Open Access Article
Open Access ArticleFluconazole conjugated-gold nanorods as an antifungal nanomedicine with low cytotoxicity against human dermal fibroblasts
Khawla M. Hamadb,
Nouf N. Mahmoud *a,
Sabaa Al-Dabasha,
Luma A. Al-Samada,
Maha Abdallahc and
Amal G. Al-Bakric
*a,
Sabaa Al-Dabasha,
Luma A. Al-Samada,
Maha Abdallahc and
Amal G. Al-Bakric
aFaculty of Pharmacy, Al-Zaytoonah University of Jordan, Amman 11733, Jordan. E-mail: nouf.mahmoud@zuj.edu.jo
bAmman Academy, Amman 11821, Jordan
cSchool of Pharmacy, The University of Jordan, Amman 11942, Jordan
First published on 8th July 2020
Abstract
Herein, a nanotechnology-based approach was adopted to develop a facile and effective nanoplatform for the treatment of superficial fungal infections. Gold nanorods (GNR) functionalized with thiolated poly ethylene glycol (PEG-SH) or thiolated PEGylated cholesterol (Chol-PEG-SH) moieties were conjugated with Fluconazole and loaded into poloxamer 407 hydrogel. The obtained nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR were characterized by optical spectroscopy, hydrodynamic size and effective surface charge. The anti-fungal activity of the nanocomplexes was investigated by estimating the minimum inhibitory concentration (MIC) and the percentage reduction of fungal viable count against Candida (C.) albicans. PEG-Fluc-GNR and Chol-Fluc-GNR resulted in 5-fold and 14-fold reduction in MIC of GNR, and in 9-fold and 12-fold reduction in MIC of Fluconazole, respectively. The average log-reduction of the viable fungal cells upon treatment with the nanocomplexes was 5 log cycles, and it ranged from 1.3–3.7 log cycles when loaded into poloxamer 407 hydrogel. Transmission electron microscope imaging of the treated C. albicans revealed an enhanced uptake of the nanoparticles into the fungus's cell wall within the first 120 min of exposure. The nanocomplexes demonstrated low cytotoxicity towards human dermal fibroblasts which represent the human skin dermal cells. Conjugating Fluconazole with GNR is a promising approach for the effective treatment of superficial fungal infections.
1. Introduction
Skin diseases are one of the most common diseases in humans and represent a huge public health problem particularly in tropical countries.1,2 Different groups of fungi are responsible for several superficial mycoses infections of skin, nails, and mouth which may spread to other regions and become widespread and negatively affect the quality of life.1 Candida (C.) albicans is the most common pathological agent of candidiasis which usually affects skin folds in axillae, genital and perigenital areas, inguinal folds, hands, fingernails, and mucous membranes.3 The conventional treatment of fungal skin infections is either topical or systemical based on the localization, severity and stage of the infection and type of causative fungi. Cutaneous candidiasis is usually treated with topical antifungal agents in creams, gels, or solutions.4 However, resistance to traditional antifungal agents has developed recently due to inadequate, irregular, overuse of drugs or occurrence of certain gene mutations which may result in failure of conventional antifungal agents.5,6 Fluconazole is most widely used to treat fungal infections caused by C. albicans due to its high bioavailability and low toxicity.7 However, long term exposure and uncontrolled clinical use of Fluconazole led to the emergence of drug-resistant strains of C. albicans.8 Some antifungal drugs may cause adverse effects upon topical or systemic applications.9,10 Therefore, novel and new approaches of treatment of fungal infections with minimum side effects would be needed to be developed. In this context, nanotechnology offers smart solutions for various skin infections and can provide a new generation of materials to fight drug-resistant microbial infections.11 Wide range of nanomaterials are known for their promising antifungal activity such as polymeric nanoparticles, metal nanoparticles, and inorganic nanoparticles. The antifungal activity of silver nanoparticles and their mechanism of action were demonstrated in several published reports.12,13 Further, chitosan nanoparticles demonstrated antifungal activity against Candida causing skin infections.14 Zinc oxide nanoparticles revealed also antifungal activity against dermatophyte infections.15Gold nanoparticles (GNP), particularly those with non-spherical shape are known for their uncommon optical properties and they attract several biomedical applications such as diagnosis, imaging, drug delivery and phototherapy-based anti-cancer and anti-microbial activities.16,17 Antifungal activity of GNP against different types of fungus species was demonstrated in the literature; for example, a recently published study showed that 0.5 mM of GNR (5 nm in diameter) were effective in inhibiting the growth of fungal cells.18 Ronavari et al. demonstrated that among the tested fungus species, only Cryptococcus neoformans was susceptible to both biosynthesized silver and GNP.19 In another study, silver nanoparticles were more toxic towards Candida albicans and Saccharomyces cerevisiae fungal strains than GNP at concentration of 1.0 mM.20 However, investigating the antifungal activity of non-spherical GNP against several fungus species was lacking in the literature. Conjugating drugs with GNP is considered a promising approach to reduce resistant of fungus strains towards drugs. Recently, indolicidin-conjugated GNP was effective against Fluconazole-resistant strains of C. albicans isolated from patients with burn infection.21
In this study, Fluconazole was conjugated with gold nanorods (GNR) modified with thiolated PEG (PEG-SH) or thiolated PEGylated cholesterol (Chol-PEG-SH) moieties and loaded into poloxamer 407 hydrogel. The antifungal activity of the nanocomplexes; PEG-Fluc-GNR or Chol-Fluc-GNR was evaluated against C. albicans by measuring the minimum inhibitory concentration (MIC) and the percentage reduction of fungal viable count. The cytotoxicity of the nanocomplexes towards human dermal fibroblasts was estimated to represent the toxicity towards human skin dermal cells.
