Open Access Article
Open Access ArticleA porous nano-adsorbent with dual functional groups for selective binding proteins with a low detection limit
Xueyan Zouaef,
Yu Zhangb,
Jinqiu Yuanc,
Zhibo Wangb,
Rui Zengc,
Kun Lid,
Yanbao Zhao*aef and
Zhijun Zhang *aef
*aef
aEngineering Research Center for Nanomaterials, Henan University, Kaifeng 475004, China. E-mail: zhangzj09@126.com; zhaoyb902@henu.edu.cn
bCollege of Chemistry and Chemical Engineering, Henan University, Kaifeng 475004, China
cInstitute of Technology, Henan University Minsheng College, Kaifeng 459000, China
dState Key Laboratory of Crop Stress Adaptation and Improvement, Kaifeng 459000, China
eNational & Local Joint Engineering Research Center for Applied Technology of Hybrid Nanomaterials, Kaifeng 459000, China
fKey Laboratory for Monitor and Remediation of Heavy Metal Polluted Soils of Henan Province, Jiyuan 459000, China
First published on 17th June 2020
Abstract
In this study, porous silica nanoparticles functionalized with a thiol group (SiO2–SH NPs) were synthesized via a one-pot method. Subsequently, iminodiacetic acid was modified, and further adsorption of Ni2+ ions was conducted to obtain a SiO2–S/NH–Ni nano-adsorbent. Then, transmission electron microscopy (TEM), scanning electron microscopy (SEM), Fourier transform infrared (FT-IR) spectroscopy, thermogravimetric analysis (TG) and X-ray diffraction (XRD) were employed to characterize its morphology and composition. The results indicate that the SiO2–S/NH–Ni nano-adsorbent is porous, has an average diameter of 77.1 nm and has a small porous structure of about 3.7 nm in the silica skeleton. The Brunauer–Emmett–Teller (BET) surface area and total pore volume were 537.2 m2 g−1 and 3.3 cm3 g−1, respectively, indicating a large BET surface area. The results indicate that the as-prepared SiO2–S/NH–Ni nano-adsorbent would be suitable to selectively and efficiently bind His-tagged proteins from an E. coli cell lysate. The SDS-PAGE results show that the as-prepared nano-adsorbent presents specifically to both His-tagged CPK4 and His-tagged TRX proteins, indicating the nano-adsorbent can be used to effectively separate His-tagged proteins and is universal to all His-tagged fusion proteins. We also found that the as-prepared nano-adsorbent exhibits a low detection limit (1.0 × 10−7 mol L−1) and a strong regeneration ability based on four regeneration experiments that were particularly suited to the separation of His-tagged proteins.
Introduction
Proteins play a variety of crucial roles in organisms, and the enrichment and purification of proteins, particularly low-abundance proteins, are currently hot topics.1–6 Histidine is a semi essential amino acid for humans that constitutes the proteins of the body. It has a very wide range of applications, such as in food, medicine and other fields.7–10 Furthermore, biotechnologies nowadays have enabled proteins to easily express with a tag, and numerous protein-purification methods are based on the specific interactions between immobilized ligands and affinity tags.11–16 Among affinity tags, histidine tags are particularly popular as they can be readily incorporated into desired proteins by the expression of the target gene in microorganism cells using commercial expression vectors.17,18 Therefore, the easy separation and purification of His-tagged proteins are very important in biology and medicine.Recently, metal-chelate affinity precipitation has been employed to separate His-tagged recombinant proteins based on the interactions between metal ions (Ni2+, Cu2+, Zn2+ and so on) and the histidine residues exposed on the surface of the proteins.19–22 Although metal-chelate affinity precipitation is easily adaptable to any protein expression system, there are some limits to using this method, such as a requirement for pretreatment, solvent consumption, and protein solubility.23–25
With the development of nanotechnology, nanomaterials are used in many fields, such as protein purification, environmental treatment, targeted drugs and so on.26–28 However, the major limitations of current nanomaterial systems are the detection limit and poor regeneration ability, which leads to low purification efficiency. In this study, porous silica with dual functional groups (–SH and –NH2 groups) was employed as an adsorbent to separate His-tagged proteins. This material can provide a larger surface area, comparable to solid particles, and improve the separation capacity. The as-prepared nano-adsorbent can be used to directly separate His-tagged proteins from the E. coli lysate.
