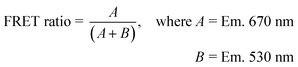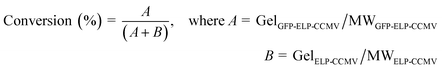Open Access Article
Open Access ArticleInsight into N-terminal localization and dynamics of engineered virus-like particles†
Daan F. M. Vervoort‡
,
Chiara Pretto‡ and
Jan C. M. van Hest *
*
Eindhoven University of Technology, Institute for Complex Molecular Systems, PO Box 513 (STO 3.41), 5600 MB Eindhoven, The Netherlands. E-mail: j.c.m.v.hest@tue.nl
First published on 22nd October 2020
Abstract
Virus-like particles composed of the cowpea chlorotic mottle virus (CCMV) capsid protein (CP) have been extensively studied as carrier systems in nanoscience. One well-established method to improve their stability under physiological conditions is to fuse a stimulus-responsive elastin-like polypeptide (ELP) to the N-terminus of the CPs. Even though the N-terminus should in principle be localized in the inner cavity of the protein cage, studies on the native CCMV revealed its accessibility on the particle surface. We verified that such phenomenon also applies to ELP-CCMVs, by exploiting the covalent functionalization of the CP N-terminal domain via a sortase A-mediated reaction. Western-blot analysis and Förster resonance energy transfer (FRET) experiments furthermore revealed this to be caused by both the external display of the N-termini and the interchange of CPs among preformed capsids. Our findings demonstrate the tunability of ELP-CCMV stability and dynamics and their potential effect on the exploitation of such protein cages as a drug delivery system.
1. Introduction
Protein cages obtained from the cowpea chlorotic mottle virus (CCMV) represent an interesting platform for the development of smart systems that can be exploited for several biologically relevant applications.1–5 CCMV can be disassembled into capsid protein dimers and reassembled, without the viral RNA, into virus-like particles (VLPs) in response to variations in the surrounding environment.6–8 The native form of the VLP, for example, disassembles at neutral pH and assembles in acidic conditions in a reversible manner.9–12 This process can be exploited for the encapsulation of different cargoes into these protein cages, for example via electrostatic interactions of the positively charged N-terminus of the capsid protein (CP) with negatively charged polyelectrolytes.13–16To broaden the scope of CCMV systems for biomedical applications, protein engineering methods have been employed to modify the internally oriented N-terminus for improving both loading capacity and stability under physiologically relevant conditions. In particular, the ELP-CCMV variant, obtained by fusion of a stimulus-responsive block co-polymer called elastin-like polypeptide (ELP, consisting of nine repeats of the Val-Pro-Gly-Xaa-Gly (VPGXG) pentapeptide) to the N-terminus of the capsid protein, results in particle formation triggered by an increase in temperature or salt concentration in solution.17 As a result, control over nanoparticle stability is achieved at neutral pH even in the absence of a specific cargo.18,19 Moreover, covalent encapsulation strategies have been successfully performed on ELP-CCMV with both enzymes and small molecules under physiological pH without additional modifications or stabilizing agents.20–22 These were achieved via either sortase A-mediated fusion of LPETG-tagged proteins to the N-terminal glycine of the CP or via CP functionalization with vinylboronic acids (VBAs) and the subsequent bioorthogonal reaction with a cargo bearing a tetrazine handle. Additionally, insertion of an N-terminal histidine tag allows for both CP purification through immobilized metal affinity chromatography (IMAC) and capsid stabilization by addition of metal ions such as nickel23,24 as a result of the binding affinity of the histidine imidazole groups towards divalent cations. For the reasons stated above, histidine-tagged ELP-CCMV are a potential candidate for various applications in nanomedicine.
Although the N-terminus is predominantly directed towards the capsid core, some studies have indicated that the N-terminus can also be orientated towards the particle surface. In particular, the N-terminal localization of the native CCMV was investigated exploiting protease digestion, mass spectrometry and crystal structure analysis.6,25,26 It was shown that this domain of the CP was partially exposed on the particle surface. These findings were in agreement with a more recent study, where the replacement of the (N-terminal) RNA-binding motif of the CP with a tumor-homing peptide not only led to the efficient assembly into VLPs but also to selective cancer cell targeting.27 Furthermore, as a result of protein–protein interactions the structure of icosahedral capsids in general, and CCMV in particular is flexible and dynamic, with capsids and capsid subunits in thermodynamic equilibrium.28,29 Consequently, association and dissociation of capsid proteins are continuous processes, even when capsids are stable in solution.
