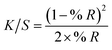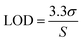Open Access Article
Open Access ArticleFabrication of a paper strip for facile and rapid detection of bovine viral diarrhea virus via signal enhancement by copper polyhedral nanoshells†
Min Woo Kim‡
 a,
Hong-Je Park‡bc,
Chan Yeong Park
a,
Hong-Je Park‡bc,
Chan Yeong Park a,
Ji Hong Kimd,
Chae Hwan Chod,
Le Minh Tu Phan
a,
Ji Hong Kimd,
Chae Hwan Chod,
Le Minh Tu Phan a,
Jong Pil Parkd,
Suresh Kumar Kailasa
a,
Jong Pil Parkd,
Suresh Kumar Kailasa e,
Chi-Ho Lee*c and
Tae Jung Park
e,
Chi-Ho Lee*c and
Tae Jung Park *a
*a
aDepartment of Chemistry, Research Institute of Chem-Bio Diagnostic Technology, Chung-Ang University, 84 Heukseok-ro, Dongjak-gu, Seoul 06974, Republic of Korea. E-mail: tjpark@cau.ac.kr
bR&D Center, Medexx Co., Ltd, 5F, #B, Korea Bio Park, 700 Daewangpangyo-ro, Bundang-gu, Seongnam-si, Gyeonggi-do 13488, Republic of Korea
cDepartment of Food Science and Biotechnology of Animal Resources, Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 05029, Republic of Korea. E-mail: leech@konkuk.ac.kr
dDepartment of Food Science and Biotechnology, Chung-Ang University, Ansung 17546, Republic of Korea
eDepartment of Applied Chemistry, Sardar Vallabhbhai National Institute of Technology, Surat-395007, Gujarat, India
First published on 12th August 2020
Abstract
The detection of bovine viral diarrhea virus (BVDV), which is a pathogen inducing fatal gastrointestinal disease in cattle, is becoming a momentous issue in the livestock farm. In that, BVDV is related to inapparent infection and various diseases with high transmissibility; it has also led to considerable economic losses. In this study, a simple dot-blotting method was devised to construct a rapid screening system for BVDV. Based on the BVDV-specific bioreceptors, it was anchored on the gold nanoparticles (AuNPs) to generate the seeding sites for signaling; then the signals were amplified by adopting the overgrowth of copper nano-polyhedral shells on AuNPs. The developed detection system shows a low detection limit of 4.4 copies per mL, and even this could be distinguished with naked eyes. These results indicate that the designed nanobiosensor possesses not only high sensitivity and selectivity but also potential usage on a point-of-care testing platform for BVDV.
1. Introduction
Bovine viral diarrhea virus (BVDV), which was first described in 1946, is one type of Pestivirus that belongs to the Flaviviridae family and causes various infectious diseases in cattle.1 As BVDV induces various acute disorders such as diarrhea, fever, respiratory disease, reproductive failure, and gastrointestinal disease with high transmissibility, there is significant financial damage for the global livestock farm.2–4 It has two recognized genotypes (BVDV-1 and BVDV-2), and each genotype is classified into cytopathic and noncytopathic biotypes. Among them, it was revealed that the noncytopathic strain of BVDVs in utero (before ∼18 weeks of gestation) is a hotbed for a large quantity of BVDV that triggers persistent infection in cattle throughout their lives.5 For these reasons, persistently infected (PI) cattle act as the primary sources for BVDV infections due to either the incapability of producing antibodies by the immune system (non-neutralizing and neutralizing) or the presence of trace levels of antibodies to BVDV.6 The amount of BVDV in the serum of PI cattle is approximately 106 cell culture infective dose per mL, and the virus is widely spread out in saliva, nasal discharge, urine, semen, and milk.7 Therefore, the development of early screening tools for BVDV that are simple, rapid, selective, and accurate is essentially required to prevent proliferation.8So far, there have been various efforts to diagnose BVDV. For example, reverse transcriptase–polymerase change reaction, insulated isothermal–polymerase change reaction, antigen-capturing enzyme-linked immunosorbent assay, microfluidic immunosensor, and the dual aptamer-based surface plasmon resonance method were researched for BVDV detection.9–14 Although these approaches exhibited high sensitivity (102 to 103 copies per mL), they still required 2–14 days for virus isolation, specific/expensive reagents, and sophisticated instruments to determine whether BVDV infections in cattle existed.
