACS Appl. Mater. Interfaces, 2013, 5, 1078 CrossRef CAS.
- (a) J.-N. Hao and B. Yan, Adv. Funct. Mater., 2017, 27, 1603856 CrossRef; (b) G.-W. Xu, Y.-P. Wu, W.-W. Dong, J. Zhao, X.-Q. Wu, D.-S. Li and Q. Zhang, Small, 2017, 13, 1602996 CrossRef; (c) J.-M. Li, R. Li and X. Li, CrystEngComm, 2018, 20, 4962 RSC; (d) M.-L. Han, G.-X. Wen, W.-W. Dong, Z.-H. Zhou, Y.-P. Wu, J. Zhao, D.-S. Li, L.-F. Ma and X. Bu, J. Mater. Chem. C, 2017, 5, 8469 RSC; (e) Z. Guo, X. Song, H. Lei, H. Wang, S. Su, H. Xu, G. Qian, H. Zhang and B. Chen, Chem. Commun., 2015, 51, 376 RSC; (f) Y.-X. Ren, X.-L. Zhao, Z.-X. Wang, Y. Pan, H.-P. Li, F.-Y. Wang, S.-F. Zhu and C.-H. Shao, RSC Adv., 2018, 8, 17497 RSC; (g) J. Zhao, Y.-N. Wang, W.-W. Dong, Y.-P. Wu, D.-S. Li and Q.-C. Zhang, Inorg. Chem., 2016, 55, 3265 CrossRef CAS; (h) L. Liu, Y. Wang, R. Lin, Z. Yao, Q. Lin, L. Wang, Z. Zhang and S. Xiang, Dalton Trans., 2018, 47, 16190 RSC; (i) H. Li, Y. Han, Z. Shao, N. Li, C. Huang and H. Hou, Dalton Trans., 2017, 46, 12201 RSC; (j) C.-T. Li, Y.-F. Zhao, H.-M. Hu, H. Zhao, X. Wang and G. Xue, Dalton Trans., 2016, 45, 15436 RSC.
- (a) X.-G. Liu, C.-L. Tao, H.-Q. Yu, B. Chen, Z. Liu, G.-P. Zhu, Z. Zhao, L. Shen and B. Z. Tang, J. Mater. Chem. C, 2018, 6, 2983 RSC; (b) N. Xu, Q. Zhang, B. Hou, Q. Cheng and G. Zhang, Inorg. Chem., 2018, 57, 13330 CrossRef CAS; (c) P. Xing, D. Wu, J. Chen, J. Song, C. Mao, Y. Gao and H. Niu, Analyst, 2019, 144, 2656 RSC; (d) H.-R. Fu, Y. Zhao, T. Xie, M.-L. Han, L.-F. Ma and S.-Q. Zang, J. Mater. Chem. C, 2018, 6, 6440 RSC; (e) H. He, Y.-Q. Xue, S.-Q. Wang, Q.-Q. Zhu, J. Chen, C.-P. Li and M. Du, Inorg. Chem., 2018, 57, 15062 CrossRef CAS; (f) Z.-S. Qin, W.-W. Dong, J. Zhao, Y.-P. Wu, Q. Zhang and D.-S. Li, Inorg. Chem. Front., 2018, 5, 120 RSC; (g) G.-N. Liu, R.-Y. Zhao, R.-D. Xu, X. Zhang, X.-N. Tang, Q.-J. Dan, Y.-W. Wei, Y.-Y. Tu, Q.-B. Bo and C. Li, Cryst. Growth Des., 2018, 18, 5441 CrossRef CAS.
- (a) K. Ren, S.-H. Wu, X.-F. Guo and H. Wang, Inorg. Chem., 2019, 58, 4223 CrossRef CAS PubMed; (b) H. Pan, S. Wang, X. Dao and Y. Ni, Inorg. Chem., 2018, 57, 1417 CrossRef CAS; (c) F. Zhang, H. Yao, Y. Zhao, X. Li, G. Zhang and Y. Yang, Talanta, 2017, 174, 660 CrossRef CAS; (d) A. H. Malik and P. K. Iyer, ACS Appl. Mater. Interfaces, 2017, 9, 4433 CrossRef CAS PubMed.
- (a) B. Wang and B. Yan, Talanta, 2019, 208, 120438 CrossRef; (b) L. Yu, H. Chen, J. Yue, X. Chen, M. Sun, J. Hou, K. A. Alamry, H. M. Marwani, X. Wang and S. Wang, Talanta, 2020, 207, 120297 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3 Search PubMed.
- N. Goel and N. Kumar, RSC Adv., 2018, 8, 10746 RSC.
- Y.-L. Xu, Y. Liu, X.-H. Liu, Y. Zhao, P. Wang, Z.-L. Wang and W.-Y. Sun, Polyhedron, 2018, 154, 350 CrossRef CAS.
- (a) J. Qin, B. Ma, X.-F. Liu, H.-L. Lu, X.-Y. Dong, S.-Q. Zang and H. Hou, J. Mater. Chem. A, 2015, 3, 12690 RSC; (b) D.-M. Chen, J.-Y. Tian, M. Chen, C.-S. Liu and M. Du, ACS Appl. Mater. Interfaces, 2016, 8, 18043 CrossRef CAS; (c) S. Wu, Y. Lin, J. Liu, W. Shi, G. Yang and P. Cheng, Adv. Funct. Mater., 2018, 8, 1707169 Search PubMed; (d) T. Wu, C. Zhang, H. Hou, P. Ge, G. Zou, W. Xu, S. Li, Z. Huang, T. Guo, M. Jing and X. Ji, Adv. Funct. Mater., 2018, 28, 1705744 CrossRef.
- M. Yu, Y. Xie, X. Wang, Y. Li and G. Li, ACS Appl. Mater. Interfaces, 2019, 11, 21201 CrossRef CAS.
- (a) H.-N. Wang, S.-Q. Jiang, Q.-Y. Lu, Z.-Y. Zhou, S.-P. Zhuo, G.-G. Shan and Z.-M. Su, RSC Adv., 2015, 5, 48881 RSC; (b) L. Wang, G. Fan, X. Xu, D. Chen, L. Wang, W. Shi and P. Cheng, J. Mater. Chem. A, 2017, 5, 5541 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Crystallographic data, FT-IR spectra, PXRD patterns, TGA curves, UV-Vis, luminescence spectra and additional figures (PDF). CCDC 1942126–1942128. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra10870j |
| This journal is © The Royal Society of Chemistry 2020 |