2. Materials and methods
2.1 Chemical synthesis of GNR suspension
GNR suspension was synthesized using a mixture of CTAB (Sigma-Aldrich Chemicals, USA) and sodium oleate (NaOL, Sigma-Aldrich Chemicals, USA) following a previous protocol.222.2 Surface functionalization of GNR with methoxy-poly(ethylene) glycol-thiol (m-PEG-SH)
To 10.0 mL of GNR suspension, a volume of 1.0 mL (10.0 mg mL−1) of m-PEG-SH solution (MW ∼ 2000 g mol−1, Sigma-Aldrich Chemicals, USA) was added and kept under stirring overnight. The obtained surface modified-GNR suspension (PEG-GNR) was centrifuged twice for 10 min at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510×g and were stored at 4 °C.
510×g and were stored at 4 °C.
2.3 Surface modification of GNR with thiolated PEGylated cholesterol (Chol-PEG-SH)
To 10.0 mL of GNR suspension, a volume of 1.0 mL (25.0 mg mL−1) of Chol-PEG-SH solution (MW ∼ 2000 g mol−1, Nanosoft Polymers, USA) was added and kept under stirring overnight. The obtained surface modified-GNR suspension (Chol-GNR) was centrifuged twice for 10 min at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510×g and the pellets were stored at 4 °C.
510×g and the pellets were stored at 4 °C.
2.4 Conjugation of fluconazole with GNR; PEG-Fluc-GNR and Chol-Fluc-GNR
A stock of Fluconazole was prepared in dimethyl sulfoxide (DMSO). Fluconazole was conjugated with PEG-GNR or Chol-PEG-GNR suspensions by adding Fluconazole (7.0 mg) into 1.0 mL (2.0 nM) of PEG-GNR or Chol-GNR suspensions. The nanocomplexes were stirred overnight and then spun down by centrifugation twice at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 × g for 8 min to remove the unbound drug. The obtained pellets of the nanocomplexes (PEG-Fluc-GNR and Chol-Fluc-GNR) were suspended in ultrapure water.
510 × g for 8 min to remove the unbound drug. The obtained pellets of the nanocomplexes (PEG-Fluc-GNR and Chol-Fluc-GNR) were suspended in ultrapure water.
2.5 Characterization of PEG-GNR; Chol-GNR; PEG-Fluc-GNR and Chol-Fluc-GNR
The prepared GNR suspensions were characterized by UV-vis absorption spectra at 200–1100 nm (UV-vis Spectrophotometer, Shimadzu UV-1800, Kyoto, Japan). The hydrodynamic size and zeta potential were conducted using Nicomp Nano Z3000 zeta potential/particle size analyzer, CA, USA. Samples of GNR with appropriate dilution (0.1–0.25 nM) were filled into dynamic light scattering (DLS) cuvettes for hydrodynamic size measurement, or folded capillary cells for zeta potential measurement at 25 °C. Mean values and standard deviations were calculated from at least three measurements. Transmission electron microscopy (TEM) imaging was performed using FEI Morgani 268, operating voltage of 60 kV, The Netherlands.The amount of the conjugated Fluconazole (mg) to GNR was measured using a well validated UV-vis absorption spectroscopy method. A standard calibration curve of Fluconazole was obtained by measuring the UV-vis absorbance of known concentrations of Fluconazole (2.0–0.03125 mg mL−1) in phosphate-buffered saline (PBS); pH 7.4 at 261 nm.