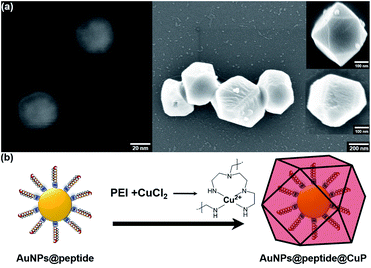
In this paper, we developed a gold nanoparticle (AuNP)-mediated dot-blot assay method to overcome these limitations. Recently, AuNPs have proven to be promising materials for colorimetric sensors, and they were widely applied in immunoassays, biochips, and aptasensors for the detection of harmful substances due to their unique optical property and high stability.15–18 The advantages of dot-blot assays are simplicity, rapidity, and not requiring skilled labor, which could make it possible to simply screen BVDV in the field.19–21 In other words, by combining this nanomaterial with the dot-blot immunoassay method, the detection performance could be highly improved.22 To enhance the optical signal reflecting the amount of BVDV in the sample, the copper polyhedral nanoshell (CuP) growth on the surface of AuNPs was introduced (Scheme 1). When Cu2+ ion–PEI–ascorbic acid (AA) is added to AuNPs that remain on the strip according to the quantity of BVDV, a CuP shell is formed via the consecutive autocatalytic-induced nucleation on the gold surface.23–25 Due to this additional step, the color changes of the strips become more clearer, and the results can easily be monitored by a smartphone camera and even naked eyes as well.
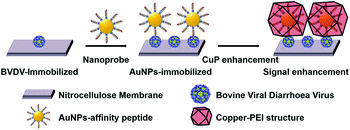 | ||
| Scheme 1 Schematic illustration for BVDV detection using affinity peptide dot-blot assay by a signal enhancement via a copper nano-polyhedral shell on the AuNPs surface. | ||
The analytical performance of the dot-blot assay was also confirmed with UV-visible (UV/Vis) reflectance spectrometry, and it was figured out that the developed dot-blot method possesses much potential to realize POCT tools for BVDV in cattle.
2. Material and methods
2.1. Chemicals
Gold (III) chloride trihydrate (HAuCl4·3H2O), polyethyleneimine (PEI, branched, MW = 800 Da), and bovine serum albumin (BSA) were purchased from Sigma Aldrich (St. Louis, MO, USA). Trisodium citrate dihydrate was obtained from BioBasic (Ontario, Canada). L-Ascorbic acid (AA) sodium salt and polyethylene glycol (PEG, MW = 8000 Da) were purchased from Alfa Aesar (Ward Hill, MA, USA). Copper chloride dihydrate (CuCl2·2H2O) was purchased from Junsei (Tokyo, Japan). Tris[hydroxymethyl]aminomethane (tris) was purchased from Bio-Rad (Hercules, CA, USA). Tween-20 was purchased from Biosesang (Seongnam, Korea). The NC membrane was obtained from Nupore (Ghaziabad, India). Deionized (DI) water at 18.2 MΩ cm was purified using a Milli-Q system (Millipore, Billerica, MA, USA). Five novel synthetic peptides were synthesized by peptron (Daejeon, Korea) and their information is listed in Table 1. All reagents were used without further purification.| Name | Sequence (N → C terminus) | MW (g mol−1) |
|---|---|---|
| BVDV #2 | CSHIASFTWYNGY | 1546.67 |
| BVDV #3 | CSVIAGHHLPWNW | 1517.73 |
| BVDV #10 | CWHYNYLTGDFVV | 1614.73 |
| BVDV #16 | CLPFMRTTPWTGF | 1554.75 |
| BVDV #NS | CKTLKTFWHHPMF | 1673.83 |
2.2. Instrumentation
The UV-vis and diffuse reflectance spectroscopy (DRS) spectra of AuNPs and CuP-modified AuNPs were measured on a UV-vis spectrophotometer (Jasco 670, Jasco, Tokyo, Japan). The zeta potential and size distribution of the AuNPs were measured using a dynamic light scattering (DLS) system (Otsuka ELSZ-1000, Otsuka Electronics, Osaka, Japan). The conjugation of BVDV peptide onto the AuNPs was confirmed from the Fourier-transform infrared (FT-IR) spectra acquired on an FT-IR apparatus (Jasco 6600FV). The X-ray diffraction (XRD) patterns were acquired on a D8-Advance instrument (Brucker AXS, Berlin, Germany). The images and size of AuNPs were recorded using field-emission scanning electron microscopy (FE-SEM) (Carl Zeiss, Cambridge, UK). ChemiDoc™ MP system (Bio-Rad, Hercules, CA, USA) and smartphone camera (Galaxy S10, Samsung Electronics, Korea) were used for digital visualization of the AuNPs and AuNPs–CuP on membranes, respectively. Subsequently, Adobe Photoshop CS6 (Adobe Systems, Mountain View, CA, USA) was used to crop these images and analyze the color intensity of the dots.2.3. Fabrication of nitrocellulose membrane with BVDV antigen-BSA-synthetic peptide modified AuNPs