These previously mentioned studies suggest that the accessibility of the CP N-terminus can be a result of capsid dynamics, which allows temporary exposure of the N-terminus, and/or a different conformational orientation of the capsid proteins, which allows the N-terminal part to fold back onto the particle surface. Therefore, a deeper understanding of the dynamic behaviour of these protein-based scaffolds, paying special attention to the N-terminus, is required. To the best of our knowledge, these tightly correlated aspects have never been studied independently. Moreover, both proposed mechanisms have yet not been investigated for the ELP-CCMV.
In this report we present our evaluation of the N-terminal accessibility of ELP-VLPs exploiting N-terminal tagging using the sortase A-mediated reaction. We subsequently performed western blot analysis using an antibody directed against the N-terminus to analyze the N-terminal localization on the particle surface. In parallel, Förster resonance energy transfer (FRET) experiments on stable capsids elucidated the dynamic behaviour of ELP-CCMV subunits. We demonstrate the complementarity of N-terminal accessibility and dynamics, which should be considered when exploiting ELP-CCMV capsids in the field of nanotechnology.
2. Experimental section
2.1. Materials
Nickel(II) chloride hexahydrate was obtained from Sigma-Aldrich. DTSSP (3,3′-dithiobis(sulfosuccinimidylpropionate)), Alexa Fluor™ 532 C5 maleimide and Anti-His6-tag mouse monoclonal antibody conjugated with HRP (MA1-21315-HRP) were purchased from ThermoFisher Scientific. Sulfo-Cyanine5 maleimide was purchased from Lumiprobe.2.2. Protein expression
General protocols for the expression of proteins used in this study and detailed buffers' composition can be found in the ESI.†2.3. Dynamic light scattering (DLS)
DLS measurements were performed on a Malvern Zetasizer Nano ZSP at 21 °C. Samples (20 μM, unless stated otherwise) were centrifuged twice prior to analysis. Buffers were filtered prior to use. All measurements were performed in triplicate and the average measurements were plotted.2.4. Size exclusion chromatography (SEC)
SEC analysis was performed on a Superose 6 increase 10/300 GL column (GE Healthcare Life Sciences). Analytical measurements were executed on an Agilent 1260 bio-inert HPLC. Samples with a protein concentration of 1.0 mg mL−1 were separated on the column with a flow rate of 0.5 mL min−1 in buffer of interest.2.5. Capsid assembly
Variants of CCMV capsids from protein expression (stored in pH 5.0 capsid buffer, Table S1, ESI†) were dialyzed for 90 minutes, unless otherwise stated, at 4 °C or 21 °C using Spectra/Por 2 Dialysis Tubing, (12–14 kDa) in the buffer of interest prior to analysis. Buffers were refreshed three times.2.6. Capsid stabilization
VY1-VY8 ELP-CCMV (T = 1) capsids in PBS (2 M NaCl) were first incubated with nickel chloride for 1 hour at room temperature (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Ni2+
1 Ni2+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP molar ratio, 45 μM CP). Samples were dialyzed against PBS at 4 °C and incubated with the crosslinker (20
CP molar ratio, 45 μM CP). Samples were dialyzed against PBS at 4 °C and incubated with the crosslinker (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DTSSP
1 DTSSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP molar ratio) for one hour at 21 °C and 600 rpm. At reaction completion, samples were dialyzed at 4 °C in PBS containing 2 mM EDTA in order to remove the metal ions. Extensive overnight dialysis in PBS was subsequently performed to remove the unreacted crosslinker.
CP molar ratio) for one hour at 21 °C and 600 rpm. At reaction completion, samples were dialyzed at 4 °C in PBS containing 2 mM EDTA in order to remove the metal ions. Extensive overnight dialysis in PBS was subsequently performed to remove the unreacted crosslinker.
2.7. Sortase A-mediated functionalization of ELP-CCMV
Standard ELP-CCMV and VY1-VY8 ELP-CCMV capsids were dialyzed at 4 °C towards PBS (500 mM NaCl) and pH 7.5 buffer (100 mM NaCl) respectively to ensure disassembly. ELP-CCMV variants and GFP were then buffer exchanged towards sortase buffer by spin-filtration (Amicon® Ultra 0.5 mL). Dimers (Standard ELP-CCMV) or capsids (VY1-VY8 ELP-CCMV), sortase A and GFP were incubated in equimolar concentrations (30 μM) at 21 °C, 400 rpm up to 24 h. Reaction mixtures at indicated time points were analyzed by SDS-PAGE using Mini-Protean TGX 4–20% gels (tris/glycine buffer) for 1 hour at 150 V. Proteins were visualized via Coomassie Brilliant Blue staining (Bio-Rad). ImageJ analysis software was used to calculate the conversion percentage using the following formula, where gel is the intensity of the protein band on the SDS-PAGE gel and MW is the molecular weight of the protein of interest:2.8. Native PAGE
Native ELP-CCMV capsids were dialyzed at 21 °C or 4 °C towards PBS (500 mM NaCl) in order to obtain dimers. VY1-VY8-ELP-CCMV capsids were dialyzed at 21 °C against PBS (500 mM NaCl) and at 4 °C in pH 7.5 buffer (100 mM NaCl) in order to get capsids and dimers respectively. Samples were then mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with native PAGE loading buffer (2×) and further diluted with PBS (500 mM NaCl) to a final concentration of 5 μM. ELP-CCMV variants (30 μL) were separated using Mini-Protean TGX 4–20% gels (tris/glycine buffer) for 1 hour at 100 V at room temperature in native PAGE running buffer or for 2 hours at 100 V in ice-cold native PAGE running buffer. Gels were stained by Coomassie Brilliant Blue (Bio-Rad) or used for the western blot analysis.
