Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeting G-quadruplex DNA with synthetic dendritic peptide: modulation of the proliferation of human cancer cells†
Soumi Biswasa,
Satyabrata Samuia,
Apurba K. Das b,
Sanjeev Pasadic,
K. Muniyappac and
Jishu Naskar
b,
Sanjeev Pasadic,
K. Muniyappac and
Jishu Naskar *a
*a
aDepartment of Biochemistry and Biophysics, University of Kalyani, Nadia, WB 741235, India. E-mail: jishunaskar@gmail.com
bDepartment of Chemistry, Indian Institute of Technology Indore, Indore 453552, India
cDepartment of Biochemistry, Indian Institute of Science, Bangalore, Karnataka 560 012, India
First published on 14th July 2020
Abstract
Telomerase, a reverse transcriptase enzyme, is found to over express in most cancer cells. It elongates the telomere region by repeated adding of TTAGGG in the 3′-end and leads to excess cell proliferation which causes cancer. G-quadruplex (G4) formation can inhibit such telomere lengthening. So, stabilization of G4 structure as well as inhibition of telomerase activity is very promising approach in targeted cancer therapy. Herein, the aptitude of a synthetic dendritic peptide, Cδ2–(YEE)–E (peptide 1), to target specifically the human telomeric G4 DNA, dAGGG(TTAGGG)3, has been evaluated. Both biochemical and biophysical techniques including gel mobility shift assay, isothermal titration calorimetry and fluorescence spectroscopy have been employed for the purpose. Circular dichroism study reveals that the targeting results an increase in thermal stability of G4 DNA. Interestingly, replacement of N-terminal tyrosine residue of peptide 1 by valine, Cδ2–(VEE)–E, (peptide 2) consequences in loss of its G4 DNA targeting ability, although both the peptides exhibit comparable affinity toward double-stranded DNA. Of note, peptide 1 causes cessation of growth of human cancer cells (HeLa and U2OS) and induces apoptosis in vitro. But it has no significant inhibitory effect on the growth of normal human embryonic kidney 293 cells. Mechanistically, Telomeric Repeat Amplification Protocol (TRAP) assay indicates that peptide 1 effectively inhibits the telomerase activity in human cell extracts. Overall, this study demonstrates the usefulness of a synthetic dendritic peptide as an inhibitor of tumor cell growth by inducing apoptosis upon targeting the telomeric G4 DNA.
Introduction
The G-quadruplexes (G4) are non-canonical DNA/RNA structures that play important roles in DNA replication,1a recombination,1b transcriptional regulation,1c maintenance of genomic stability1d and aging.1e The G4 forming sequences occur throughout the human genome, but they are most prevalent in the telomeres,2a immunoglobulin switch regions2b and in promoters of proto-oncogenes.2c The telomerase activity is suppressed in most human somatic cells, except in stem cells and lymphocytes,3a but it becomes up-regulated in most tumor cells.3b The unfolded single-stranded DNA is required for optimal telomerase activity; whereas, G-quartet formation inhibits the telomerase activity.4 For this reason, G4 structures are considered to be the potential biomedical targets to impede cancer progression. Consequently, an escalating interest has been grown with the development of G4 binding small molecules for therapeutic purpose.5a,b In recent years, various small molecules targeting G4 structure have been designed, synthesized and evaluated for their antitumor activities against various human cancer cell lines.6a–d The most interesting synthetic molecules as the telomerase inhibitor and telomeric G4 DNA binding ligand belong to the class of planar, substituted aromatic compounds. Some examples of such substituted aromatic compounds are isoquinoline alkaloids (berberine, palmatine, coralyne and sanguinarine),7a quarfloxine,7b RHPS4,7c Phen-DC3,7d BRACO-19,7e tetra-substituted naphthalene diimides,7f tri-substituted indolo[3,2-b]quinolines,7g and carbazole based compounds.7h–j Various metal complexes8a–g have been found to stabilize the G4 structure. Among these, it is found that the square planar metal-complex serves as the optimal candidates for stabilization of G4 DNA. For example, Pt(II) squares are selective and effective human telomeric G4 DNA binders and potential cancer therapeutics.8g Several small molecules, including porphyrin,9a carbocynamide dyes,9b and piperine9c are reported to bind with the quadruplex structures. Naturally occurring molecules derived from vegetables, nuts, wine, bacteria etc. also can serve as G4 stabilizing ligand.10In this study, we explore the ability of a synthetic dendritic peptide as ligand to stabilize the G4 DNA structure. Peptides are comparatively less explored toward this end. Peptides have several advantages over proteins, antibodies and other small molecules due to their small size, ease of synthesis and purification, cell permeability, tumor penetrating ability and improved biocompatibility.11a,b Previous studies have demonstrated that several dodecameric peptides stabilize the G4 structure derived from c-MYC promoter and also induce apoptosis in the cancer cells.12a Selective recognition of human telomeric G4 DNA by designed peptides composed of glutamic acid and tryptophan residues has been reported recently.12b In literature, there are various reports of cyclic peptides as well which have significant potential to target the G4 structure.12c,d Here, we demonstrate the ability of a synthetic dendritic peptide, Cδ2–(YEE)–E, to stabilize the human telomeric G4 DNA structure and also address the potential to inhibit the proliferation of human cancer cells in vitro.