2.6 In vitro release of Fluconazole from PEG-Fluc-GNR and Chol-Fluc-GNR complexes
A volume of 1.0 mL of PEG-Fluc-GNR or Chol-Fluc-GNR was transferred into a dialysis bag (MWCO 12–14 kD, Spectrum Lab, USA) which was placed in a glass vial containing 20.0 mL of PBS (pH 7.4). Then, the vials were placed into a shaking water bath (37 °C, 120 rpm) (GFL 1083, Burgwedel, Germany). A volume of 0.5 mL of the release media was removed at specific time intervals (1, 2, 4, 6 and 24 h) and replaced with a fresh PBS (pH 7.4) to keep the sink condition. The concentration (mg mL−1) and the amount (mg) of Fluconazole in the removed media was calculated by measuring the optical absorbance at each time point at 261 nm against a standard calibration curve of Fluconazole in PBS (pH 7.4). The release profile of Fluconazole was presented as percentage of released cumulative amount vs. time.2.7 Loading of GNR suspensions into poloxamer 407 hydrogel
GNR suspensions (PEG-GNR, Chol-GNR, PEG-Fluc-GNR, Chol-Fluc-GNR) were loaded into poloxamer 407 polymer (18%, BSAF, Germany). The GNR-hydrogels were kept at 4 °C overnight, and then were sterilized by UV light for 45 min before use.The GNR-hydrogels were characterized by their colloidal color, UV-vis absorption spectra hydrodynamic size and zeta potential. The sol–gel transition temperature of the GNR-hydrogels were measured following a previously described method.23
2.8 Measurement of the anti-fungal activity of GNR suspensions and the nanocomplexes against C. albicans
The inoculum was maintained on Sabouraud Dextrose Agar (SDA, Biolab, Hungary) and the culture was stored at 4 °C. The 48 h-old C. albicans culture was standardized using 0.5 McFarland standard, which was used to prepare ca. 1 × 106 CFU mL−1 inoculum. The inoculum suspension was used within 15 min of preparation.
A volume of 150 μL of Muller Hinton broth medium (Biolab, Hungary) was distributed to each well in the row, then, a volume of 150 μL (4.0 nM) of the treatment (PEG-GNR; Chol-GNR; PEG-Fluc-GNR or Chol-Fluc-GNR) was added to the first well only. Two-fold serial dilutions were carried out across the wells of the plate. Each well was subsequently inoculated with 15 μL of C. albicans and the plates were incubated at 37 °C temperature for 48 h. After incubation, the MIC value was determined as the lowest concentrations of the treatment showing no growth of the microorganism. Subsequently, the viable count of fungus (CFU mL−1) within the well of lowest concentration showing no turbidity was estimated using the standard spread plate count method as described previously.17 Briefly, a volume of 100 μL from the clear well was serially ten-fold diluted in normal saline. A volume of 100 μL of each dilution was spread onto nutrient SDA and the plates were incubated overnight at 37 °C.
The fungal viable count (CFU mL−1) was calculated from the number of fungal colonies and the dilution factor (eqn (1)). The percentage reduction in the fungal viable count and the log-reduction in fungal viability were calculated from the fungal viable count and the initial inoculum size (eqn (2)).
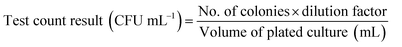 | (1) |
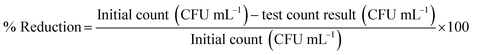 | (2) |
2.9 Cytotoxicity of the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR against human dermal fibroblast
For MTT assay, the medium from the wells was removed carefully after incubation and 100 μL of fresh medium and 10 μL of MTT (5 mg mL−1) were added into each well. The plates were incubated for 4 h in 5% CO2 incubator for cytotoxicity. After incubation, the medium from the wells was removed carefully and 100 μL of DMSO was added to each well and mixed well by shaking for 10–15 min. The viable cells were measured by the development of purple color due to formation of formazan crystals. The absorbance was recorded at 570 nm by an ELISA plate reader, and the cellular viability percentage of the treated cells was calculated relative to the cellular viability of the control untreated cells.
3. Results and discussion
3.1 Synthesis and characterization of GNR and their surface modification with Fluconazole
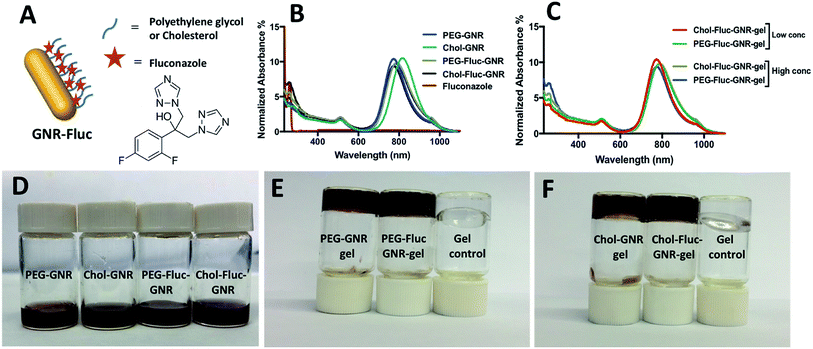
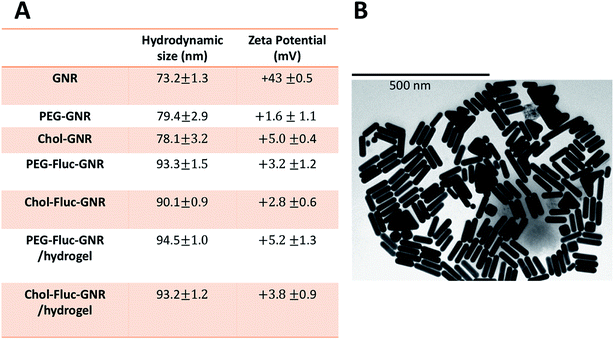