1 ratio with native PAGE loading buffer (2×) and further diluted with PBS (500 mM NaCl) to a final concentration of 5 μM. ELP-CCMV variants (30 μL) were separated using Mini-Protean TGX 4–20% gels (tris/glycine buffer) for 1 hour at 100 V at room temperature in native PAGE running buffer or for 2 hours at 100 V in ice-cold native PAGE running buffer. Gels were stained by Coomassie Brilliant Blue (Bio-Rad) or used for the western blot analysis.
2.9 Western blot
Proteins of native PAGE gels were transferred on a nitrocellulose membrane in blotting buffer using a blotting system (Tetra Blotting Module, Bio-Rad) for 1 hour at 100 V. The membrane was incubated with blocking buffer for 40 minutes at room temperature. The blot was probed with Anti-His6-tag mouse monoclonal antibody conjugated with HRP (MA1-21315-HRP 1:1000) overnight at 4 °C. Excess of antibody was removed by washing the membrane 3 times with PBST. 3,3′,5,5′-Tetramethylbenzidine (TMB) was added to visualize antibody binding.2.10. Evaluation of ELP-CCMV dynamics using FRET
VY1-VY8 ELP-CCMV capsids (T = 3) were dialyzed towards pH 7.5 buffer (100 mM NaCl) to ensure disassembly. Capsid proteins were then dialyzed towards PBS (2 M NaCl) to ensure ELP-induced capsid assembly. Capsid labeling with either sulfo-Cyanine5 maleimide or Alexa Fluor 532 maleimide was performed according to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 molar ratio (capsid protein (50 μM): dye (250 μM)) in PBS (2 M NaCl) for 2 hours at 21 °C and 600 rpm. Extensive dialysis towards PBS (2 M NaCl) was performed to remove the excess of unlabeled dye. Labeled and unlabeled capsids were dialyzed against pH 7.5 (100 mM NaCl) in order to get dimers. The following samples were then mixed in a 1
5 molar ratio (capsid protein (50 μM): dye (250 μM)) in PBS (2 M NaCl) for 2 hours at 21 °C and 600 rpm. Extensive dialysis towards PBS (2 M NaCl) was performed to remove the excess of unlabeled dye. Labeled and unlabeled capsids were dialyzed against pH 7.5 (100 mM NaCl) in order to get dimers. The following samples were then mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio: (a) unlabeled and Cy5-labeled, (b) unlabeled and Alexa532-labeled and (c) Cy5-labeled and Alexa532-labeled. Subsequently, samples were buffer-exchanged using centrifugal filters (Amicon® Ultra 0.5 mL) towards PBS (2 M NaCl) to force ELP-induced capsid formation. Additionally, Cy5-labeled, Alexa532-labeled and unlabeled capsid proteins were assembled under the same conditions. Cy5-labeled and Alexa-labeled capsids were then crosslinked as previously described (2.6 capsid stabilization). In order to perform the FRET assay, the following capsids were analyzed: (i) capsids containing 50% unlabeled CPs and 50% Alexa-labeled CPs, (ii) capsids containing 50% unlabeled CPs and 50% Cy5-labeled CPs, (iii) preformed capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs (to ensure the maximum FRET per capsid), (iv) capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs, (v) stabilized capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs and (vi) unlabeled capsids. Using a SPARK® 10 M multimode microplate reader (Tecan), an emission scan (530–800 nm) was performed with an excitation wavelength of 485/20 nm. The ratiometric FRET-ratio was calculated according to the following equation, where the emission of the acceptor (Cy5) and donor (Alexa532) were measured at 670 nm and 530 nm respectively:
1 ratio: (a) unlabeled and Cy5-labeled, (b) unlabeled and Alexa532-labeled and (c) Cy5-labeled and Alexa532-labeled. Subsequently, samples were buffer-exchanged using centrifugal filters (Amicon® Ultra 0.5 mL) towards PBS (2 M NaCl) to force ELP-induced capsid formation. Additionally, Cy5-labeled, Alexa532-labeled and unlabeled capsid proteins were assembled under the same conditions. Cy5-labeled and Alexa-labeled capsids were then crosslinked as previously described (2.6 capsid stabilization). In order to perform the FRET assay, the following capsids were analyzed: (i) capsids containing 50% unlabeled CPs and 50% Alexa-labeled CPs, (ii) capsids containing 50% unlabeled CPs and 50% Cy5-labeled CPs, (iii) preformed capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs (to ensure the maximum FRET per capsid), (iv) capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs, (v) stabilized capsids containing 50% Cy5-labeled CPs and 50% Alexa-labeled CPs and (vi) unlabeled capsids. Using a SPARK® 10 M multimode microplate reader (Tecan), an emission scan (530–800 nm) was performed with an excitation wavelength of 485/20 nm. The ratiometric FRET-ratio was calculated according to the following equation, where the emission of the acceptor (Cy5) and donor (Alexa532) were measured at 670 nm and 530 nm respectively:All data points were normalized to the maximum FRET (sample iii).