2.4. Immobilization of antigen on the NC paper strips
The nitrocellulose (NC) membrane was adopted as a platform for the modification of BVDV and reaction with the AuNPs@peptides. Above all, to regulate the reaction site, the membrane was coated with a solid printing wax. To remove other impurities, the membrane was treated with TBST buffer (20 mM Tris; pH 7.5, 150 mM NaCl, 0.1% (v/v) Tween 20, filter sterilized) for 10 min and then completely dried at 37 °C. BVDV solutions of different concentrations (0, 101, 102, 103, 104, and 105 copies per mL, 0.5 μL) were added dropwise onto the uncoated sites of each membrane, and they were anchored by incubating for 30 min at 37 °C. Next, 5% (w/v) BSA solution was cast onto the BVDV-bounded sites to prevent unwanted binding while the peptide-conjugated AuNPs were reacting with BVDV. After incubation for 60 min, it was washed with TBST buffer thrice.2.5. Dot-blot assay using AuNPs
Onto the various concentration of BVDV-immobilized NC membranes, 4 mL of AuNPs@peptide conjugated with different bioreceptors was added and incubated for 1 h with mild shaking. After the reaction, the unbounded AuNPs@peptide was discarded, and then the membranes were thoroughly washed thrice with DI water.2.6. CuP-mediated signal enhancement process
In order to amplify the color of the dot on the NC membrane, Cu2+ ion, PEI and AA were introduced on the surface of the BVDV conjugated NC membrane. Specifically, 4 mL of 0.1 M CuCl2 and 4 mL of 0.5% (w/v) PEI were applied to the membrane, and the color of the membrane gradually turned from colorless to deep-red. After the pre-reaction, 20 mL of 0.25 M AA sodium salt was mixed with the above solution for 10 min, and the color changed from light blue to deep navy (AuNPs@peptide@CuP). Finally, the modified NC membrane strips were washed with DI water and dried. The color of the NC membranes was measured by ChemiDoc™ MP and a smartphone camera and analyzed using UV-vis and DRS. The numerical color intensity of the dots was obtained using Adobe Photoshop CS6.2.7. Selectivity test
Solutions of several viruses (BVDV, norovirus, rotavirus, and dengue virus) were prepared at 104 copies per mL and bound on the NCs, as described in Section 2.4. Various viruses were tested with four milliliters of the peptide-conjugated AuNPs in the presence of Cu2+ ion–PEI–AA as per the above described procedure. After the reaction, the membranes were washed thoroughly thrice with TBST.2.8. Calculation of evaluation test
To calculate the isotropic absorbance (averaged over all directions) from the UV-vis reflectance value, we used the ‘Kubelka–Munk’ transformation equation. As this equation convert % R, which is inversely proportional to the concentration, to a curve proportional, it makes the results intuitive and easy to comprehend.28In the above equation, % R is the UV-vis reflectance value, and the K/S value is composed of absorption (K) and scattering (S) characteristics of the surface environment of the membrane. When applying the DRS results from different concentrations of BVDV to the above equation, the K/S value increased gradually according to the initially treated amount of BVDV on the membrane. The limit of detection (LOD) for the BVDV-spiked samples was calculated by the 3σ-rule.29
where σ is the standard deviation at blank (n = 3), and S is the slope of the calibration curve. Based on the equation, the performance of the candidates for BVDV-specific peptides was determined.
3. Results and discussion
3.1. Characterization and confirmation of novel peptide modified AuNPs and the growth of CuP
The size, morphology, and zeta potential of AuNPs are shown in Fig. S1.† The FE-SEM images indicate that the AuNPs are well dispersed and their size is 40 nm (Fig. S1a†). The measured zeta potential of AuNPs was −49.10 mV owing to the assembled citrates on the particles, and it contributed to the high dispersity and uniform size of AuNPs (Fig. S1b and c†). To verify the conjugation between the synthetic peptides and the AuNPs, UV-visible spectroscopy and zeta potential analysis were performed (Fig. S2†). After immobilization, the absorbance band of AuNPs was slightly red-shifted due to the change in the surface plasmon resonance (SPR), and the surface charge moved toward a less negative value due to the attached peptides. These results show that the BVDV-specific peptide is well-conjugated on the AuNPs. The formation of CuP on the novel synthetic peptides conjugated AuNPs–NC membrane was confirmed by using FE-SEM (Fig. S3†). As the PEI molecules possessing a significant number of amine groups chelate with the metal ion and form complexes with Cu2+, the chelated complexes decrease the free ion concentration and stabilize the chemical potential of the reactants. As a result, Cu2+ ion was reduced to Cu0 on the surfaces of AuNPs@peptide in the presence of AA. Furthermore, it can be noticed that the formation of CuP enlarges the size of AuNPs via self-nucleation and favours the growth of the CuP complex on the surface of the AuNPs (Fig. 1). To verify the signal enhancement by CuP, it was compared to that of the LOD values between AuNPs@peptide and AuNPs@peptide@CuP, respectively (Fig. S4†). As a result, this additional procedure played a pivotal role in not only dramatically elevating the signal intensity but also lowering the LOD about 1500 times. Moreover, the dynamic range and reliability of detection are highly improved.To validate the conjugation of BVDV peptides on AuNPs and the growth of CuP on the AuNP surface, their FT-IR spectra were recorded and compared (Fig. S5a†). Commonly, whole samples represent the stretching vibration of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of citrates at 1617 cm−1. For the AuNPs@peptide sample, this peak is slightly shifted and new peaks were noticed at 954, 1022, 1317, 1410 and 1650 cm−1 due to the stretching vibrations of C
O group of citrates at 1617 cm−1. For the AuNPs@peptide sample, this peak is slightly shifted and new peaks were noticed at 954, 1022, 1317, 1410 and 1650 cm−1 due to the stretching vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of alkene, C–N of histidine, tryptophan, and proline, and N–H groups of peptides, respectively, and it confirmed that the AuNPs were successfully modified with BVDV affinity peptide. However, these phenomena greatly diminished or shifted after growing CuP onto the AuNP surfaces. Their differences can also be seen in UV-vis spectra (Fig. S5b†). The SPR band of AuNPs was present at 521 nm, whereas that of AuNPs@peptide occurred at 527 nm. However, the larg red-shift occurred due to the modification of AuNPs with BVDV affinity peptide without PEI.
C bending of alkene, C–N of histidine, tryptophan, and proline, and N–H groups of peptides, respectively, and it confirmed that the AuNPs were successfully modified with BVDV affinity peptide. However, these phenomena greatly diminished or shifted after growing CuP onto the AuNP surfaces. Their differences can also be seen in UV-vis spectra (Fig. S5b†). The SPR band of AuNPs was present at 521 nm, whereas that of AuNPs@peptide occurred at 527 nm. However, the larg red-shift occurred due to the modification of AuNPs with BVDV affinity peptide without PEI.
In the case of AuNPs@peptide@CuP, large Mie light scattering-dominant interactions were observed because the property of SPR is closely related to the size and spatial orientation of nanoparticles (Fig. S5b†).30
The formation of CuP on AuNPs was characterized by XRD (Fig. S6†). The XRD peaks of AuNPs were exhibited at (111), (200), (220) and (311) with the relative intensities of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4
0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2
0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.15, respectively. The XRD data strongly revealed that the fabricated AuNPs are in uniform size and shape.31 The XRD peaks of AuNPs@peptide@CuP appeared at (111), (200), and (220), and it reflects crystallized Cu, not copper oxides.32 The XRD peak of AuNPs did not appear after the formation of CuP on AuNPs as it was surrounded by thick CuP shells (∼400 nm). Following the above results, each pattern matched well with the library data and indicated that there were no impurities.