The synthesized GNR has two typical absorption peaks at 536 nm and 783 nm for transverse and longitudinal plasmons, respectively. The surface of the synthesized GNR were functionalized with PEG-thiol and Chol-PEG-SH to obtain PEG-GNR and Chol-GNR, respectively. The longitudinal optical spectra of PEG-GNR and Chol-GNR were slightly shifted upon surface functionalization and they showed no obvious broadening or tailing which suggest their excellent colloidal stability (Fig. 1B). The hydrodynamic sizes of PEG-GNR and Chol-GNR were ∼79 nm and ∼78, respectively (Fig. 2A). The zeta potentials of PEG-GNR and Chol-GNR were dropped from +43 mV before coating to +1.6 mV and +5.0 mV after coating, respectively, which suggest their successful surface functionalization (Fig. 2A). The ligands used for functionalization of GNR are thiolated in order to enhance their attachment to the surface of the nanorods by S–Au covalent bond.24 Attachment of Chol-PEG-SH to the surface of GNR was confirmed previously by 1H-NMR method.25The attachment of hydrophobic ligands into aqueous nano-systems is considered a big challenge. In this study, Fluconazole, which is the most common drug used for treatment of fungal infections, was attached to the surface of GNR using PEG or cholesterol moieties (Fig. 1A). Fluconazole is supposed to form hydrophobic interactions with either PEG or cholesterol moieties that are grafted on the surface of GNR.
The optical spectrum of Fluconazole in PBS buffer demonstrated a sharp peak at ∼261 nm. The optical spectra of PEG-Fluc-GNR and Chol-Fluc-GNR demonstrated typical plasmon peaks without broadening or tailing, and they showed an extra peak at ∼262 nm which is corresponding to the loaded Fluconazole (Fig. 1B). However, the amount of Fluconazole loaded into both GNR preparations was different. The conjugated amount of Fluconazole into GNR was estimated using a validated optical absorption spectroscopy method against a standard calibration curve of known concentrations of Fluconazole (2.0–0.03125 mg mL−1) in PBS (pH 7.4), R2 = 0.981. The drug loading efficiency percentage was estimated by dividing the amount of Fluconazole loaded into GNR by the initial amount of Fluconazole added to the GNR suspension. The amount of Fluconazole loaded into 1.0 mL of PEG-GNR and Chol-GNR was ∼4.34 ± 0.3 mg and 2.32 ± 0.53 mg, with a drug loading efficiency percentage of 62.5% and 33.0%, respectively. The hydrodynamic size of PEG-Fluc-GNR and Chol-Fluc-GNR was increased to ∼93.3 nm and ∼90.1 nm, respectively upon loading with Fluconazole (Fig. 2A). The shape and size of the nanorods were confirmed by TEM imaging of PEGylated GNR. The image in Fig. 2B demonstrates a length and width of 74.6 ± 0.4 nm and 18.2 ± 0.5 nm, respectively of one nanorod.
The optical spectra of PEG-Fluc-GNR and Chol-Fluc-GNR loaded into poloxamer 407 hydrogel indicate excellent colloidal stability of the nanorods upon loading into hydrogel, where the plasmon peaks do not demonstrate broadening or tailing (Fig. 1C). The hydrodynamic size and zeta potential of the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR loaded into hydrogel did not demonstrate significant changes which suggest their excellent colloidal stability upon loading into hydrogel (Fig. 2A). The sol–gel transition temperature of GNR preparations loaded into poloxamer 407 hydrogel was ∼35.3 °C.
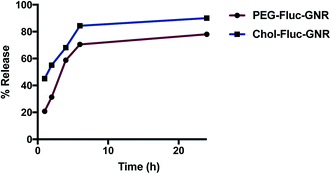
The release profile of Fluconazole from PEG-Fluc-GNR or Chol-Fluc-GNR nanocomplexes demonstrates a fast release of the drug over the first 6 h of incubation. However, Fluconazole conjugated with Chol-GNR showed a slightly faster release than that observed from PEG-Fluc-GNR (Fig. 3). Such a release pattern may affect the antifungal activity of the nanocomplexes as described below.
3.2 Antifungal activity of PEG-GNR, Chol-PEG-GNR, and the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR
Fluconazole, a third generation triazole antifungal drug, has broad spectrum activity towards systemic and topical fungal infections,26 and it acts by inhibiting the fungal cytochrome P-450 enzyme.27 Topical skin delivery of Fluconazole is considered as an effective approach for local therapy of fungal infections, however, its efficiency to penetrate the skin is challenging.Fluconazole is a moderate lipophilic compound,28 and its conjugation to nanoparticles is supposed to improve it solubility and enhancing its delivery into the skin layers.