Results and discussion
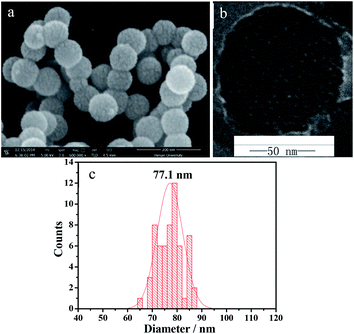
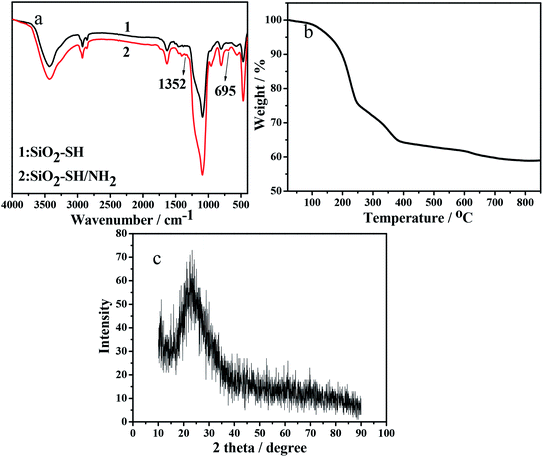
Fig. 1 shows the SEM (a) and TEM (b) images of the as-synthesized SiO2–S/NH–Ni sample. It can be seen in Fig. 1a that the main products were spherical in shape and presented uniform spheres. Moreover, we could see that the as-prepared SiO2–S/NH–Ni sample displayed a narrow size distribution and had an average diameter of 77.1 nm (Fig. 1c). Evidently, the surfaces of these nanospheres were rough, which may be due to the oriented growth and crystallinity.27 As shown in Fig. 1b, the obtained SiO2–S/NH–Ni sample displays a small porous structure, with about 3.7 nm, in the silica skeleton.Fig. 2a shows the FT-IR absorption spectra of the as-prepared SiO2–SH NPs (curve 1) and SiO2–SH/NH2 NPs (curve 2). In the FT-IR spectra, the peak at 3430 cm−1 is attributed to the –OH stretching vibration of porous silica spheres. The peaks at around 1105, 807 and 471 cm−1 were due to Si–O–Si stretching vibrations. The strong peaks at 2939 and 2836 cm−1 belong to the stretching vibrations of the –CH2– bonds in MPS, which indicates the presence of –SH in the SiO2–SH NPs (Fig. 2a curve 1). It is clear that the curve of the SiO2–SH/NH2 NPs is very similar to that of the SiO2–SH NPs, which indicates that the main component of SiO2–SH/NH2 NPs is SiO2. The presence of the N–H bending vibration at around 695 cm−1 confirms the incorporation of the imino group of IDA.29
Fig. 2b shows the TG curve of the SiO2–SH/NH2 NPs. It can be seen that the sample displays successive weight loss from room temperature to 850 °C. The initial mass loss below 250 °C can be attributed to the desorption of adsorbed water from the surface of the sample powder, and the following mass loss was related to the further release of the inner adsorbed or crystal water (total mass loss 25%). The major mass loss (about 15.5%) took place in the range of 250–600 °C, which might be attributed to the thermal decomposition of the –SH and –NH2 groups of the as-prepared SiO2–SH/NH2 NPs. This indicates that there are two groups (–SH and –NH2) on the surface of the as-synthesized NPs. Fig. 2c shows the XRD pattern of the as-prepared SiO2–SH/NH2 NPs. The XRD pattern exhibits an obvious broad diffraction peak at around 23°, which was indexed to the scattering of amorphous SiO2 (JCPDS 76-0933).30
The surface and pore properties of the as-prepared SiO2–SH/NH2 NPs was examined via N2 sorption, as shown in Fig. 3. It can be seen in Fig. 3a that the N2 adsorption–desorption isotherms are characteristic of type IV curves with the H4 hysteresis loop in the relative-pressure range of 0.04–0.3, which indicates the presence of well-defined mesoporous structures with narrow slit-like pores.31,32 It can be seen that the pore size distributions calculated using the BJH method were 3.3 and 3.7 nm (Fig. 3b). Moreover, the Brunauer–Emmett–Teller (BET) surface area and total pore volume were 537.2 m2 g−1 and 3.3 cm3 g−1, respectively, which represents a large BET surface area that is suitable for separating proteins.
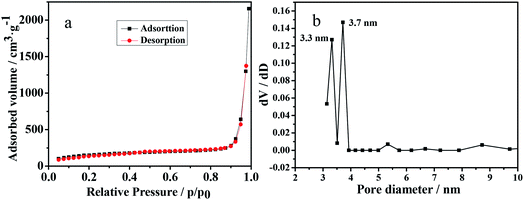 | ||
| Fig. 3 N2 adsorption–desorption isotherm (a) and the pore size distribution (b) of the SiO2–S/NH–Ni NPs. | ||
Fig. 4 shows a scheme of the preparation of the SiO2–S/NH–Ni nano-adsorbent and their separation of His-tagged proteins. It can be seen that there are three steps required to synthesize the SiO2–S/NH–Ni nano-adsorbent. First, SiO2–SH was prepared by the hydrolysis of TEOS and MPS via a one-pot method. Second, the IDA molecules were modified on the surfaces of the SiO2–SH NPs. Third, the SiO2–S/NH–Ni nano-adsorbent was obtained by chelating Ni2+. Then, the nano-adsorbent can be used to separate His-tagged proteins from the E. coli lysate, and the obtained SiO2–S/NH–Ni/His-tagged proteins can be eluted by imidazole. Subsequently, the His-tagged proteins can be obtained, and the remaining SiO2–S/NH–Ni nano-adsorbent could be reused by regeneration with EDTA and NiCl2 solutions. The results show that the as-prepared SiO2–S/NH–Ni nano-adsorbent has a strong regeneration ability, particularly suited to the separation of the His-tagged fusion proteins.
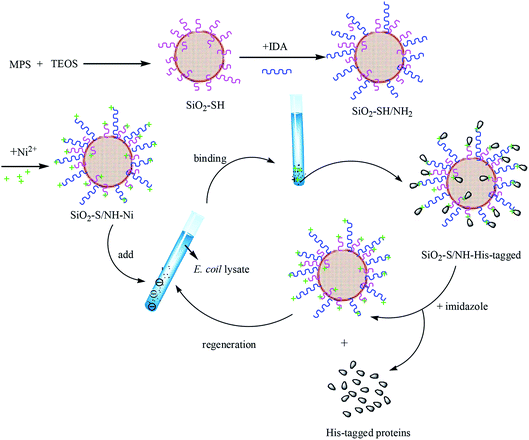 | ||
| Fig. 4 Scheme for the preparation of the SiO2–S/NH–Ni samples and their separation of His-tagged proteins. | ||
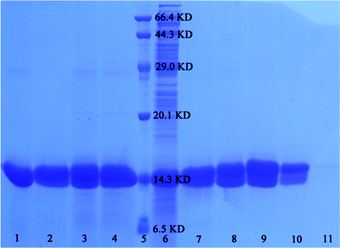
To estimate the separation effect of the as-prepared samples, SDS-PAGE was employed as a general method for assessing the target proteins. Fig. 5 shows the SDS-PAGE analysis of His-tagged proteins that were purified by the SiO2–S/NH–Ni nano-adsorbent. Lanes 1–4 contain the fractions washed off from the SiO2–S/NH–Ni nano-adsorbent with different amounts of imidazole (lane 1, 0.5 mol L−1; lane 2, 1 mol L−1; lane 3, 2 mol L−1; lane 4, 3 mol L−1). Lane 5 contains the marker, and lane 6 contains the E. coli lysate. It was been found that the SiO2–S/NH–Ni nano-adsorbent could efficiently enrich target proteins from cell lysate. By changing the concentration of imidazole (0.5–3 mol L−1) and keeping the others unchanged, the quantity of disassociated proteins remained basically the same with increment in the concentration of imidazole (Fig. 5 lane 1–4), so 0.5 mol L−1 imidazole was used in subsequent experiments.
Keeping the imidazole concentration constant (0.5 mol L−1), the effect of the concentration of the target proteins was investigated, as shown in Fig. 5. The concentrations in the lanes 7–10 are as follows: lane 7, 1.0 × 10−4 mol L−1; lane 8, 1.0 × 10−5 mol L−1; lane 9, 1.0 × 10−6 mol L−1; lane 10, 1.0 × 10−7 mol L−1; lane 11, 1.0 × 10−8 mol L−1. When the concentrations of the TRX proteins were 1.0 × 10−4, 1.0 × 10−5, 1.0 × 10−6, 1.0 × 10−7 and 1.0 × 10−8 mol L−1, the target proteins could be detected effectively by the as-prepared SiO2–S/NH–Ni nano-adsorbent, which shows that the samples can be used to detect target proteins with a detection limit lower than 1.0 × 10−7 mol L−1.
In order to investigate the reusability properties of the as-prepared SiO2–S/NH–Ni nano-adsorbent, we separated the target proteins four times. Fig. 6 shows the SDS-PAGE for the four recycling times using lane 1, Ni-NTA agarose; lane 2, marker; lane 3, His-tagged TRX E. coli lysate; lane 5, 1st; lane 6, 2nd; lane 7, 3rd; lane 8, 4th. The adsorbent could be repeatedly used in the experiment if it was treated with EDTA and NiCl2 as needed. It can be seen that the specificity and affinity of the SiO2–S/NH–Ni nano-adsorbent remained unaffected after four recycling times. The binding capacities of the target protein were 9.8, 9.8, 9.6 and 9.2 mmol g−1.
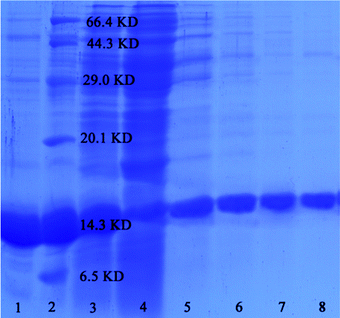 | ||
| Fig. 6 SDS-PAGE analysis of His-tagged proteins separated by the as-prepared SiO2–S/NH–Ni nano-adsorbent. | ||
In order to verify the universality of the SiO2–S/NH–Ni nano-adsorbent, we used the adsorbent to separate His-tagged Ca2+-dependent protein kinase 4 (His-tagged CPK4) proteins from the E. coli lysate, as shown in Fig. 6. The lanes are as follow: lane 3, His-tagged TRX E. coli lysate; lane 4, His-tagged CPK4 E. coli lysate; lane 9, His-tagged TRX separated by the SiO2–S/NH–Ni nano-adsorbent; lane 10, His-tagged CPK4 separated by the SiO2–S/NH–Ni nano-adsorbent. It is clear that the His-tagged CPK4 proteins could be separated specifically from the E. coli lysate, and there was no nonspecific binding. These results indicate that the as-prepared SiO2–S/NH–Ni nano-adsorbent could be used effectively in His-tagged protein affinity separation or purification, and it is universally applicable to all His-tagged fusion proteins. These results are due to the Ni of the as-prepared SiO2–S/NH–Ni nano-adsorbent being specific to histidine and the specific binding being independent of the type of fusion proteins.