Results and discussions
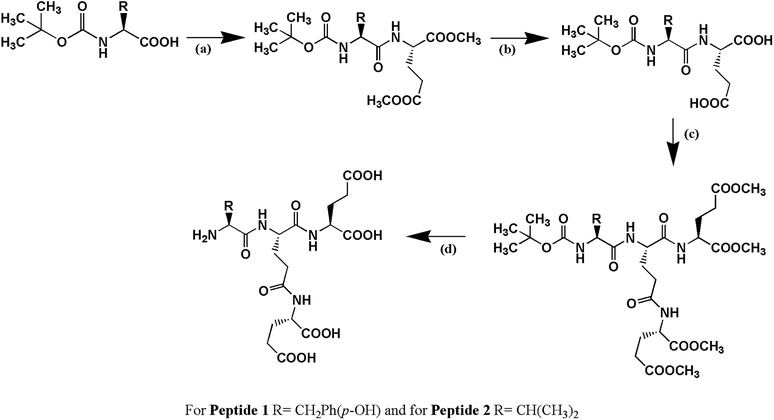
Peptide 1, Cδ2–(YEE)–E, has been designed in such a way that it holds two factors (a) hydrogen bonding interaction and (b) π–π interaction simultaneously which play very important role in peptide–DNA interaction. Peptide 2, Cδ2–(VEE)–E, has been designed to address whether only hydrogen bonding interaction is enough to stabilize the G4 structure. Both the peptides have been synthesized by solution phase racemization free fragment condensation strategy and final compounds have been characterized by NMR spectroscopy (1H and 13C) and mass spectrometry (Scheme 1).Electrophoretic gel mobility shift assay
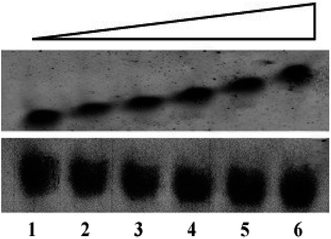
Electrophoretic mobility shift assay (EMSA) has been performed to anticipate the interaction of peptides with G4 DNA. The assay has been carried out by incubating 30 μM of G4 DNA with increasing concentrations (0, 15, 30, 45, 60 and 120 μM) of peptides for 30 min at room temperature and the DNA binding ability of the peptides is assessed by shift (retardation) in the electrophoretic migration of G4 DNA. As shown in Fig. 1 (upper panel), addition of increasing concentrations of peptide 1 led to the significant reduction in electrophoretic migration of G4 DNA (lane 2–6) compared to unbound DNA (lane 1). On the other hand, no significant reduction in the electrophoretic migration of G4 DNA is observed in the presence of peptide 2 (Fig. 1, lower panel). Thus the EMS assay indicates that the reduction in electrophoretic migration of G4 DNA in presence of peptide 1 occurs probably due to the binding interaction of peptide 1 with G4 DNA.Isothermal titration calorimetry study
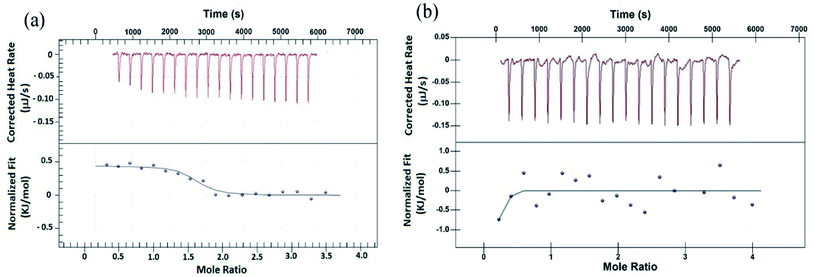
To validate the results from EMS assay, binding of peptides to G4 DNA has been studied by isothermal titration calorimetry (ITC). ITC experiment has been carried out keeping G4 DNA (64 μM) into the cell and peptide (700 μM) into the syringe. Fig. 2 shows the characteristic ITC profiles for peptide–G4 DNA interaction. The ITC titration data of peptide 1 (Fig. 2a) yields the association constant (Ka) 1.10 × 106 M−1, enthalpy change (ΔH) 0.443 kJ M−1 and entropy change (ΔS) 0.1168 kJ M−1 K−1. Interestingly, under similar condition peptide 2 shows no characteristic G4 DNA binding signature (Fig. 2b). Further, to check whether these peptides bind to double stranded DNA (ds-DNA), ITC measurements have been carried out with ds26, a model ds-DNA (5′CAATCGGATCGAATTCGATCCGATTG3′). From the results, the association constants (Ka) of peptide 1 and 2 with ds26 are found to be 9.60 × 103 M−1 and 3.36 × 103 M−1 respectively (Fig. S9†).Fluorescence spectroscopy
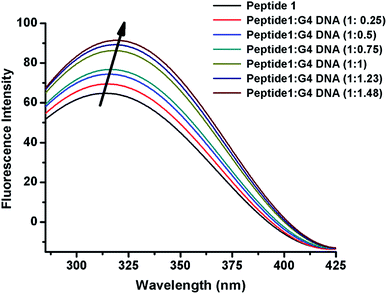
The interaction of peptide 1 with G4 DNA has also been monitored by fluorescence spectroscopy. Peptide 1 shows the characteristic emission band at 305 nm upon excitation at 275 nm. From Fig. 3 and S12,† it is found that the intensity of the emission band increases upon progressive addition of G4 DNA, indicating the binding interaction of peptide 1 with G4 DNA. Here it is important to note that, unlike peptide 1, peptide 2 lacks intrinsic fluorescence property, which prevents us to perform similar study with it.Time correlated single photon counting (TCSPC)
The TCSPC experiment has been carried out to access the insight about the interaction behavior of peptide 1 with G4 DNA. The experiment has been done keeping the peptide 1 concentration fixed at 5.5 μM and G4 DNA concentration is varied from 0 to 10.5 μM. From the experiment, it is found that the decay pattern of peptide 1 is tri-exponential and the average life time (τavg) is 4.93 ns. However, upon progressive addition of G4 DNA up to 10.5 μM, τavg value of peptide 1 decreases slowly from 4.93 to 3.09 ns (Fig. 4 and Table 1). From this result, it can be concluded that the decrease in τavg is attributed to the change in micro-environment of tyrosine residue of peptide 1 upon complexation with G4 DNA.13a,b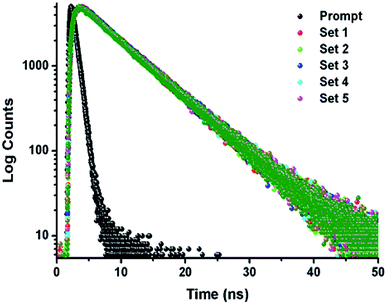 | ||
| Fig. 4 Time-resolved fluorescence spectra of peptide 1 in the absence and presence of increasing concentrations of G4 DNA in buffer (10 mM Tris and 100 mM NaCl) of pH = 7.3 at 25 °C. | ||
| Sample | [G4 DNA] (μM) | [Peptide] (μM) | χ2 | τ1 | τ2 | τ3 | q1 | q2 | q3 | Average life time τavg (ns) |
|---|---|---|---|---|---|---|---|---|---|---|
| Set-1 | 0 | 5.5 | 1.033 | 3.11 × 10−9 | 6.6 × 10−9 | 3.1 × 10−10 | 0.12 | 0.68 | 0.20 | 4.93 |
| Set-2 | 2.5 | 1.004 | 3.08 × 10−9 | 6.61 × 10−9 | 3.0 × 10−10 | 0.12 | 0.67 | 0.21 | 4.86 | |
| Set-3 | 4.5 | 0.995 | 2.24 × 10−9 | 6.5 × 10−9 | 1.5 × 10−10 | 0.08 | 0.59 | 0.33 | 4.07 | |
| Set-4 | 8.5 | 1.006 | 1.80 × 10−9 | 6.54 × 10−9 | 8.1 × 10−11 | 0.08 | 0.53 | 0.55 | 3.65 | |
| Set-5 | 10.5 | 1.014 | 1.89 × 10−9 | 6.54 × 10−9 | 9.7 × 10−11 | 0.06 | 0.45 | 0.49 | 3.09 |
Thiazole orange (TO) displacement assay
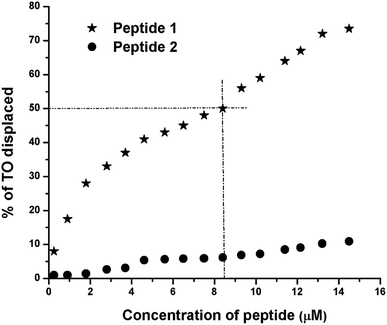
The TO displacement assay is extensively used to study G4-binding properties of specific ligands. Into aqueous buffer, TO exhibits week fluorescence property. But upon binding to G4 DNA, TO–G4 DNA complex exhibits a significant increase in fluorescence intensity compared to the free TO.14 Thus, the affinity of a given ligand toward G4 DNA can be assessed by measuring its ability to displace TO from the TO–G4 DNA complex. From the results of TO displacement assay, it is found that ∼8.4 μM peptide 1 is able to displace ∼50% TO from the TO–G4 DNA complex (Fig. 5). In contrast, an identical concentration of peptide 2 could displace only ∼6.5% TO, suggesting that peptide 1 has much higher binding affinity than peptide 2 toward G4 DNA.Circular dichroism (CD) spectroscopy
CD spectroscopy is a very sensitive technique to investigate ligand induced conformational changes of nucleic acids. In aqueous buffer containing Na+ ions, human telomeric dAGGG(TTAGGG)3 DNA adopts anti-parallel G4 structure which shows a strong positive band at 295 nm as well as a negative band at 260 nm followed by a shoulder at 245 nm.15 In presence of increasing concentrations of peptides 1 and 2 (Fig. S10†), the intensity of the positive band (at 295 nm) decreases whereas the intensity of the negative band (at 260 nm) increases. These changes indicate that the conformation of the G4 DNA gets perturbed subsequent to the binding of both peptides.CD melting studies
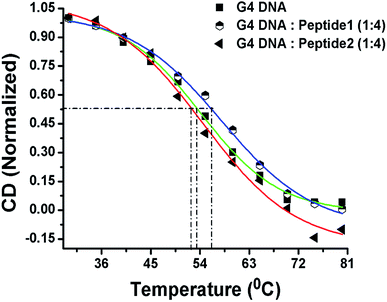
Now, in order to compare the stability of human telomeric G4 DNA in the absence and presence of peptides, thermal melting study of G4 DNA is performed using CD spectroscopy. The melting curve is constructed by plotting the molar ellipticity (at 295 nm) against temperature.16 The melting profile of G4 DNA in the presence of peptide 1 reveals (Fig. 6) an increase in the melting temperature (Tm) of ∼3 °C compared to free G4 DNA. On the other hand, the Tm value is found to decrease ∼1 °C in the presence of peptide 2. Thus, CD melting studies clearly indicate that the peptide 1 increases the thermal stability of G4 DNA but peptide 2 doesn't.Peptide 1 inhibits cellular telomerase
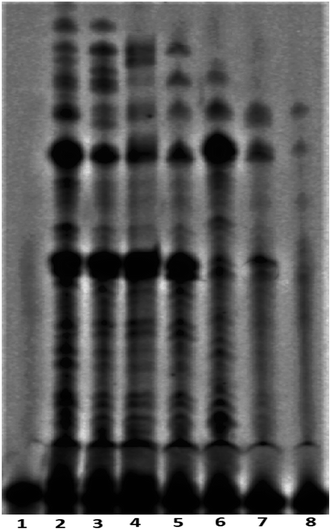
The telomerase activity is found to be up-regulated (nearly 90%) in cancer cells compared to normal differentiated somatic cells.17 To gain a better understanding of the mechanism of action exerted by peptide 1, the mode of telomerase inhibition has been assayed using PCR-based Telomeric Repeat Amplification Protocol (TRAP) assay with HeLa nuclear extract.8a,b The TRAP assay has been performed using the concentrations of peptide 1 ranging from 0.25 to 2.0 mM. As shown in Fig. 7, the increasing concentrations of peptide 1 (lane 3–8) inhibit the telomerase activity in a dose-dependent manner in vitro.Cytotoxic effect of peptide 1 on cancer and non-cancer cells
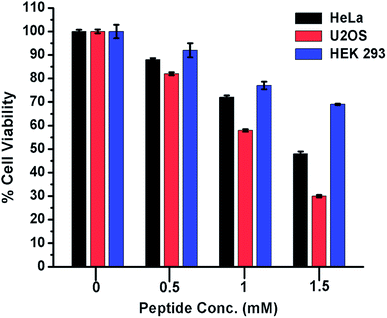
From the foregoing results, it is evident that peptide 1 confers the thermodynamic stability to G4 DNA. The ligands, those stabilize the G4 DNA display the anticancer property.5,18 Therefore, to check whether peptide 1 could modulate the proliferation of cancer cells, HeLa and U2OS cells have been selected for the cytotoxicity analysis in vitro. The human embryonic kidney (HEK 293) cell line served as a negative control. The assay has been performed by treating the cells with various concentrations of peptide 1 (0, 0.5, 1.0 and 1.5 mM) for 72 h. As the result, it is found that peptide 1 exhibits much higher cytotoxicity on cancer cells compared to non-cancer cell under the similar conditions (Fig. 8). The IC50 values are1.48 mM and 1.1 mM respectively for HeLa and U2OS cells. Interestingly, at 1.5 mM concentration of peptide 1, ∼70% of HEK 293 cells are found to be viable. Further, to examine the effect of peptide 1 on cancer and non-cancer cells, phase contrast microscopic study has been performed. The study reveals that the effect of peptide 1 to inhibit the proliferation of HeLa cells and subsequent cell death is much higher compared to HEK 293 cells (Fig. S11†). These results suggest that peptide 1 exerts significant cytotoxic effect on cancer cells, but has very little effect on the non-cancer cells.Long-term cell viability assay
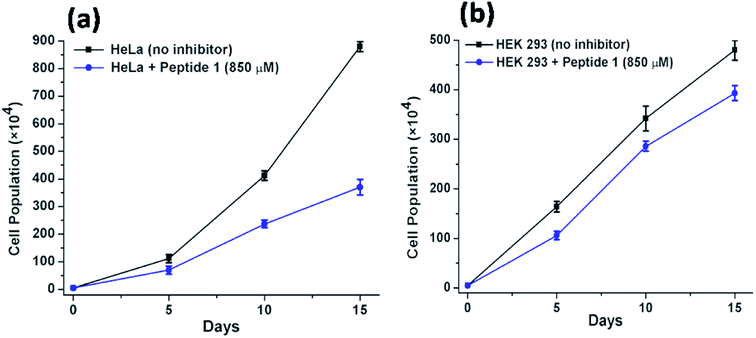
Further to investigate the cytotoxic and growth inhibitory effect of a sub-inhibitory dose of peptide 1, the cell proliferation assay has been performed over a longer period of time. For this purpose, the HeLa and HEK 293 cells have been treated with 850 μM of peptide 1 for 15 days. Upon treatment, it is found that (Fig. 9) the proliferation of HeLa cells lessens from (880 ± 18) × 104 to (370 ± 28) × 104. In contrast, the proliferation of HEK 293 cells under similar condition is found to lessen from (480 ± 20) × 104 to (393 ± 15) × 104. Thus, the studies clearly establish that the inhibition efficiency of peptide 1 on cancer cells is significantly higher than that in the normal cells regardless of dose and time of treatment.Study of mechanism of cell death
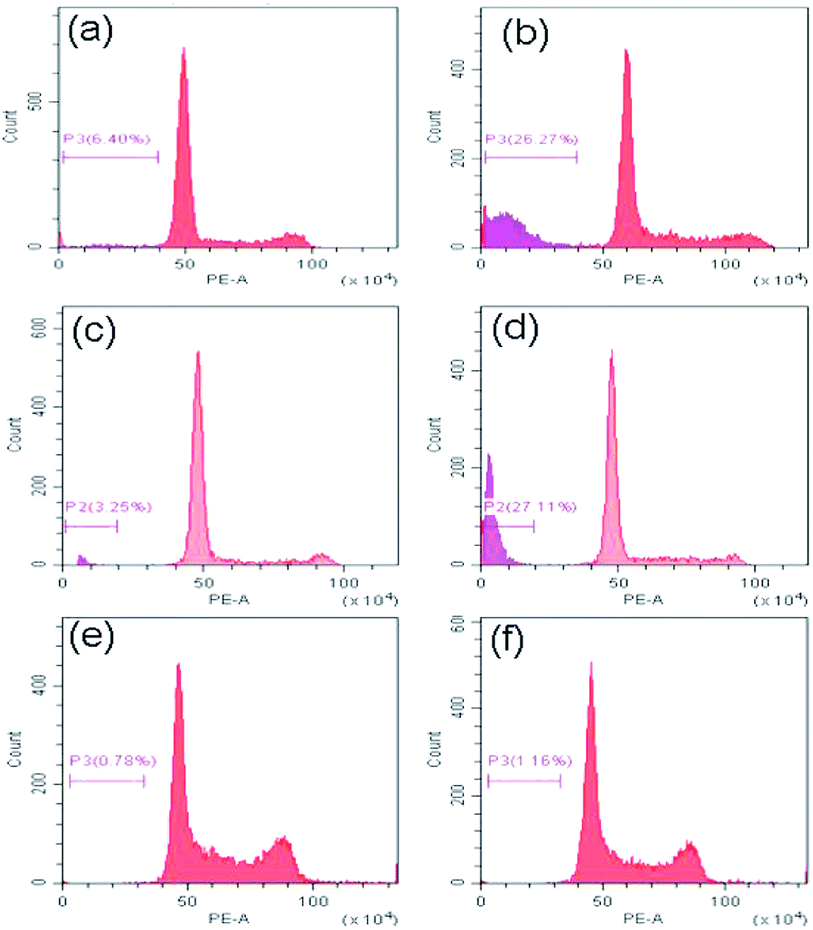
Now to ascertain, whether peptide 1 induced cell death is due to apoptosis, the cell cycle analysis has been performed by staining HeLa, U2OS and HEK 293 cells with propidium iodide. The results (Fig. 10) show that the sub-G1 population in the treated cells increases significantly in case of HeLa and U2OS with respect to untreated control which clearly indicates the cell death is through apoptosis.Conclusion
In this study, we describe the interaction of two synthetic dendritic peptides with human telomeric G4 DNA. The results establish that the peptide 1, Cδ2–(YEE)–E, containing tyrosine as the N-terminal residue binds robustly to G4 DNA. The binding affinity is drastically reduced in peptide 2, Cδ2–(VEE)–E where N-terminal tyrosine residue has been replaced by valine. CD measurements reveal that binding of peptide 1 increases the thermal stability of G4 DNA. Importantly, peptide 1 exhibits significant telomerase inhibition property along with profound cytotoxic effect selectively on cancer cells compared to non-cancer cells. These results underscore the utility of a short, synthetic peptide as potential anti-tumor agent targeting telomeric G4 DNA.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work is supported by Fellowships to Soumi Biswas and Satyabrata Samui from the Department of Biotechnology, New Delhi, and University of Kalyani respectively. Dr Jishu Naskar indebted to the Department of Science and Technology (DST), New Delhi, for FIST and PURSE programs, University of Kalyani. UGC for the SAP (DRS-II) program to the Department of Biochemistry and Biophysics, University of Kalyani, and the Department of Biotechnology (DBT), New Delhi, for financial support (BT/PR14646/NNT/28/989/2015). We thank Dr Soma Mukherjee, Dept. of Environmental Science, University of Kalyani, for allowing us to use spectrofluorimeter and the Division of Biological Sciences, Indian Institute of Science for the FACS facility. Dr Apurba K. Das sincerely acknowledges SIC, IIT Indore for providing instrumental facilities. K. M. is the recipient of CSIR Bhatnagar Fellowship from the Council of Scientific and Industrial Research, New Delhi.References
- (a) M. L. Bochman, K. Paeschke and V. A. Zakian, DNA secondary structures: stability and function of G-quadruplex structures, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed; (b) M. L. Duquette, P. Handa, J. A. Vincent, A. F. Taylor and N. Maizels, Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA, Genes Dev., 2004, 18, 1618–1629 CrossRef CAS PubMed; (c) D. Yang and K. Okamoto, Structural insights into G-quadruplexes: towards new anticancer Drugs, Future Med. Chem., 2010, 2, 619–646 CrossRef CAS PubMed; (d) J. A. Hackett, D. M. Feldser and C. W. Greider, Telomere Dysfunction Increases Mutation Rate and Genomic Instability, Cell, 2001, 106, 275–286 CrossRef CAS PubMed; (e) A. G. Bodnar, M. Ouellette, M. Frolkis, S. E. Holt, C.-P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner and W. E. Wright, Extension of Life-Span by Introduction of Telomerase into Normal Human Cells, Science, 1998, 279, 349–352 CrossRef CAS PubMed.
- (a) T.-M. Ou, Y.-J. Lu, J.-H. Tan, Z.-S. Huang, K.-Y. Wong and L.-Q. Gu, G-Quadruplexes: Targets in Anticancer Drug Design, ChemMedChem, 2008, 3, 690–713 CrossRef CAS PubMed; (b) J. Carvalho, P. Nottelet, J.-L. Mergny, J. A. Queiroz, G. F. Salgado and C. Cruz, Study of the interaction between indole-based compounds and biologically relevant G-quadruplexes, Biochimie, 2017, 135, 186–195 CrossRef CAS PubMed; (c) J. Carvalho, J. Ferreira, P. Pereira, E. Coutinho, A. Guédin, P. Nottelet, G. F. Salgado, J.-L. Mergny, J. A. Queiroz, F. Sousa, E. J. Cabrita and C. Cruz, Stabilization of novel immunoglobulin switch regions G-quadruplexes by naphthalene and quinoline-based ligands, Tetrahedron, 2016, 72, 1229–1237, DOI:10.1016/j.tet.2016.01.022.
- (a) E. Hiyama and K. Hiyama, Telomere and telomerase in stem cells, Br. J. Cancer, 2007, 96, 1020–1024 CrossRef CAS PubMed; (b) N. W. Kim, M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. C. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich and J. W. Shay, Specific Association of Human Telomerase Activity with Immortal Cells and Cancer, Science, 1994, 266, 2011–2015 CrossRef CAS PubMed.
- S. Neidle and G. Parkinson, Telomere Maintenance as a Target For Anticancer Drug Discovery, Nat. Rev. Drug Discovery, 2002, 1, 383–393 CrossRef CAS PubMed.
- (a) S. Asamitsu, S. Obata, Z. Yu, T. Bando and H. Sugiyama, Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy, Molecules, 2019, 24, 429 CrossRef PubMed; (b) S. M. Haider, G. N. Parkinson and S. Neidle, Structure of a G-quadruplex–Ligand Complex, J. Mol. Biol., 2003, 326, 117–125 CrossRef CAS PubMed.
- (a) S. Balasubramanian and S. Neidle, G-quadruplex nucleic acids as therapeutic targets, Curr. Opin. Chem. Biol., 2009, 13, 345–353 CrossRef CAS PubMed; (b) S. Neidle and M. A. Read, G-Quadruplexes as Therapeutic Targets, Biopolymers, 2001, 56, 195–208 CrossRef CAS; (c) A. Arola and R. Vilar, Stabilisation of G-Quadruplex DNA by Small Molecules, Curr. Top. Med. Chem., 2008, 8, 1405–1415 CrossRef CAS PubMed; (d) V. Dhamodharan, S. Harikrishna, A. C. Bhasikuttan and P. I. Pradeepkumar, Topology Specific Stabilization of Promoter over Telomeric G-Quadruplex DNAs by Bisbenzimidazole Carboxamide Derivatives, ACS Chem. Biol., 2015, 10, 821 CrossRef CAS PubMed.
- (a) K. Bhadra and G. S. Kumar, Interaction of berberine, palmatine, coralyne, and sanguinarine to quadruplex DNA: a comparative spectroscopic and calorimetric study, Biochim. Biophys. Acta, 2011, 1810, 485–496 CrossRef CAS PubMed; (b) D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten, J. K. C. Lim, D. V. Hoff, K. Anderes and W. G. Rice, Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis, Cancer Res., 2009, 69, 7653–7661 CrossRef CAS PubMed; (c) A. Rizzo, S. Iachettini, P. Zizza, C. Cingolani, M. Porru, S. Artuso, M. Stevens, M. Hummersone, A. Biroccio, E. Salvati and C. Leonetti, Identification of novel RHPS4-derivative ligands with improved toxicological profiles and telomere-targeting activities, J. Exp. Clin. Cancer Res., 2014, 33, 81 CrossRef; (d) W. J. Chung, B. Heddi, F. Hamon, M.-P. Teulade-Fichou and A. T. Phan, Solution Structure of a G-quadruplex Bound to the Bisquinolinium Compound Phen-DC3, Angew. Chem., Int. Ed., 2014, 53, 999–1002 CrossRef CAS; (e) A. M. Burger, F. Dai, C. M. Schultes, A. P. Reszka, M. J. Moore, J. A. Double and S. Neidle, The G-Quadruplex-Interactive Molecule BRACO-19 Inhibits Tumor Growth, Consistent with Telomere Targeting and Interference with Telomerase Function, Cancer Res., 2005, 65, 1489–1496 CrossRef CAS PubMed; (f) F. Cuenca, O. Greciano, M. Gunaratnam, S. Haider, D. Munnur, R. Nanjunda, W. D. Wilson and S. Neidle, Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands, Bioorg. Med. Chem. Lett., 2008, 18, 1668–1673 CrossRef CAS; (g) J. Lavrado, P. M. Borralho, S. A. Ohnmacht, R. E. Castro, C. M. P. Rodrigues, R. Moreira, D. J. V. A. dos Santos, S. Neidle and A. Paulo, Synthesis, G-Quadruplex Stabilisation, Docking Studies, and Effect on Cancer Cells of Indolo [3,2-b] quinolines with One, Two, or Three Basic Side Chains, ChemMedChem, 2013, 8, 1648–1661 CAS; (h) T. Das, D. Panda, P. Saha and J. Dash, Small Molecule Driven Stabilization of Promoter G-quadruplexes and Transcriptional Regulation of c-MYC, Bioconjugate Chem., 2018, 29(8), 2636–2645, DOI:10.1021/acs.bioconjchem.8b00338; (i) B. Maji, K. Kumar, M. Kaulage, K. Muniyappa and S. Bhattacharya, Design and Synthesis of New Benzimidazole−Carbazole Conjugates for the Stabilization of Human Telomeric DNA, Telomerase Inhibition, and Their Selective Action on Cancer Cells, J. Med. Chem., 2014, 57, 6973–6988 CrossRef CAS PubMed; (j) A. Głuszynska, B. Juskowiak, M. Kuta-Siejkowska, M. Hoffmann and S. Haider, Carbazole Derivatives' Binding to c-KIT G-Quadruplex DNA, Molecules, 2018, 23, 1134, DOI:10.3390/molecules23051134.
- (a) A. Ali, M. Kamra, S. Roy, K. Muniyappa and S. Bhattacharya, Novel Oligopyrrole Carboxamide based Nickel(II) and Palladium(II) Salens, Their Targeting of Human G-Quadruplex DNA, and Selective Cancer Cell Toxicity, Chem.–Asian J., 2016, 11, 2542–2554 CrossRef CAS PubMed; (b) A. Ali, M. Kamra, S. Roy, K. Muniyappa and S. Bhattacharya, Enhanced G-Quadruplex DNA Stabilization and Telomerase Inhibition by Novel Fluorescein Derived Salen and Salphen Based Ni(II) and Pd(II) Complexes, Bioconjugate Chem, 2017, 28, 341–352 CrossRef CAS PubMed; (c) A. W. Kleij, Zinc-centred salen complexes: versatile and accessible supramolecular building motifs, Dalton Trans, 2009, 4635–4639 RSC; (d) A. Łęczkowska, J. Gonzalez-Garcia, C. Perez-Arnaiz, B. Garcia, A. J. P. White and R. Vilar, Binding studies of metal-salphen and metal-bipyridine complexes towards G-quadruplex DNA, Chem.–Eur. J., 2018, 24, 11785–11794 CrossRef PubMed; (e) C.-Q. Zhou, T.-C. Liao, Z.-Q. Li, J. Gonzalez-Garcia, M. Reynolds, M. Zou and R. Vilar, Dinickel–Salphen Complexes as Binders of Human Telomeric Dimeric G-Quadruplexes, Chem.–Eur. J., 2017, 23, 4713–4722 CrossRef CAS PubMed; (f) L. Xu, X. Chen, J. Wu, J. Wang, L. Ji and H. Chao, Dinuclear Ruthenium(II) Complexes That Induce and Stabilise G-Quadruplex DNA, Chem.–Eur. J., 2015, 21, 4008–4020 CrossRef CAS PubMed; (g) X. H. Zheng, Y.-F. Zhong, C.-P. Tan, L.-N. Ji and Z.-W. Mao, Pt(II) squares as selective and effective human telomeric G-quadruplex binders and potential cancer therapeutics, Dalton Trans., 2012, 41, 11807–11812 RSC.
- (a) C. Wei, G. Jia, J. Yuan, Z. Feng and C. Li, A Spectroscopic Study on the Interactions of Porphyrin with G-Quadruplex DNAs, Biochemistry, 2006, 45, 6681–6691 CrossRef CAS PubMed; (b) S. M. Kerwin, D. Sun, J. T. Kern, A. Rangan and P. W. Thomas, G-Quadruplex DNA Binding by a Series of Carbocyanine Dyes, Bioorg. Med. Chem. Lett., 2001, 11, 2411–2414 CrossRef CAS PubMed; (c) A. Tawani, A. Amanullah, A. Mishra and A. Kumar, Evidences for Piperine inhibiting cancer by targeting human G-quadruplex DNA sequences, Sci. Rep., 2016, 6, 39239–39251 CrossRef CAS PubMed.
- B. Sengupta, B. Pahari, L. Blackmon and P. K. Sengupta, Prospect of Bioflavonoid Fisetin as a Quadruplex DNA Ligand: A Biophysical Approach, PLoS One, 2013, 8, e65383 CrossRef CAS PubMed.
- (a) C. Borghouts, C. Kunz and B. Groner, Current strategies for the development of peptide-based anti-cancer therapeutics, J. Pept. Sci., 2005, 11, 713–726 CrossRef CAS PubMed; (b) S. Biswas, S. Samui, A. Chakraborty, S. Biswas, D. De, U. Ghosh, A. K. Das and J. Naskar, Insight into the binding of a non-toxic, self-assembling aromatic tripeptide with ct-DNA: spectroscopic and viscositic studies, Biochem. Biophys. Rep., 2017, 11, 112–118 Search PubMed.
- (a) P. Sengupta, N. Banerjee, T. Roychowdhury, A. Dutta, S. Chattopadhyay and S. Chatterjee, Site-specific amino acid substitution in dodecameric peptides determines the stability and unfolding of c-MYC quadruplex promoting apoptosis in cancer cells, Nucleic Acids Res., 2018, 46, 9932–9950 CrossRef CAS PubMed; (b) S. Tyagi, S. Saxena, N. Kundu, T. Sharma, A. Chakraborty, S. Kaur, D. Miyoshi and J. Shankaraswamy, Selective recognition of human telomeric G-quadruplex with designed peptide via hydrogen bonding followed by base stacking interactions, RSC Adv., 2019, 9, 40255–40262 RSC; (c) K. H. Ngo, R. Yang, P. Das, G. K. T. Nguyen, K. W. Lim, J. P. Tam, B. Wu and A. T. Phan, Cyclization of a G4-specific peptide enhances its stability and G-quadruplex binding affinity, Chem. Commun., 2020, 56, 1082–1084 RSC; (d) K. C. Liu, K. Röder, C. Mayer, S. Adhikari, D. J. Wales and S. Balasubramanian, Affinity-selected bicyclic peptide G-quadruplex ligands mimic a protein-like binding mechanism, J. Am. Chem. Soc., 2020, 142(18), 8367–8373 CrossRef CAS PubMed.
- (a) S. Biswas, S. Samui, S. Biswas, A. K. Das and J. Naskar, Molecular recognition of double-stranded DNA by a synthetic, homoaromatic tripeptide (YYY): the spectroscopic and calorimetric study, Int. J. Biol. Macromol., 2019, 123, 221–227 CrossRef CAS PubMed; (b) B. K. Paul and N. Guchhait, Exploring the Strength, Mode, Dynamics, and Kinetics of Binding Interaction of a Cationic Biological Photosensitizer with DNA: Implication on Dissociation of the Drug–DNA Complex via Detergent Sequestration, J. Phys. Chem. B, 2011, 115, 11938–11949 CrossRef CAS PubMed.
- E. Largy, F. Hamon and M.-P. Teulade-Fichou, Development of a high-throughput G4-FID assay for screening and evaluation of small molecules binding quadruplex nucleic acid structures, Anal. Bioanal. Chem., 2011, 400, 3419–3427 CrossRef CAS PubMed.
- E. M. Rezler, J. Seenisamy, S. Bashyam, M.-Y. Kim, E. White, W. D. Wilson and L. H. Hurley, Telomestatin and Diseleno Sapphyrin Bind Selectively to Two Different Forms of the Human Telomeric G-Quadruplex Structure, J. Am. Chem. Soc., 2005, 127, 9439–9447 CrossRef CAS PubMed.
- A. K Jain, A. Paul, B. Maji, K. Muniyappa and S. Bhattacharya, Dimeric 1, 3-Phenylene-bis(piperazinyl benzimidazole)s: Synthesis and Structure−Activity Investigations on their Binding with Human Telomeric G-Quadruplex DNA and Telomerase Inhibition Properties, J. Med. Chem., 2012, 55, 2981–2993 CrossRef PubMed.
- W. N. Keith, T. R. Jeffry Evans and R. M. Glasspool, Telomerase and cancer: time to move from a promising target to a clinical reality, J. Pathol., 2001, 195, 404–414 CrossRef CAS PubMed.
- A. Sengupta, A. Ganguly and S. Chowdhury, Promise of G-Quadruplex Structure Binding Ligands as Epigenetic Modifiers with Anti-Cancer Effects, Molecules, 2019, 24, 582, DOI:10.3390/molecules24030582.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04780e |
| This journal is © The Royal Society of Chemistry 2020 |