The antifungal activity of the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR was investigated in vitro by measuring the MIC of the nanocomplexes against C. albicans and compare them to that of the unloaded GNR suspensions. The MIC for PEG-GNR and Chol-GNR was 0.125 nM and 0.25 nM, respectively (Table 1). Conjugating Fluconazole with PEG-GNR or Chol-GNR dramatically reduced the MIC value of GNR to be 0.025 nM (5-fold) and 0.018 nM (14-fold), respectively, and corresponding to 5-log reduction of fungal viable count (Table 1). Upon conjugation of PEG-GNR or Chol-GNR with Fluconazole, the MIC value of Fluconazole was reduced by 9-fold and 12-fold, respectively. Although the loading percentage of Fluconazole using Chol-GNR (Chol-Fluc-GNR) was less than that using PEG-GNR (PEG-Fluc-GNR), Chol-Fluc-GNR demonstrates higher anti-fungal activity than that of PEG-Fluc-GNR. This may be related to the pattern of Fluconazole release from both preparations (Fig. 3).
| Treatment | MIC GNR (nM)/Fluc (mg mL−1) | Reduction of bacterial viable count (%) | Average log reduction |
|---|---|---|---|
| PEG-GNR | 0.125 | 99.96 | 3.34 |
| Chol-GNR | 0.25 | 72.0 | 0.55 |
| PEG-Fluc-GNR | 0.025/0.041 ± 0.007 | 99.999 | 5.0 |
| Chol-Fluc-GNR | 0.018/0.032 ± 0.009 | 99.999 | 5.0 |
| Fluconazole | 0.375 ± 0.176 | 91.21 | 1.0 |
From the above results, we propose that coupling of Fluconazole with GNR drastically enhanced the antifungal activity of the obtained nanocomplexes and resulted in significant reduction of the viable fungal cells. In addition to the toxic effect of GNR towards C. albicans, we propose that conjugating Fluconazole with GNR enhanced the delivery efficiency of Fluconazole to the cell wall of the fungal cells and accelerated their cellular uptake as demonstrated from the TEM imaging below.
Combination of Fluconazole with other nano-systems was investigated in the literature; for example, physical mixing of Fluconazole with silver nanoparticles has produced synergistic effect against ATCC 26790 strain of C. albicans.29 Further, copper oxide was combined with Fluconazole to enhance the anti-fungal activity of the mixture against C. albicans, however, the MIC value of Fluconazole in the mixture was ∼0.2 mg mL−1, which is higher than that observed in our study.30 Several studies demonstrated that incorporating Fluconazole with several nano-systems such as graphene dioxide, chitosan nanoparticles, solid lipid nanoparticles or poly(lactic-co-glycolic acid) (PLGA) was proved to enhance the antimycotic activity of Fluconazole against different strains of C. albicans.31–34 On the other hand, curcumin conjugated with silver nanoparticles and selenium nanoparticles demonstrated antifungal activity against fluconazole-resistant Candida isolates.35,36 However, utilization of GNP, and particularly non-spherical shapes such as GNR as antifungal agents is limited in the literature. Recently, Rahimi et al. have demonstrated the antifungal activity of indolicidin upon conjugation with gold nanospheres.21 In our work, we designed a stable Fluconazole-conjugated GNR loaded into hydrogel as a topical antifungal agent with enhanced antimycotic activity.
3.3 Antifungal activity of PEG-GNR and Chol-GNR, and the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR loaded into poloxamer 407 hydrogel
Loading the GNR preparations into poloxamer 407 hydrogel enhances their topical delivery. Thermo-responsive hydrogels containing GNP demonstrated antibacterial activity and enhanced the contact time with the infected areas.37 We have previously demonstrated the effective antibacterial activity of GNR loaded into poloxamer 407 hydrogel against planktonic and biofilm of Pseudomonas aeruginosa.38The viable count of C. albicans was estimated upon exposure to GNR preparations (for 3 h) at high concentration (2 nM) and at the MIC of that GNR preparation loaded into poloxamer 407 hydrogel. Table 2 indicates that exposure of C. albicans to the nanocomplexes; PEG-Fluc-GNR or Chol-Fluc-GNR, loaded into hydrogel was resulted in 1.3–3.7 log-reduction in viable C. albicans depending on the concentration of the nanocomplexes.