3. Results and discussion
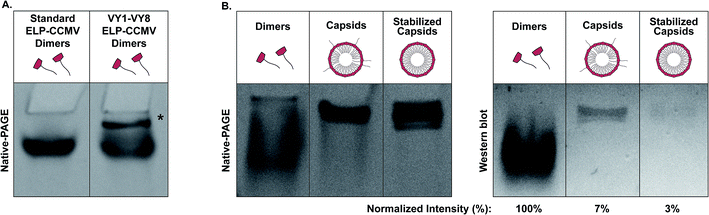
The “standard” ELP-CCMV contained nine ELP pentapeptide repeats (VPGXG), with four valines, four leucines and one glycine as guest residue X (in guest residue order, starting from the N terminus: VLVLVLGVL).17 The CP assembled into stable capsids at physiological pH, although a high concentration of sodium chloride was necessary (∼2 M). Lowering the salt concentration to 500 mM fully disassembled the capsids into dimers. In order to further stabilize the ELP-CCMV towards physiological salt concentration, a library of variants was developed by replacing the first and eighth guest residues with amino acids of higher hydrophobicity.18 Substitution of valine to tyrosine resulted in a variant, named VY1-VY8 ELP-CCMV, that assembled at 500 mM NaCl and disassembled at 100 mM NaCl.19 These capsids remained even stable at nearly physiological conditions (pH 7.5, 150 mM NaCl and 37 °C) up to seven hours. A more hydrophobic variant, VW1-VW8 ELP-CCMV, developed by substitution of the first and eighth valine guest residue with tryptophan, remained stable at these conditions up to 24 hours. In fact, this variant seemed to lack reversible disassembly behaviour. Data even suggested that VW1-VW8 ELP-CCMV was already assembled into capsids prior to purification through immobilized metal affinity chromatography. Since purification of these capsids can only be achieved when the N-terminal His6-tag is accessible, this data represents the first indication that ELP-CCMV has similar N-terminal accessibility compared to native CCMV. The N-terminus availability could be a result of the N-terminal domain backfolding on the surface of preformed engineered capsids and/or be a consequence of the capsid proteins exchanging and intermixing over time, as illustrated in Fig. 1. In order to verify whether either of these two mechanisms applies for ELP-CCMV, a sortase A-mediated reaction was performed. As the ELP-CCMV construct contains an N-terminal glycine, it is amenable to undergo the enzymatic tagging with appropriately LPETG-labeled proteins. Here, standard ELP-CCMV and VY1-VY8 ELP-CCMV were used, as VW1-VW8 ELP-CCMV's inability to disassemble made it less relevant for drug delivery applications. Additionally, stabilized capsids of VY1-VY8 ELP-CCMV were generated as follows. First, nickel ions were added to the capsids in order to direct the N-termini towards the particle core. Second, the capsids were partially crosslinked with the homobifunctional crosslinker DTSSP (3,3′-dithiobis(sulfosuccinimidylpropionate)) to hinder dynamic intermixing of CPs (Fig. S5, ESI†).30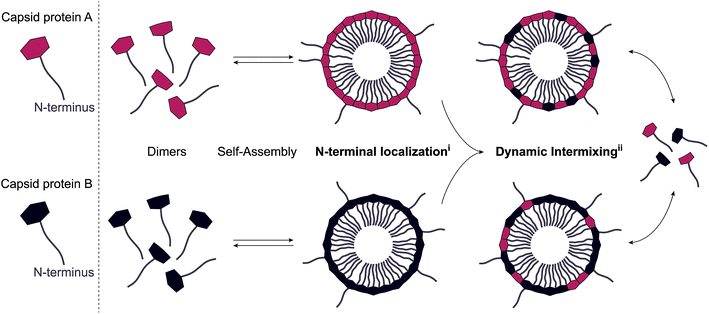 | ||
| Fig. 1 Schematic representation of the two proposed mechanisms, (i) surface exposed N-terminal localization, (ii) dynamic intermixing. | ||
This dual-step modification allowed us to limit both proposed mechanisms. A schematic overview of all samples used in this work and their relative stability (depending on the salt concentration) is reported in Table 1. The conjugation of an LPETG-tagged green fluorescent protein (GFP) was executed at neutral pH in the presence of 500 mM of sodium chloride. In this way, we were able to perform the reaction on dimers, capsids and stabilized capsids under the same environmental conditions (Table 1). SDS-PAGE analysis demonstrated successful conjugation of GFP to standard ELP-dimers after 24 hours of incubation according to a conjugation yield of 33% (based on ImageJ analysis of the band intensity) as shown in Fig. 2A. Interestingly, successful conjugation (5%), albeit less pronounced, was also observed when performing the same reaction on VY1-VY8 ELP-capsids. These findings perfectly correlate to our previous data related to protein purification, demonstrating N-terminal accessibility of ELP-CCMV VLPs. As expected, 0% conjugation efficiency was recorded performing the reaction on stabilized capsids. DLS analysis of standard ELP-dimers as well as VY1-VY8 ELP-capsids and stabilized capsids was performed after 24 hours of incubation under the same conditions used for the sortase A reaction to confirm the assembly state of tested samples (Fig. 2B).
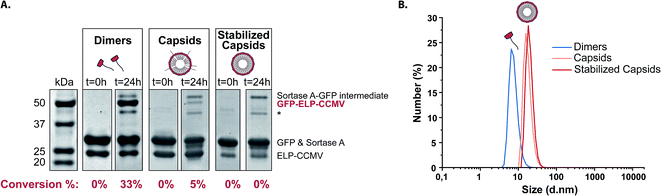 | ||
| Fig. 2 (A) SDS-PAGE analysis of GFP conjugation. Proteins were visualized with Coomassie staining. Bands associated with unmodified CP appeared at 22 kDa, sortase A overlapped with GFP at about 27 kDa and GFP–CP conjugates appeared at about 49 kDa. GFP–sortase A intermediate appeared about 50 kDa. Conversion percentages were calculated using ImageJ analysis (Experimental Section 2.7). *Impurity in sortase A (Fig. S3, ESI†). (B) DLS analysis of standard ELP dimers, VY1-VY8-ELP capsids and stabilized capsids after 24 hours of incubation under the same conditions used for sortase A reaction. | ||
Since the sortase A-mediated reaction confirmed the accessibility of the N terminus, we subsequently investigated both processes independently. We first focused our attention on the N-terminal localization on preformed capsids. In order to do so, we performed native PAGE and western blot analysis using an anti-His6 HRP-labeled antibody against the N-terminal His6-tag. Since dimers and capsids run differently on the native gel it is not only possible to clearly distinguish between these two assemblies but also to analyze the accessibility of the N-terminus due to antibody recognition in a highly sensitive manner. Moreover, the relative bulkiness of the antibody does not allow binding to the N-termini when they are located in the particle core. Since full dimerization of the more stable VY1-VY8 ELP-CCMV was not achieved at 4 °C, even in the presence of 100 mM of sodium chloride (Fig. 3A), the analysis of the antibody binding capacity was performed towards standard ELP-dimers, VY1-VY8 ELP-CCMV capsids as well as stabilized capsids at 21 °C in the presence of 500 mM of NaCl (Fig. 3B). We normalized the binding efficiency to the maximum value achievable with the dimers, as all N-termini of the CP should be exposed and available for antibody binding. By analyzing the capsids via this approach, a normalized binding efficiency of about 7% was measured. This result demonstrated that the antibody is capable of interacting with the His6-tag of preformed particles. Since the antibody as well as the CP concentration was constant under both conditions, the binding observed at capsid level has to be attributed to the exposure of the N-terminus on the particle surface. In the case of stabilized capsids, a lower binding efficiency was recorded (3%) indicating that the N-terminal accessibility was reduced by over two-fold compared to normal capsids.
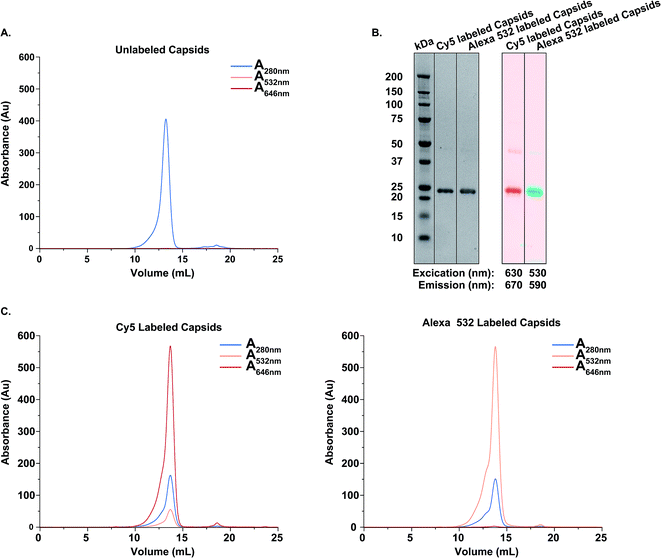
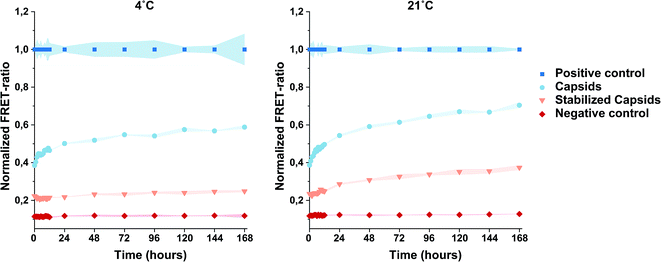
Next, we analyzed the dynamic process involving CP intermixing by using FRET. To this aim, a FRET pair consisting of Cy5 and AlexaFluor-532 was selected. VY1-VY8 ELP-CCMV capsids were prepared in PBS (2 M NaCl) and their assembly state confirmed by DLS (Fig. S6, ESI†) and SEC (Fig. 4A). Subsequently, capsids were independently functionalized with one of the fluorophores via maleimide-coupling. Labeling efficiency was analyzed according to manufacturer instructions (Table S4, ESI†) and confirmed by reducing SDS-PAGE analysis (Fig. 4B) and size exclusion chromatography (Fig. 4C). After purification, samples for FRET analysis were prepared. By mixing capsids labeled with either Cy5 or AlexaFluor-532 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, the resulting FRET signal can be directly associated to the CPs intermixing, thus allowing us to monitor the dynamics of the system. Stabilized capsids, whose dynamic behaviour was expected to be hindered, were prepared in the same way. As a positive control, both fluorescently labeled VY1-VY8 ELP-CCMV particles were first disassembled into dimers (100 mM NaCl, Table 1), mixed together in equimolar ratio and finally forced into capsid formation by increasing the salt concentration in solution. This allowed us to obtain capsids bearing both dyes in equimolar amounts and therefore achieving the maximum FRET signal (positive control).
1 molar ratio, the resulting FRET signal can be directly associated to the CPs intermixing, thus allowing us to monitor the dynamics of the system. Stabilized capsids, whose dynamic behaviour was expected to be hindered, were prepared in the same way. As a positive control, both fluorescently labeled VY1-VY8 ELP-CCMV particles were first disassembled into dimers (100 mM NaCl, Table 1), mixed together in equimolar ratio and finally forced into capsid formation by increasing the salt concentration in solution. This allowed us to obtain capsids bearing both dyes in equimolar amounts and therefore achieving the maximum FRET signal (positive control).
A mixture of unlabeled VY1-VY8 ELP-CCMV and AlexaFluor-532 labeled VY1-VY8 ELP-CCMV (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) was prepared as a negative control. The dynamic behavior of nanoparticles was monitored for one week by measuring the FRET emission spectra at both 4 °C and 21 °C. These temperatures were selected, in combination with a high salt concentration (2 M NaCl), to guarantee that neither particle aggregation nor protein unfolding may occur. A significant increase of the normalized FRET ratio was observed for capsids at both temperatures, especially within the first 24 hours (Fig. 5). As expected, the analysis of the negative control, consisting of capsids functionalized with the fluorophore donor only (AlexaFluor-532), resulted in negligible background signal. These data clearly demonstrated CP intermixing over time. The trend of the curves related to intermingled capsids, and in particular at 21 °C, suggests that more time compared to the positive control is required for capsids to reach an equilibrium and with that the maximum FRET value. When analyzing the behaviour of labeled particles after stabilization, a different trend was observed. In this case, CPs' dynamics was indeed limited since the increase in the FRET ratio was significantly slower. Additionally, it was shown that also the temperature plays a role in this process. Indeed, higher normalized FRET ratios observed for both capsids and stabilized capsids at 21 °C indicate that the dynamics of these systems and therefore CP exchange was faster compared to the experiment performed at 4 °C. All data reported indicate dynamic behaviour of the CPs, which is more clearly observed within the first 24 hours of incubation. Moreover, the flattening of the slope of the FRET curves over time, observed for both capsids and stabilized capsids, indicate that a dynamic equilibrium was reached.
1 molar ratio) was prepared as a negative control. The dynamic behavior of nanoparticles was monitored for one week by measuring the FRET emission spectra at both 4 °C and 21 °C. These temperatures were selected, in combination with a high salt concentration (2 M NaCl), to guarantee that neither particle aggregation nor protein unfolding may occur. A significant increase of the normalized FRET ratio was observed for capsids at both temperatures, especially within the first 24 hours (Fig. 5). As expected, the analysis of the negative control, consisting of capsids functionalized with the fluorophore donor only (AlexaFluor-532), resulted in negligible background signal. These data clearly demonstrated CP intermixing over time. The trend of the curves related to intermingled capsids, and in particular at 21 °C, suggests that more time compared to the positive control is required for capsids to reach an equilibrium and with that the maximum FRET value. When analyzing the behaviour of labeled particles after stabilization, a different trend was observed. In this case, CPs' dynamics was indeed limited since the increase in the FRET ratio was significantly slower. Additionally, it was shown that also the temperature plays a role in this process. Indeed, higher normalized FRET ratios observed for both capsids and stabilized capsids at 21 °C indicate that the dynamics of these systems and therefore CP exchange was faster compared to the experiment performed at 4 °C. All data reported indicate dynamic behaviour of the CPs, which is more clearly observed within the first 24 hours of incubation. Moreover, the flattening of the slope of the FRET curves over time, observed for both capsids and stabilized capsids, indicate that a dynamic equilibrium was reached.
4. Conclusions
From our data, it becomes clear that the N-terminal localization on CCMV-VLPs and dynamic intermixing of CPs represent highly interconnected processes. Both aspects need to be considered when developing a drug delivery system or a smart nanoreactor for biological applications based on these protein cages. First, the N-terminal position is not only important when considering cargo loading but also when performing particle functionalization. In particular, its partial external localization may indicate that a specific payload, either covalently linked or electrostatically bound to this portion of the protein, could be partially exposed on the particle surface. On the other hand, the presence of free N-termini on the capsid exterior could represent an additional handle for further particle functionalization after cargo loading.Our results clearly demonstrate that even in the case of ELP-induced assembly, where salt is used to trigger ELP hydrophobic interactions and orientation towards the inner cavity of the capsids, some of these N-terminal residues are still exposed on the particle surface. At the same time, we have shown that CP intermixing is an ongoing dynamic process. Even though ELP-CCMVs display higher stability under nearly physiological conditions, the dynamic behaviour of ELP-CPs is not completely inhibited. When designing systems for drug delivery, stabilizing particles with a crosslinker which hampers the dynamics could prove to be a necessary measure to prevent early cargo leakage. Therefore, in order to use these particles in biomedical applications the balance between stability and dynamicity should be carefully tuned.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was financially supported by OcuTher and FMS gravitation program. The authors acknowledge Wiggert Altenburg for the supportive role in the cloning and expression of the LPETG-tagged superfolder GFP. We also thank Suzanne Timmermans, Wiggert Altenburg and Glenn Cremers for the fruitful discussions.References
- M. Brasch, I. K. Voets, M. S. T. Koay and J. J. L. M. Cornelissen, Faraday Discuss., 2013, 166, 47–57 RSC
.
- T. Douglas and M. Young, Nature, 1998, 393, 152–155 CrossRef CAS
.
- P. Lam and N. F. Steinmetz, Biomater. Sci., 2019, 7, 3138–3142 RSC
.
- L. Sánchez-Sánchez, R. D. Cadena-Nava, L. A. Palomares, J. Ruiz-Garcia, M. S. T. Koay, J. J. M. T. Cornelissen and R. Vazquez-Duhalt, Enzyme Microb. Technol., 2014, 60, 24–31 CrossRef
.
- J. A. Tejeda-Rodríguez, A. Núñez, F. Soto, V. García-Gradilla, R. Cadena-Nava, J. Wang and R. Vazquez-Duhalt, ChemNanoMat, 2019, 5, 194–200 CrossRef
.
- J. A. Speir, S. Munshi, G. Wang, T. S. Baker and J. E. Johnson, Structure, 1995, 3, 63–78 CrossRef CAS
.
- L. Lavelle, J. P. Michel and M. Gingery, J. Virol. Methods, 2007, 146, 311–316 CrossRef CAS
.
- K. W. Adolph, J. Gen. Virol., 1975, 28, 147–154 CrossRef
.
- D. L. D. Caspar and A. Klung, in Cold Spring Harb. Symp. Quant. Biol., Cold Spring Harbor Laboratory Press, New York, United States, 1962, vol. 27, pp. 1–24 Search PubMed
.
- L. Lavelle, M. Gingery, M. Phillips, W. M. Gelbart, C. M. Knobler, R. D. Cadena-Nava, J. R. Vega-Acosta, L. A. Pinedo-Torres and J. Ruiz-Garcia, J. Phys. Chem. B, 2009, 113, 3813–3819 CrossRef CAS
.
- K. W. Adolph and P. J. G. Butler, J. Mol. Biol., 1974, 88, 327–341 CrossRef CAS
.
- G. W. Wagner, Virology, 1968, 756, 748–756 CrossRef
.
- R. F. Garmann, M. Comas-Garcia, M. S. T. Koay, J. J. L. M. Cornelissen, C. M. Knobler and W. M. Gelbart, J. Virol., 2014, 88, 10472–10479 CrossRef
.
- Y. Hu, R. Zandi, A. Anavitarte, C. M. Knobler and W. M. Gelbart, Biophys. J., 2008, 94, 1428–1436 CrossRef CAS
.
- I. J. Minten, Y. Ma, M. A. Hempenius, G. J. Vancso, R. J. M. Nolte and J. J. L. M. Cornelissen, Org. Biomol. Chem., 2009, 7, 4685–4688 RSC
.
- M. V. de Ruiter, R. M. van der Hee, A. J. M. Driessen, E. D. Keurhorst, M. Hamid and J. J. L. M. Cornelissen, J. Controlled Release, 2019, 307, 342–354 CrossRef CAS
.
- M. B. Van Eldijk, J. C. Y. Wang, I. J. Minten, C. Li, A. Zlotnick, R. J. M. Nolte, J. J. L. M. Cornelissen and J. C. M. Van Hest, J. Am. Chem. Soc., 2012, 134, 18506–18509 CrossRef CAS
.
- L. Schoonen, R. J. M. Maas, R. J. M. Nolte and J. C. M. van Hest, Tetrahedron, 2017, 73, 4968–4971 CrossRef CAS
.
- S. B. P. E. Timmermans, D. F. M. Vervoort, L. Schoonen, R. J. M. Nolte and J. C. M. van Hest, Chem.–Asian J., 2018, 13, 3518–3525 CrossRef CAS
.
- L. Schoonen, J. Pille, A. Borrmann, R. J. M. Nolte and J. C. M. Van Hest, Bioconjugate Chem., 2015, 26, 2429–2434 CrossRef CAS
.
- L. Schoonen, R. J. M. Nolte and J. C. M. Van Hest, Nanoscale, 2016, 8, 14467–14472 RSC
.
- L. Schoonen, S. Eising, M. B. Van Eldijk, J. Bresseleers, M. Van Der Pijl, R. J. M. Nolte, K. M. Bonger and J. C. M. Van Hest, Bioconjugate Chem., 2018, 29, 1186–1193 CrossRef CAS
.
- I. J. Minten, K. D. M. Wilke, L. J. A. Hendriks, J. C. M. Van Hest, R. J. M. Nolte and J. J. L. M. Cornelissen, Small, 2011, 7, 911–919 CrossRef CAS
.
- L. Schoonen, S. Maassen, R. J. M. Nolte and J. C. M. Van Hest, Biomacromolecules, 2017, 18, 3492–3497 CrossRef CAS
.
- J. A. Speir, B. Bothner, C. Qu, D. A. Willits, M. J. Young and J. E. Johnson, J. Virol., 2006, 80, 3582–3591 CrossRef CAS
.
- L. O. Liepold, J. Revis, M. Allen, L. Oltrogge, M. Young and T. Douglas, Phys. Biol., 2005, 2, S166–S172 CrossRef CAS
.
- Y. Wu, J. Li, H. Yang, J. Seoung, H. Lim, G. Kim and H. Shin, Biotechnol. Bioprocess Eng., 2017, 708, 700–708 CrossRef
.
- J. D. Perlmutter and M. F. Hagan, Annu. Rev. Phys. Chem., 2015, 66, 217–239 CrossRef CAS
.
- S. Katen and A. Zlotnick, in Methods Enzymol, ed. M. L. Johnson, J. M. Holt and G. K. Ackers, Elsevier, San Diego, United States, 2009, vol. 455, pp. 395–417 Search PubMed
.
- C. Pretto and J. C. M. Van Hest, Bioconjugate Chem., 2019, 30, 3069–3077 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07612k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |