Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Monitoring intracellular metal ion complexation with an acetylene-tagged ligand by Raman spectroscopy†
Seiya Takemuraa,
Hikaru Watanabea,
Tatsuya Nishiharaa,
Akimitsu Okamoto b and
Kazuhito Tanabe
b and
Kazuhito Tanabe *a
*a
aDepartment of Chemistry and Biological Science, College of Science and Engineering, Aoyama Gakuin University, 5-10-1 Fuchinobe, Chuo-ku, Sagamihara, 252-5258, Japan. E-mail: tanabe.kazuhito@chem.aoyama.ac.jp; Fax: +81-42-759-6493; Tel: +81-42-759-6229
bResearch Center for Advanced Science and Technology, The University of Tokyo, 4-6-1 Komaba, Meguro-ku, Tokyo, 153-8904, Japan
First published on 1st October 2020
Abstract
We propose to monitor molecular vibrations to identify metal ion–ligand complexation by means of Raman spectroscopy, which has been applied to track vibrational modes of molecules and to obtain a structural fingerprint. We prepared ligand molecules for Zn2+ ion complexation with a dipycolylaminoethyl aniline (DPEA) skeleton and phenylacetylene unit as the Raman tag which showed a typical band around 2200 cm−1. Among the labeled ligands synthesized in this study, A-DPEA showed a strong band attributed to the acetylene unit at 2212 cm−1, while the addition of Zn2+ ion resulted in a band shift to 2220 cm−1 due to complex formation. The addition of other metal ions and titration experiments showed that A-DPEA bound with Zn2+ selectively with a dissociation constant (Kd) that was estimated to be 0.22 μM. We also conducted cellular experiments and found that complexation between A-DPEA and Zn2+ also occurred in cells, with a shift in the Raman signal of the ligand from 2212 to 2215 cm−1. Thus, complex formation of the metal ion was identified by monitoring the Raman band shift.
Complex formation with metal cations can determine the function of biomolecules. For instance, proteins with their many reactive side chains have the potential capacity to bind several metal ions and become activated or disactivated.1,2 DNA or RNA molecules show enzyme-like properties that can be induced by complexation with metal cations to form second-order structures;3–5 indeed, these molecules have been used as building blocks for nanostructures.6,7 One of the most useful applications of complex formation is as molecular probes for metal cations in biological systems.8–10 A number of ligands bearing reporter units have been designed for sensing of metal cations in living cells or tissue. Complexation is an important and indispensable chemical reaction in living organisms, and therefore, the tracking systems for complex formation have been required to understand their biological roles.
Herein, we make a proposal, in which molecular vibrations are monitored to determine complexation by Raman spectroscopy, which is a standard technique for characterization of vibrational modes of molecules.11–15 Recent studies have shown that Raman spectroscopy can provide structural fingerprints of target molecules;16–18 thus, we expected that complex formation of metal cations would affect the vibrational modes of its ligand molecules to be determined. Heretofore, a variety of procedures are available to monitor the complexation of metal cations. Representative methods include absorption, fluorescence emission, and NMR spectroscopic techniques to track complexation.19–23 Although these spectroscopic methods are useful for characterization, low compatibility of absorption spectra for biological application and the requirement for large-sized substituents for fluorescence emission and a large number of molecules for NMR analysis are issues for these approaches. In this context, we considered that monitoring vibrational modes by Raman spectroscopy may represent a key technology that could be used to identify complexation and to resolve conventional issues because of the advantages that this technique offers, such as the need for small substituents and small volumes (ca. 1 μL) to obtain the spectra.
To identify metal ion complexation, we designed ligand molecules for Zn2+ ion, the complexation of which could be tracked by monitoring their Raman spectra. We employed a (dipycolylamino)ethyl aniline (DPEA) derivative24 as a ligand for Zn2+ and modified it with phenylacetylene as a Raman tag because phenylacetylene exhibits a characteristic Raman band even in complex living cell environments.14,16 We prepared two modified DPEAs and assessed their complex formation (Fig. 1). Eventually, we could trace the complexation of A-DPEA with Zn2+ by monitoring the Raman band shift. In addition, the complexation of A-DPEA in living cells was also successfully monitored.
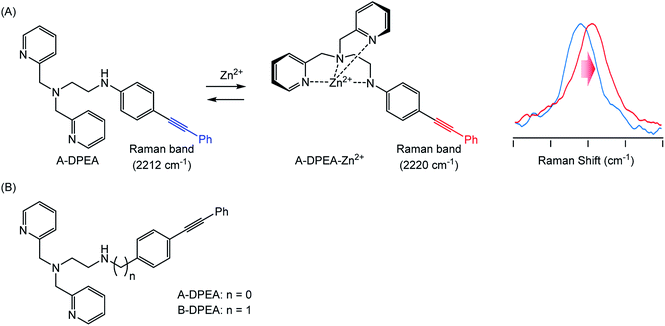 | ||
| Fig. 1 (A) Complexation of A-DPEA and Zn2+ ion that results in change of Raman spectra. (B) Ligands bearing Raman tag used in this study. | ||
The synthesis of the ligands bearing a phenylacetylene unit is illustrated in Scheme 1. A-DPEA was synthesized by Sonogashira coupling between phenylacetylene and aromatic bromide 1, which was prepared according to a reported procedure.24 We also synthesized a control compound, B-DPEA, in which the phenyl ring and alkylamine were separated by a methylene linkage, by reductive amination of diphenylacetylene 2.
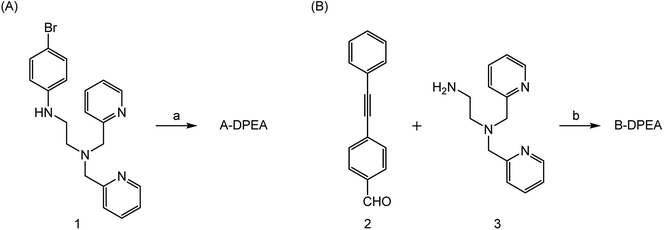 | ||
| Scheme 1 Synthesis of A-DPEA (A) and B-DPEA (B). Reagents and conditions. (a) Phenylacetylene, Pd(PPh3)4, CuI, Et3N, THF, 70 °C, 6%; (b) NaBH4, CH2Cl2–MeOH, 50 °C, 43%. | ||
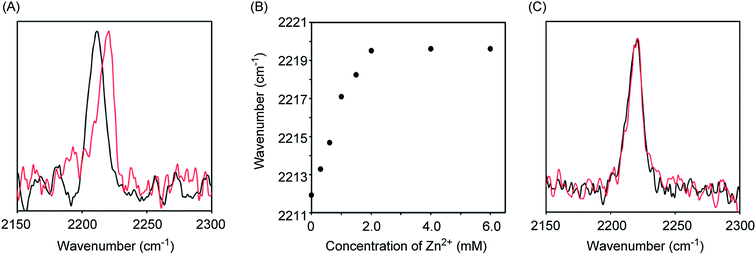
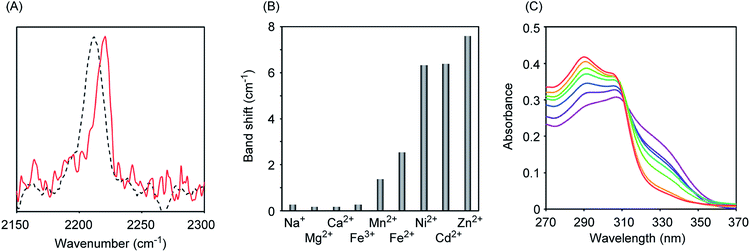
Initially, we measured the Raman spectra of A-DPEA using excitation at 532 nm. As shown in Fig. 2A, A-DPEA showed a typical band arising from the acetylene group at 2212 cm−1 under neutral conditions. As the concentration of Zn2+ was increased, the band shifted gradually from 2212 to 2220 cm−1 and fully shifted when ca. 2 equivalent of Zn2+ was added (Fig. 2B). These results indicate that the binding of Zn2+ caused a shift of the vibrational frequency of the acetylene unit in A-DPEA. In separate experiments, similar treatment of the control compound, B-DPEA, was conducted, but a negligible change of the Raman band at 2221 cm−1 was observed (Fig. 2C). Thus, conjugation between the acetylene unit and binding unit for Zn2+ is required for the band shift in the Raman spectra. To assess the complex formation of A-DPEA with Zn2+ further, we evaluated the reversibility of the signal change by the addition of a competing agent. Ethylenediaminetetraacetic acid (EDTA), which is a multidentate ligand for Zn2+, was added to the complex of A-DPEA and Zn2+, and changes of the Raman spectra were monitored (Fig. 3A). The signal of the acetylene unit was shifted from 2220 to 2212 cm−1, because the EDTA led to dissociation of Zn2+ from A-DPEA. Thus, the binding of Zn2+ with A-DPEA is responsible for the band shift in the Raman spectra. We next evaluated the selectivity of A-DPEA toward other metal ions (Fig. 3B). Measurement of Raman spectra revealed that addition of Na+, Mg2+, Ca2+ and Fe3+ did not lead to any band shift. Addition of Mn2+ and Fe2+ resulted in a slight band shift, and addition of Ni2+ and Cd2+ led to a moderate band shift. Thus, Zn2+ was selectively bind with A-DPEA, but several metal ions also showed moderate affinity toward A-DPEA, according to Irving–Williams order.25
We next measured the absorption spectra of A-DPEA to verify its binding with Zn2+. As shown in Fig. 3C, we observed absorption at 310 nm in the absence of Zn2+, which is a typical band for the noncomplexed ligand. Addition of Zn2+ resulted in a decrease of the absorption at 330 nm, while an increase of that at 290 nm was observed. An isosbestic point was located, which indicates a quantitative change from A-DPEA to the A-DPEA–Zn2+ complex. The apparent dissociation constant, Kd was determined to be 0.22 μM. Thus, A-DPEA retained the binding ability with Zn2+, although its affinity for Zn2+ was lower than that of the previous modified ligand for Zn2+ with the DPEA skeleton,19 probably due to the electron-withdrawing properties of the phenylacetylene unit. This low binding affinity of A-DPEA may require the excess amount of Zn2+ to complete shift of Raman band in Fig. 2B.
To further assess the function of A-DPEA, we measured the Raman spectra of A-DPEA in the presence of cell lysate. A-DPEA was dissolved in a lysate of a human cell line of fibrosarcoma HT-1080 and lung carcinoma cell line A549. Intense Raman band was observed around 2212 cm−1 in both lysates, while addition of Zn2+ led to band shift (Fig. S1†). These results are strong indication that complexation of A-DPEA was clearly monitored even in the complicated cellular environments.
For a better understanding of the function of A-DPEA in living cells, we assessed the complexation of Zn2+ in a human fibroblast fibrosarcoma cell line, HT1080. The cells were cultured in the presence of A-DPEA for 30 min and, after washing the cells, Raman spectra of the cells were measured. As shown in Fig. 4, the signal arising from the acetylene unit at 2212 cm−1 was observed from the cells, while a negligible signal was observed extracellularly. These results strongly indicate that A-DPEA was smoothly taken into the cells and that there are no significant interactions between A-DPEA and cells, because A-DPEA showed the signal at same wavenumber in the extracellular experiments (Fig. 2A). The administration of Zn2+ into the cells led to a band shift from 2212 to 2215 cm−1. This relatively small band shift (3 cm−1) is probably due to a low concentration of Zn2+ in the cells, which contributed to the complexation. Thus, the A-DPEA bound with Zn2+ in the cells, and it is reasonable to conclude that A-DPEA acts as a reporter molecule for Zn2+ complexation even in the cells that can be monitored by Raman spectra.
Conclusion
Complexation between ligand and Zn2+ cations was tracked by monitoring changes in molecular vibration. The Zn2+-binding ligand, A-DPEA, was prepared, to which a phenylacetylene unit was introduced as a reporter tag for the measurement of Raman spectra. A-DPEA showed a band at 2212 cm−1 typical of acetylene. Complexation of A-DPEA with Zn2+ resulted in a Raman band shift, whereupon the wavenumber was increased by 8 cm−1. The addition of EDTA as a competing chelate agent restored the signal to the original wavenumber, and thus the band shift was attributed to the complexation of Zn2+. We also applied the present system to monitor the complexation of Zn2+ in living cells. A-DPEA was smoothly taken into the cells and the Raman signal of the acetylene unit was easily detected from the cells. The administration of Zn2+ to the cells resulted in a band shift due to its complexation in the cells.In this report, we found that complexation affected molecular vibration of the ligand molecules. This change which attributed to interaction between metal ions and ligand molecules is attractive for the direct observation of complexation, although the low sensitivity and selectivity of the present system should be improved. Resonance Raman spectroscopy or surface enhanced Raman spectroscopy (SERS) may be useful to enhance the sensitivity and intensity of Raman signal, and the improvement of the ligand structure for high performance is in progress.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by fund for research unit in Aoyama Gakuin University Research Institute (for K. T.) and Grant-in-Aid for Scientific Research (for K. T. grant number 20H02863).References
- W. Maret, Adv. Nutr., 2013, 4, 82 CrossRef CAS.
- G. Rotilio, Inorg. Chim. Acta, 1980, 40, X49 CrossRef.
- W. Zhou and J. Liu, Metallomics, 2018, 10, 30 RSC.
- A. Sett, S. Das and U. Bora, Appl. Biochem. Biotechnol., 2014, 174, 1073 CrossRef CAS.
- B. J. Pages, D. L. Ang, E. P. Wright and J. R. Aldrich-Wright, Dalton Trans., 2015, 44, 3505 RSC.
- M. Xiao, X. Qu, L. Li and H. Pei, Methods Mol. Biol., 2018, 1811, 137 CrossRef CAS.
- I. Berger, A. G. Blanco, R. Boelens, J. Cavarelli, M. Coll, G. E. Folkers, Y. Nie, V. Pogenberg, P. Schultz, M. Wilmanns, D. Moras and A. Poterszman, J. Struct. Biol., 2011, 175, 135 CrossRef CAS.
- S. Mizukami, H. Matsushita and K. Kikuchi, Kagaku Furontia, 2010, 22, 15 CAS.
- Q. Zhang, J. Wang, A. M. Kirillov, W. Dou, C. Xu, C. Xu, L. Yang, R. Fang and W. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 23976 CrossRef CAS.
- Y. Imai, Y. Nakano, T. Kawai and J. Yuasa, Angew. Chem., Int. Ed., 2018, 57, 8973 CrossRef CAS.
- P. Larkin, Infrared and Raman Spectroscopy, Elsevier, 2011 Search PubMed.
- B. Hernández, F. Pflüger, S. G. Kruglik and M. Fhomi, Chem. Phys., 2013, 425, 104 CrossRef.
- A. T. Tu, Raman Spectroscopy in Biology: Principles and Applications, John Wiley & Sons Inc., New York, 1982 Search PubMed.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102 CrossRef CAS.
- L. Wei, F. Hu, Y. Shen, Z. Chen, Y. Yu, C.-C. Lin, M. C. Wang and W. Min, Nat. Methods, 2014, 11, 410 CrossRef CAS.
- A. F. Palonpon, M. Sodeoka and K. Fujita, Curr. Opin. Chem. Biol., 2013, 17, 708 CrossRef CAS.
- R. Kurihara, Y. Ikemura and K. Tanabe, Bioorg. Med. Chem. Lett., 2016, 26, 4892 CrossRef CAS.
- S. Yamaguchi, T. Matsushita, S. Izuta, S. Katada, M. Ura, T. Ikeda, G. Hayashi, Y. Suzuki, Y. Kobayashi, K. Tokunaga, Y. Ozeki and A. Okamoto, Sci. Rep., 2017, 7, 41007 CrossRef CAS.
- T. Hirano, K. Kikuchi, Y. Urano, T. Higuchi and T. Nagano, J. Am. Chem. Soc., 2000, 122, 12399 CrossRef CAS.
- K. Komatsu, K. Kikuchi, H. Kojima, Y. Urano and T. Nagano, J. Am. Chem. Soc., 2005, 127, 10197 CrossRef CAS.
- X. Tian, S. Hussain, C. de Pace, L. Ruiz-Perez and G. Battaglia, Chem.–Asian J., 2019, 14, 509 CAS.
- Y. Suzuki and K. Yokoyama, Biosensors, 2015, 5, 337 CrossRef CAS.
- S. Cortes, E. Brucher, C. F. G. C. Geraldes and A. D. Sherry, Inorg. Chem., 1990, 29, 5 CrossRef CAS.
- G. Zhang, S. Bi, L. Song, F. Wang, J. Yu and L. Wang, Dyes Pigm., 2013, 99, 779 CrossRef CAS.
- H. Irving and R. J. P. Williams, Nature, 1948, 162, 746 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedure, 1H and 13C NMR data of compounds and Raman spectra in cell lysate. See DOI: 10.1039/d0ra06329k |
| This journal is © The Royal Society of Chemistry 2020 |