| Treatment in poloxamer 407 hydrogels | Average reduction of fungal viable count (%) | Average log reduction |
|---|---|---|
| PEG-GNR | ||
| 2.0 nM | 89.1 ± 0.06 | 0.95 |
| 0.125 nM | 82.5 ± 0.03 | 0.75 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Chol-GNR | ||
| 2.0 nM | 85.4 ± 0.02 | 0.83 |
| 0.25 nM | 81.1 ± 0.002 | 0.72 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| PEG-Fluc-GNR | ||
| 2.0 nM | 99.3 ± 0.003 | 2.15 |
| 0.025 nM | 95.7 ± 0.01 | 1.36 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Chol-Fluc-GNR | ||
| 2.0 nM | 99.98 ± 0.005 | 3.7 |
| 0.018 nM | 96.8 ± 0.006 | 1.5 |
| Poloxamer 408 hydrogel | 72.8 ± 0.002 | 0.55 |
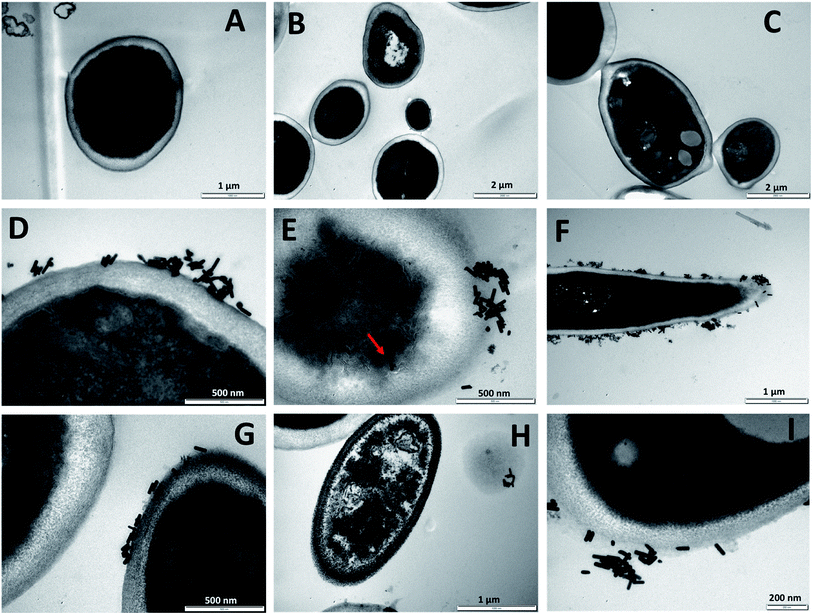
3.4 Imaging of C. albicans treated with PEG-Fluc-GNR by transmission electron microscope (TEM)
In order to understand the mechanism of interaction of the nanocomplexe; PEG-Fluc-GNR with C. albicans, TEM imaging of the treated C. albicans was performed. Fig. 4A–C demonstrate untreated C. albicans which indicate the normal shape of C. albicans with well-intact cell wall. GNR conjugated with Fluconazole were highly up-taken into the cell wall of the C. albicans within the first 120 min of exposure (Fig. 4D–I).The uptake of the nanocomplex into the cell wall of the C. albicans was resulted in partial collapse and disintegration of the cytoplasm material as indicated in Fig. 4E–G. A complete collapse of the fungal cell was observed upon treatment with the nanocomplex as indicated in Fig. 4H (right). We propose that loading of Fluconazole into GNR has enhanced its delivery and uptake into the cells and accordingly enhanced its cytotoxicity. However, the contribution of GNR themselves to the observed cytotoxicity could not be excluded.
3.5 Cytotoxicity of the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR against human dermal fibroblasts
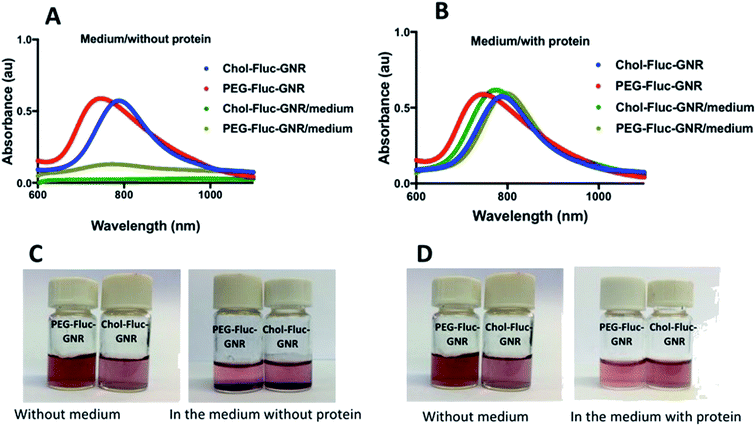
The possible cytotoxicity of the nanocomplexes; PEG-Fluc-GNR and Chol-Fluc-GNR was investigated towards human dermal fibroblasts which represent the human skin cells. The colloidal stability of the nanocomplexes upon mixing with the tissue culture medium was investigated with and without addition of FBS.Fig. 5A represents the optical spectra of the nanocomplexes upon mixing with the tissue culture medium without addition of FBS; the spectra showed obvious broadening and tailing of the peaks, which are strongly correlated with severe aggregation of the nanoparticles. However, addition of FBS to the tissue culture medium drastically enhanced the colloidal stability of the nanocomplexes as demonstrated by their typical optical spectra without significant broadening or tailing (Fig. 5B). The aggregation of the nanoparticles upon mixing with FBS-free tissue culture media was presented in photos in Fig. 5C, while Fig. 5D represents photos of the nanoparticles upon mixing with FBS-containing tissue culture medium as there was no aggregation of the nanoparticles. We propose that the presence of FBS in the tissue culture medium greatly stabilized the nanoparticles and prevented their aggregation during conducting the cellular viability test. The colloidal stability of the nanoparticles upon mixing with tissue culture media is considered as a “hidden factor”, and it is usually ignored in the majority of studies related to the nano–bio interfaces. According to this stability study, the cellular viability of the human dermal fibroblasts was conducted using FBS-containing tissue culture medium to maintain the stability of the nanoparticles, and to prevent any possible misinterpretation of the findings.
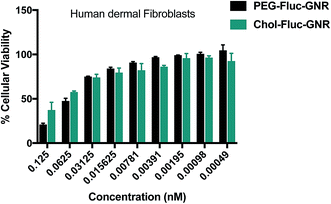
Fig. 6 represents the cellular viability percentages of human dermal fibroblasts upon treatment with the nanocomplexes; over a concentration range of 0.125–0.00049 nM of GNR, for 24 h. The cytotoxicity of the nanocomplexes against skin dermal cells is concentration-dependent. The cellular viability percentage was more than 75% over a concentration range of 0.03125–0.00049 nM, that includes the MIC concentrations against C. albicans.
Based on these results, conjugating of GNR with Fluconazole has significantly reduced the MIC for both GNR and Fluconazole against C. albicans and has low cytotoxicity towards human skin cells represented by human dermal fibroblasts.
4. Conclusions
Nanotechnology-based drug delivery is considered a cornerstone in nanomedicine. Loading of Fluconazole, which is the most common antifungal agent, into GNR functionalized with thiolated PEG or thiolated-PEGylated cholesterol drastically enhanced the antifungal activity of the nanocomplexes against C. albicans by increasing the delivery and uptake of the drug by the cells. The GNR loaded with Fluconazole showed low cytotoxicity against human skin cells represented by human dermal fibroblasts. Loading Fluconazole into surface functionalized GNR could be considered a promising approach to enhance the activity of Fluconazole towards topical fungal infections and to reduce its adverse reactions and resistance.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge the Deanship of Scientific Research and Graduate Studies at Al-Zaytoonah University of Jordan for financial funding.References
- R. Hay, Medicine, 2017, 45, 707–710 CrossRef.
- K. T. Clebak and M. A. Malone, Prim. Care Clin. Off. Pract., 2018, 45, 433–454 CrossRef.
- S. Grover and P. Roy, Med. J. Armed Forces India, 2003, 59, 114–116 CrossRef.
- A. K. Gupta and K. A. Foley, J. Fungi, 2015, 1, 13–29 CrossRef.
- S. Y. Chon, H. Q. Doan, R. M. Mays, S. M. Singh, R. A. Gordon and S. K. Tyring, Dermatol. Ther., 2012, 25, 55–69 CrossRef.
- V. Pai, A. Ganavalli and N. N. Kikkeri, Indian J. Dermatol., 2018, 63, 361–368 CrossRef.
- S. Sarkar, P. Uppuluri, C. G. Pierce and J. L. Lopez-Ribot, Antimicrob. Agents Chemother., 2014, 58, 1183–1186 CrossRef PubMed.
- W. Youngsaye, C. L. Hartland, B. J. Morgan, A. Ting, P. P. Nag, B. Vincent, C. A. Mosher, J. A. Bittker, S. Dandapani, M. Palmer, L. Whitesell, S. Lindquist, S. L. Schreiber and B. Munoz, Beilstein J. Org. Chem., 2013, 9, 1501–1507 CrossRef PubMed.
- D. F. Lewis, A. D. Rodrigues, C. Ioannides and D. V. Parke, J. Biochem. Toxicol., 1989, 4, 231–234 CrossRef CAS PubMed.
- X. S. Ramírez-Gómez, S. N. Jimenez-García, V. Campos, E. R. Miranda, G. Herrera-Pérez and R. Vargas-Bernal, 2018, Clinical Relevance of Medicinal Plants and Foods of Vegetal Origin on the Activity of Cytochrome P450 Search PubMed.
- X. Zhu, A. F. Radovic-Moreno, J. Wu, R. Langer and J. Shi, Nano Today, 2014, 9, 478–498 CrossRef CAS PubMed.
- H. H. Lara, D. G. Romero-Urbina, C. Pierce, J. L. Lopez-Ribot, M. J. Arellano-Jimenez and M. Jose-Yacaman, J. Nanobiotechnol., 2015, 13, 91 CrossRef PubMed.
- K.-J. Kim, W. S. Sung, B. K. Suh, S.-K. Moon, J.-S. Choi, J. G. Kim and D. G. Lee, BioMetals, 2009, 22, 235–242 CrossRef CAS PubMed.
- Y. Y. Albasarah, S. Somavarapu, P. Stapleton and K. M. Taylor, J. Pharm. Pharmacol., 2010, 62, 821–828 CrossRef CAS.
- N. Tiwari, R. Pandit, S. Gaikwad, A. Gade and M. Rai, IET Nanobiotechnol., 2017, 11, 205–211 CrossRef PubMed.
- P. K. Jain, X. Huang, I. H. El-Sayed and M. A. El-Sayed, Acc. Chem. Res., 2008, 41, 1578–1586 CrossRef CAS PubMed.
- N. N. Mahmoud, A. M. Alkilany, E. A. Khalil and A. G. Al-Bakri, Sci. Rep., 2018, 8, 6881 CrossRef.
- M. Nidhin, D. Saneha, S. Hans, A. Varghese, Z. Fatima and S. Hameed, Mater. Res. Bull., 2019, 119, 110563 CrossRef CAS.
- A. Ronavari, N. Igaz, M. K. Gopisetty, B. Szerencses, D. Kovacs, C. Papp, C. Vagvolgyi, I. M. Boros, Z. Konya, M. Kiricsi and I. Pfeiffer, Int. J. Nanomed., 2018, 13, 695–703 CrossRef CAS.
- U. T. Khatoon, G. V. S. N. Rao, M. K. Mohan, A. Ramanaviciene and A. Ramanavicius, J. Environ. Chem. Eng., 2018, 6, 5837–5844 CrossRef CAS.
- H. Rahimi, S. Roudbarmohammadi, H. H. Delavari and M. Roudbary, Int. J. Nanomed., 2019, 14, 5323–5338 CrossRef CAS PubMed.
- X. Ye, C. Zheng, J. Chen, Y. Gao and C. B. Murray, Nano Lett., 2013, 13, 765–771 CrossRef CAS PubMed.
- M. J. Bhandwalkar and A. M. Avachat, AAPS PharmSciTech, 2013, 14, 101–110 CrossRef CAS PubMed.
- H. Hinterwirth, S. Kappel, T. Waitz, T. Prohaska, W. Lindner and M. Lammerhofer, ACS Nano, 2013, 7, 1129–1136 CrossRef CAS PubMed.
- N. N. Mahmoud, A. A. Alhusban, J. I. Ali, A. G. Al-Bakri, R. Hamed and E. A. Khalil, Sci. Rep., 2019, 9, 5796 CrossRef PubMed.
- M. M. Abdel-Mottaleb, N. D. Mortada, A. A. El-Shamy and G. A. Awad, Drug Dev. Ind. Pharm., 2009, 35, 311–320 CrossRef CAS PubMed.
- N. Longley, C. Muzoora, K. Taseera, J. Mwesigye, J. Rwebembera, A. Chakera, E. Wall, I. Andia, S. Jaffar and T. S. Harrison, Clinical infectious diseases, an official publication of the Infectious Diseases Society of America, 2008, vol. 47, pp. 1556–1561 Search PubMed.
- F. X. Mathy, D. Ntivunwa, R. K. Verbeeck and V. Preat, J. Pharm. Sci., 2005, 94, 770–780 CrossRef CAS PubMed.
- C. Longhi, J. P. Santos, A. T. Morey, P. D. Marcato, N. Durán, P. Pinge-Filho, G. Nakazato, S. F. Yamada-Ogatta and L. M. Yamauchi, Med. Mycol., 2015, 54, 428–432 CrossRef PubMed.
- I. S. Weitz, M. Maoz, D. Panitz, S. Eichler and E. Segal, J. Nanopart. Res., 2015, 17, 342 CrossRef.
- S. Asadi Shahi, S. Roudbar Mohammadi, M. Roudbary and H. Delavari, Prog. Biomater., 2019, 8, 43–50 CrossRef PubMed.
- H. A. El Rabey, F. M. Almutairi, A. I. Alalawy, M. A. Al-Duais, M. I. Sakran, N. S. Zidan and A. A. Tayel, Int. J. Biol. Macromol., 2019, 141, 511–516 CrossRef CAS PubMed.
- N. Gómez-Sequeda, R. Torres and C. Ortiz, Nanotechnol., Sci. Appl., 2017, 10, 95–104 CrossRef PubMed.
- M. Moazeni, M. Saeedi, H. Kelidari, M. Nabili and A. Davari, Curr. Top. Med. Mycol., 2019, 5, 8–13 Search PubMed.
- S. Paul, K. Mohanram and I. Kannan, Ayu, 2018, 39, 182–186 CrossRef PubMed.
- N. Parsameher, S. Rezaei, S. Khodavasiy, S. Salari, S. Hadizade, M. Kord and S. A. Ayatollahi Mousavi, Curr. Top. Med. Mycol., 2017, 3, 16–20 CrossRef PubMed.
- N. N. Mahmoud, S. Hikmat, D. Abu Ghith, M. Hajeer, L. Hamadneh, D. Qattan and E. A. Khalil, Int. J. Pharm., 2019, 565, 174–186 CrossRef CAS PubMed.
- A. G. Al-Bakri and N. N. Mahmoud, Molecules, 2019, 24, 2661 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2020 |