Experimental
Materials
Ni-NTA agarose was purchased from QIAGEN. Phosphate buffer saline solutions (abridged as PBS; concentration 0.1 mol L−1, pH = 8.0; concentration 0.01 mol L−1, pH = 7.4) were purchased from Sigma-Aldrich, America. 3-Glycidyloxy-propyltrimethoxysilane (GPTMS) and iminodiacetic acid (IDA) were purchased from Aladdin. Tetraethyl orthosilicate (TEOS) was from Tianjin Fuchen Chemicals. 3-Mercaptopropyltrimethoxysilane (MPS) was supplied from Alfa-Aesar. Hexadecyl-trimethylammonium bromide (CTAB) was from Sinopharm Chemicals. Triethanolamine (TEA), ethylene diamine tetraacetic acid (denoted as EDTA), absolute alcohol and aqueous ammonia (28 wt%) were purchased from Tianjin Kermel Chemicals.Synthesis of the SiO2–S/NH–Ni nano-adsorbent
In a typical synthesis, 93.75 μmol of CTAB was added into 19 mL of an ethanol solution (v(CH3CH2OH)/v(H2O) = 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and stirred for a few minutes. Then, 1.1 mL of TEA was also added into the above solution. Subsequently, 1.54 mL of the mixed solution of TEOS and MPS (v/v = 10
1) and stirred for a few minutes. Then, 1.1 mL of TEA was also added into the above solution. Subsequently, 1.54 mL of the mixed solution of TEOS and MPS (v/v = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was dropped slowly into the above solution at 60 °C (28 μL s−1) and reacted for 3 h. Then, the precipitate was treated with an ethanol–HCl solution for 12 h to remove the CTAB template and was then washed with deionized water three times and dried for 48 h at 60 °C to obtain SiO2–SH NPs.
1) was dropped slowly into the above solution at 60 °C (28 μL s−1) and reacted for 3 h. Then, the precipitate was treated with an ethanol–HCl solution for 12 h to remove the CTAB template and was then washed with deionized water three times and dried for 48 h at 60 °C to obtain SiO2–SH NPs.
Then, 20 mL of an IDA solution (0.085 g mL−1, pH = 11) was added into a flask, and the solution was kept at 0 °C in an ice-bath. Then, 2 mL of GPTMS was added under stirring conditions, and the mixture was reacted at 65 °C for 12 h. Subsequently, 0.2 g of SiO2–SH NPs was dispersed in 6 mL of the GPTMS-IDA solution (pH = 2), and the suspension was sustained at 90 °C for 3 h. Finally, the suspension was centrifuged, washed and dried to get SiO2–SH/NH2 NPs. For chelating the Ni2+ ions, 5 mg of SiO2–SH/NH2 NPs was dispersed in 50 mL of a 0.2 mol L−1 NiCl2 solution and reacted at 25 °C for 24 h. Then, the SiO2–S/NH–Ni nano-adsorbent was washed by deionized water for several times and stored at 4 °C.
Affinity separation of His-tagged proteins
After being washed with a 0.02 mol L−1 Tris–HCl binding buffer three times, the SiO2–S/NH–Ni nano-adsorbent was dispersed directly into 1 mL of E. coli lysate and shaken for 2 h at a rotation speed of 70 rpm at 4 °C. Then, the SiO2–S/NH–Ni nano-adsorbent with the captured His-tagged proteins was washed with Tris–HCl buffer three times to remove any residual uncaptured proteins. Finally, the SiO2–S/NH–Ni nano-adsorbent with the captured His-tagged proteins was eluted with 0.3 mL of 0.05 mol L−1 imidazole solution. The SiO2–S/NH–Ni nano-adsorbent can be recovered and reused by washing sequentially with 0.1 mol L−1 EDTA and 0.2 mol L−1 NiCl2 solutions.Characterization
Transmission electron microscopy (TEM, JEM-2010), scanning electron microscopy (SEM, JSM 5600LV), Fourier transform infrared spectroscopy (FT-IR, AVATAR360) and thermogravimetric analysis (TG, EXSTAR 6000) were employed to detect the morphology and composition of the as-prepared nanomaterials. The surface area was measured via the Brunauer–Emmett–Teller method (BET, QUADRASORB). The separated His-tagged proteins were detected with sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE, Power PAC 300). The concentration of the target proteins was analyzed at 280 nm on a UV-Vis spectrophotometer (UV-Vis, nanodrop 2000c).Conclusions
In this study, thiol-functionalized porous silica nanoparticles were synthesized via a one-pot method. Then, the SiO2–SH NPs were further modified by iminodiacetic acid and chelating Ni2+ ions to get the porous affinity absorbent, which could then be applied to separate His-tagged proteins. The results showed that the as-prepared absorbent had good specificity and regeneration ability for use with His-tagged fusion proteins. Moreover, the SiO2–SH NP absorbent had high sensitivity to His-tagged proteins, and the detection limit for the target proteins was lower than 1.0 × 10−7 mol L−1.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support provided by the National Natural Science Foundation of China (grant no. 21571051), the Science and Technology Department of Henan Province (in the name of “Major Science and Technology Project”, grant no. 181100310600), Henan University (in the name of “Interdisciplinary Research for First-Class Discipline Construction Project”, grant no. 2019YLXKJC04), Henan University Minsheng College (in the name of “Innovative Experimental and Practical Projects for College Students”, grant no. MSCXSY2019007 and MSCXSY2019016).Notes and references
- W. Li, F. A. Zhao, W. P. Fang, D. Y. Xie and J. N. Hou, Front. Plant Sci., 2015, 6, 732–745 Search PubMed.
- L. Q. Chai, J. H. Meng, J. Gao, Y. H. Xu and X. W. Wang, Fish Shellfish Immunol., 2018, 80, 155–164 CrossRef CAS.
- J. F. Hua, S. Zhang, J. J. Cui, D. J. Wang and C. Y. Wang, J. Insect Physiol., 2013, 59, 690–696 CrossRef CAS.
- M. Y. Chen, K. Li, H. P. Li, C. P. Song and Y. C. Miao, Sci. Rep., 2017, 7, 44743–44757 CrossRef.
- S. S. Liang, L. Xiao, P. H. Huang, Z. Y. Cheng, F. L. Chen, Y. C. Miao, Y. F. Fu and Q. S. Chen, Plant Cell Rep., 2019, 38, 1263–1271 CrossRef PubMed.
- L. Li, M. J. Hou, L. Cao, Y. Xia and Z. G. Shen, Environ. Exp. Bot., 2018, 155, 313–320 CrossRef CAS.
- B. K. Oh, S. Park, J. E. Millstone, S. W. Lee, K. B. Lee and C. A. Mirkin, J. Am. Chem. Soc., 2006, 128, 11825 CrossRef CAS PubMed.
- J. H. Lee, Y. M. Huh, Y. W. Jun, J. W. Seo, J. T. Jang, H. T. Song, S. Kim, E. J. Cho, H. G. Yoon, J. S. Suh and J. Cheon, Nat. Med., 2007, 13, 95–99 CrossRef CAS PubMed.
- D. Driss, F. Bhiri, R. Ghorbel and S. E. Chaabouni, Protein Expression Purif., 2012, 83, 8–14 CrossRef CAS PubMed.
- M. Mielecki, J. Wojtasikb, M. Zborowskab, K. Kurzatkowska, K. Grzelak, W. Dehaen, J. Radecki and H. Radecka, Electrochim. Acta, 2013, 96, 147–154 CrossRef CAS.
- K. J. Lei, Y. Lin, J. Ren, L. Bai and Y. C. Miao, Plant Cell Physiol., 2016, 57, 192–203 CrossRef CAS PubMed.
- M. Perrier, M. Gary-Bobo, L. Lartigue, D. Brevet, A. Alain Morère, M. Garcia, P. Maillard, L. Raehm, Y. Guari, J. Larionova, J. O. Durand, O. Mongin and M. J. Blanchard-Desce, J. Nanopart. Res., 2013, 15, 1602–1618 CrossRef.
- P. C. Wang, Y. Y. Du, Y. J. Hou, Y. Zhao and C. C. P. Hsu, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 613–618 CrossRef CAS PubMed.
- D. R. Bae, S. J. Lee, S. W. Han, J. M. Lim, D. M. Kang and J. H. Jung, Chem. Mater., 2008, 20, 3809–3813 CrossRef CAS.
- X. Zhao, J. Wang, J. Yuan, X. L. Wang and Q. P. Zhao, New Phytol., 2015, 207, 211–224 CrossRef CAS PubMed.
- N. Li, X. Sun and M. Q. J. Wang, J. Appl. Entomol., 2017, 141, 751–757 CrossRef CAS.
- Y. B. Yin, G. M. Wei, X. Y. Zou and Y. B. Zhao, Sens. Actuators, B, 2015, 209, 5108–5511 Search PubMed.
- X. Y. Zou, K. Li, Y. B. Yin, Y. B. Zhao, Y. Zhang, B. J. Li, S. S. Yao and C. P. Song, Mater. Sci. Eng., C, 2014, 34, 468–473 CrossRef CAS PubMed.
- F. Xu, J. H. Geiger, G. L. Baker and M. L. Bruening, Langmuir, 2011, 27, 3106–3112 CrossRef CAS PubMed.
- X. Y. Zou, K. Li, Y. B. Yin, Y. B. Zhao, Y. Zhang, B. J. Li, S. S. Yao and C. P. Song, Mater. Sci. Eng., C, 2014, 34, 468–473 CrossRef CAS PubMed.
- W. J. Fang, X. L. Chen and N. F. Zheng, J. Mater. Chem., 2010, 20, 8624–8630 RSC.
- W. B. Yap, B. T. Tey, N. B. M. Alitheen and W. S. Tan, J. Chromatogr. A, 2010, 1217, 3473–3480 CrossRef CAS PubMed.
- C. J. Xu, K. M. Xu, H. W. Gu, X. F. Zhong, Z. H. Guo, R. K. Zheng, X. X. Zhang and B. Xu, J. Am. Chem. Soc., 2004, 126, 3392–3393 CrossRef CAS PubMed.
- K. S. Lee and I. S. Lee, Chem. Commun., 2008, 14, 709–711 RSC.
- B. Mizrahi, S. Irusta, M. Mckenna, C. Stefanescu, L. Yedidsion, M. Myint, R. Langer and D. S. Kohane, Adv. Mater., 2011, 23, H258–H262 CrossRef CAS PubMed.
- Y. Y. Wang, Y. D. Liu, W. H. Zhan, K. X. Zheng, J. N. Wang, C. S. Zhang and R. H. Chen, Sci. Total. Environ., 2020, 729, 139060 CrossRef CAS PubMed.
- X. Y. Zou, K. Li, Y. B. Zhao, Y. Zhang, B. J. Li and C. P. Song, J. Mater. Chem. B, 2013, 1, 5108–5113 RSC.
- Y. Y. Wang, Y. D. Liu, W. H. Zhan, K. X. Zheng, M. M. Lian, C. S. Zhang, X. L. Ruan and T. Li, Ecotoxicol. Environ. Saf., 2020, 197, 110600 CrossRef CAS.
- M. F. Jeng, A. P. Campbell, T. Begley, A. Holmgren, D. A. Case, P. E. Wright and H. J Dyson, Structure, 1994, 2, 853–868 CrossRef CAS.
- H. Zhu, B. Lee, S. Dai and S. H. Overbury, Langmuir, 2003, 19, 3974–3980 CrossRef CAS.
- S. S. Wu, X. Huang and X. Z. Du, J. Mater. Chem. B, 2015, 3, 1426–1432 RSC.
- S. Q. Liu, M. Y. Wei, J. C. Rao, H. X. Wang and H. Zhao, Mater. Lett., 2011, 65, 2083–2085 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2020 |