0.15, respectively. The XRD data strongly revealed that the fabricated AuNPs are in uniform size and shape.31 The XRD peaks of AuNPs@peptide@CuP appeared at (111), (200), and (220), and it reflects crystallized Cu, not copper oxides.32 The XRD peak of AuNPs did not appear after the formation of CuP on AuNPs as it was surrounded by thick CuP shells (∼400 nm). Following the above results, each pattern matched well with the library data and indicated that there were no impurities.
3.2. Preparation and optimization of the BVDV detection system (BVDV immobilization/growth of CuP)
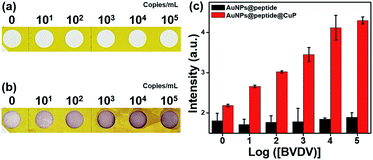
The optimization of the immunoreaction time between BVDV and AuNPs@peptide was investigated at different reaction times (10–120 min) to enhance the signalling effect. The effective immobilization time was studied under the fixed amount of BVDV (104 copies per mL) immobilized NC membrane. As shown in Fig. S7,† the signal of AuNPs steadily increased along with the conjugation time up to 60 min and then decreased slightly. Based on these observations, 60 min was chosen as the optimum reaction time for BVDV and AuNPs@peptide reaction. Moreover, since the function of CuP is a key point of the signal enhancement and assists detecting ultra-trace levels of BVDV, the influence of the reaction time (from 1 to 20 min) for the growth of CuP was investigated (Fig. S8†). According to the reaction time, the optical intensity increased until 10 min and saturated, thus, that time was selected. By measuring UV-vis DRS of the signal intensity, the appearance of deep-red color was directly proportional to the concentration of the initially immobilized amount of BVDV onto the NC membrane (Fig. S9†). Through this series of optimization experiments, the detection performance for BVDV was further enhanced (Fig. 2).3.3. Validation of the peptide candidates through optical performance for BVDV
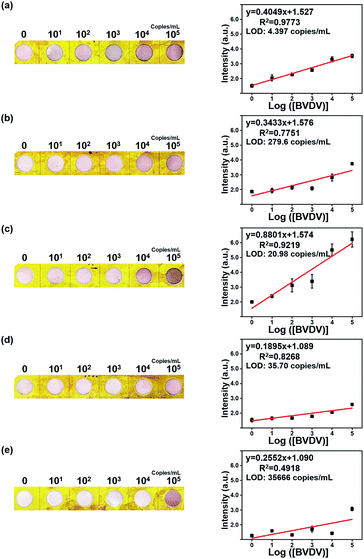
To define the analytical efficiency and detection limit of the five different BVDV-specific peptides (BVDV #2, #3, #10, #16, and #NS), a series of BVDV samples (0, 101, 102, 103, 104 and 105 copies per mL) were quantified by the developed dot-blot assay under the optimized conditions. As shown in Fig. 3, the dot color of the modified NC membrane was changed to deep-red, and the color intensity is proportional to the concentration of BVDV. To authenticate the analytical efficiency of the present dot-blot assay, its reflectance value of the dots of the NC membrane was recorded via DRS and then converted using the ‘Kubelka–Munk’ equation.33,34 As shown in Fig. S9,† the dot-blot intensity of the NC membrane at 550 nm gradually decreased with an increase in the concentration of BVDV, and the intensity of dots at 550 nm was inversely proportional to the concentration of BVDV. Based on these results, the corresponding regression correlation coefficient (R2) values and the regression curve of each peptide were obtained and are listed in Table S1.† Among the five BVDV affinity peptides, BVDV #2 peptide–AuNPs modified NC membrane exhibited higher sensitivity than others. The analytical performance of the present method was compared with those of the traditional molecular- and immuno-diagnostic methods for BVDV (Table S2†). When compared with the conventional detection methods such as RT-PCR and ELISA, this dot-blot assay presented high sensitivity with a long linear range.3.4. Evaluation of selectivity of the novel peptide for BVDV
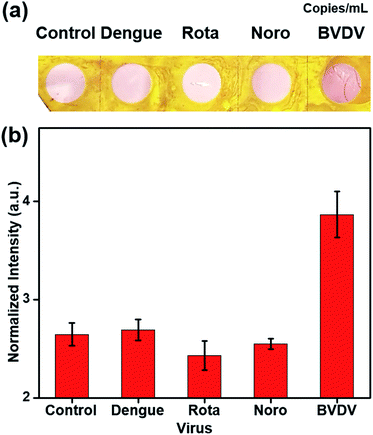
The selectivity was evaluated to determine whether the five novel synthetic peptides have affinity towards only BVDV, and not other viruses (dengue virus, rotavirus, and norovirus). These selectivity tests were performed using the same process as those of peptide BVDV #2 and the virus (104 copies per mL, and 0 copies per mL as a negative control) to confirm the affinity peptide performance (Fig. 4). There were no color changes observed when BVDV affinity peptide–AuNPs–NCs were treated with dengue, rota, and norovirus in the presence of Cu2+ ion–PEI–AA. These results show that the affinity peptides exhibited high selectivity to detect BVDV and enabled the development of a sensitive and portable detection method.4. Conclusions
In this study, a dot-blot method was developed for the rapid detection of BVDV by adopting novel BVDV recognizing peptides and the formation of a CuP nanoshell on the surface of AuNPs. The peptide-based optical biosensor provided a distinctive performance in detecting the target, and the CuP nanoshell served as the quantitative diagnosis of the virus by emphasizing the appearance of the pink dot that was visible to the naked eye. Except for NS, the peptide candidates showed high detecting performance. BVDV #2 shows the highest detection limit of up to 4.397 copies per mL. The development of the affinity peptide-based method with the signal enhancing process can make it possible to detect and diagnose the virus, even with a small amount of the virus sample. Consequently, this viral diagnosis system can be utilized for developing a POCT system and can contribute to the prevention of virus infections in cattle.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Animal Disease Management Technology Development Program, Ministry of Agriculture, Food and Rural Affairs (120090-2) and by the Chung-Ang University Research Scholarship Grants in 2019-2020.References
- P. Olafson, Cornell Vet., 1946, 36, 205–213 CAS.
- C. Tidona and G. Darai, The Springer index of viruses, Springer Science & Business Media, 2011 Search PubMed.
- R. W. Fulton, B. Hessman, B. J. Johnson, J. F. Ridpath, J. T. Saliki, L. J. Burge, D. Sjeklocha, A. W. Confer, R. A. Funk and M. E. Payton, J. Am. Vet. Med. Assoc., 2006, 228, 578–584 CrossRef.
- R. A. Palomares, D. J. Hurley, A. R. Woolums, J. E. Parrish and K. V. Brock, Comp. Immunol. Microb., 2014, 37, 331–338 CrossRef PubMed.
- R. E. Werdin, T. R. Ames, S. M. Goyal and G. P. DeVries, J. Vet. Diagn. Invest., 1989, 1, 57–61 CrossRef CAS.
- S. R. Lanyon, F. I. Hill, M. P. Reichel and J. Brownlie, Vet. J., 2014, 199, 201–209 CrossRef PubMed.
- H. Houe, Vet. Clin. North Am.: Large Anim. Pract., 1995, 11, 521–547 CAS.
- J. T. Saliki and E. J. Dubovi, Vet. Clin. North Am.: Large Anim. Pract., 2004, 20, 69–83 Search PubMed.
- O. Lung, J. Pasick, M. Fisher, C. Buchanan, A. Erickson and A. Ambagala, Transboundary Emerging Dis., 2016, 63, e395–e402 CrossRef CAS PubMed.
- B. C. Heinze, J.-Y. Song, C.-H. Lee, A. Najam and J.-Y. Yoon, Sens. Actuators, B, 2009, 138, 491–496 CrossRef CAS.
- X. Zhang, T. Diraviyam, X. Li, G. Yao and A. Michael, Biosci. Biotechnol. Biochem., 2016, 80, 2467–2472 CrossRef CAS PubMed.
- J.-W. Park, S. J. Lee, E.-J. Choi, J. Kim, J.-Y. Song and M. B. Gu, Biosens. Bioelectron., 2014, 51, 324–329 CrossRef CAS PubMed.
- X. Mi, E. M. Lucier, D. G. Turpeinen, E. L. L. Yeo, J. C. Y. Kah and C. L. Heldt, Analyst, 2019, 144, 5486–5496 RSC.
- E. J. Dubovi, Biologicals, 2013, 41, 8–13 CrossRef PubMed.
- S. K. Kailasa, J. R. Koduru, M. L. Desai, T. J. Park, R. K. Singhal and H. Basu, TrAC, Trends Anal. Chem., 2018, 105, 106–120 CrossRef CAS.
- F. Di Nardo, S. Cavalera, C. Baggiani, C. Giovannoli and L. Anfossi, ACS Appl. Mater. Interfaces, 2019, 11, 32758–32768 CrossRef CAS.
- Z.-M. Lyu, X.-L. Zhou, X.-N. Wang, P. Li, L. Xu and E.-H. Liu, Sens. Actuators, B, 2019, 284, 437–443 CrossRef CAS.
- M. Solra, R. Bala, N. Wangoo, G. K. Soni, M. Kumar and R. K. Sharma, Chem. Commun., 2020, 56, 289–292 RSC.
- K. M. Raval, V. Ghormade, P. Rajamohanan, H. Choudhary, S. M. Rudramurthy, A. Chakrabarti and K. Paknikar, J. Med. Microbiol., 2019, 68, 1341–1352 CrossRef CAS PubMed.
- S. H. Win, P. Wongchitrat, S. Kladsomboon, P. A. Dharmasaroja and A. Apilux, Sci. Asia, 2019, 45, 260–267 CAS.
- Q.-Q. Zhuang, H.-H. Deng, S.-B. He, H.-P. Peng, Z. Lin, X.-H. Xia and W. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 31729–31734 CrossRef CAS PubMed.
- H.-A. Joung, Z. S. Ballard, A. Ma, D. K. Tseng, H. Teshome, S. Burakowski, O. B. Garner, D. Di Carlo and A. Ozcan, Lab Chip, 2019, 19, 1027–1034 RSC.
- N. R. Jana, L. Gearheart and C. J. Murphy, Chem. Mater., 2001, 13, 2313–2322 CrossRef CAS.
- A. Tao, P. Sinsermsuksakul and P. Yang, Angew. Chem., Int. Ed., 2006, 45, 4597–4601 CrossRef CAS PubMed.
- T. M. D. Dang, T. T. T. Le, E. Fribourg-Blanc and M. C. Dang, Adv. Nat. Sci. Nanosci., 2011, 2, 015009 CrossRef.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- S. H. Baek, M. W. Kim, C. Y. Park, C.-S. Choi, S. K. Kailasa, J. P. Park and T. J. Park, Biosens. Bioelectron., 2019, 123, 223–229 CrossRef CAS.
- P. Kubelka, Z. Phys., 1931, 12, 593–601 Search PubMed.
- A. Shrivastava and V. B. Gupta, Chron. Young Sci., 2011, 2, 21 CrossRef.
- J. H. Kim, J. E. Park, M. Lin, S. Kim, G. H. Kim, S. Park, G. Ko and J. M. Nam, Adv. Mater., 2017, 29, 1702945 CrossRef PubMed.
- D. Seo, J. C. Park and H. Song, J. Am. Chem. Soc., 2006, 128, 14863–14870 CrossRef CAS PubMed.
- D. Mott, J. Galkowski, L. Wang, J. Luo and C.-J. Zhong, Langmuir, 2007, 23, 5740–5745 CrossRef CAS PubMed.
- B. Judd Deane and W. Gunter, Color in Business, Science and Industry, John Wiley & Sons, New York, 1975 Search PubMed.
- P. Kubelka, F. Munk and Z. Tech, J. Opt. Soc. Am. A, 1948, 38, 8 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental optimized data. See DOI: 10.1039/d0ra03677c |
| ‡ Co-first authors and contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |