Open Access Article
Open Access ArticleSynthesis of aryl azide chain-end functionalized N-linked glycan polymers and their photo-labelling of specific protein†
Ka Keung Chana,
Qiaoshi Leib,
Jinshan Tang b and
Xue-Long Sun
b and
Xue-Long Sun *a
*a
aDepartment of Chemistry, Chemical and Biomedical Engineering, Center for Gene Regulation in Health and Disease (GRHD), Cleveland State University, 2121 Euclid Avenue, Cleveland, Ohio 44115, USA. E-mail: x.sun55@csuohio.edu
bInstitute of Traditional Chinese Medicine & Natural Products, College of Pharmacy, Jinan University, West 601, Huangpu Avenue, Guangzhou, People's Republic of China
First published on 19th October 2020
Abstract
We report a straightforward synthesis of aryl azide chain-end functionalized N-linked glycan polymers and its application for affinity-assisted photo-labelling of specific protein. The aryl azide chain-end functionalized N-glycan polymers, including N-galactosyl, N-glucosyl, and N-lactosyl polymer, were synthesized from free glycan via glycosylamine intermediates followed by acrylation and polymerization via cyanoxyl-mediated free radical polymerization (CMFRP) in a one-pot fashion. The aryl azide chain-end functionalized N-glycan polymers were characterized by 1H NMR and IR spectroscopy. The affinity-assisted photo-labeling capabilities of the aryl azide N-glycan polymers were demonstrated with aryl azide N-lactosyl polymer as a ligand for β-galactose-specific lectin from Arachis hypogaea (PNA) after UV irradiation and confirmed by SDS-PAGE with silver staining. Overall, the aryl azide chain-end functionalized N-linked glycan polymers will be useful multivalent ligands for specific protein labelling and functionality studies.
Cell surface carbohydrates, existing as glycoproteins, glycolipids, and proteoglycans, often serve as receptors to relay critical interactions and trigger crucial signalling events in many biological processes, such as cell–cell interactions, pathogen/host interactions, tumor metastasis, immune responses, and many more.1 Glycoscience research specifically concerning the carbohydrate–protein interactions can provide an abundance of opportunities to discover the molecular mechanisms of biological processes, potential therapeutic targets, and diagnostic mechanisms for various diseases.1 Therefore, sensitive profiling of carbohydrate–protein interactions is the initial and key step for basic research, clinical diagnostics, and therapeutic applications. In fact, the diversity and complexity of glycan structures, together with their crucial roles in many physiological or pathological processes, require the development of effective tools for the comprehensive analysis of carbohydrate–protein interactions.2 Multifunctional glyco-ligands with other functional groups, such as labelling and probing units, facilitate the proteome-wide identification of various carbohydrate-binding proteins.3–5 Photo-crosslinking functional groups, such as aryl azides, benzophenones and diazirines, can generate highly reactive species that react with adjacent molecules, resulting in direct covalent modifications, which can be used for target imaging, isolation and identification applications.3–7
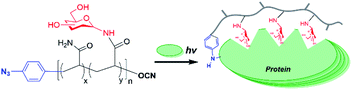
Glycopolymers, polymers with glycan pendant groups, have been widely used as multivalent glyco-ligand as they mimic native glycoprotein structure and functions.8 Glycopolymers with both glycans and photoactivatable crosslinkers pending on the polymer backbone have been used for target protein labelling, but have substantial problems for both accessibility to the target and efficiency of crosslinking due to hinderances from each unit.5 Chain-end functionalized glycopolymers have been demonstrated in many potential applications, such as in microarray,9 cells surface re-engineering10 and glyco-engineering11 as the chain functional group facilitates site-specific and oriented glycopolymer immobilization as well as end-to-end protein–glycopolymer conjugation. Herein, we proposed an aryl azide chain-end functionalized glycopolymer as multifunctional glyco-probe for specific protein labelling and functionality study (Fig. 1). Specifically, the multivalent sugars pending the polymer backbone will facilitate specific binding to the target protein, while the single chain-end aryl azide will serve as photoactivatable crosslinker for labelling of the bound protein.
To construct efficient glycopolymers, glycan linkage must be taken into consideration when mimicking native glycosylation, which affects significant protein properties, namely stability, folding, molecular recognition, and immunogenicity. The common types of glycosylation are N-linked and O-linked. N-Linked glycans possess both intrinsic and extrinsic functions, where intrinsically, they provide structural components to the cell membrane and extracellular matrix; while extrinsically, they direct the trafficking of glycoproteins and mediate immune signalling.1,12–14 Also, the importance of N-linked glycosylation has been becoming increasingly evidential with therapeutic proteins in the market, such as Etanercept, Infliximab and Rituximab, which are all N-glycosylated glycoproteins.12,14–16 Therefore, N-linked glycan polymers that closely mimic the natural N-glycan conjugates will be effective multivalent glyco-ligands for specific protein targeting and functionality study. Herein, we report an aryl azide chain-end functionalized N-linked glycan polymer as glyco-probe for specific protein labeling and functionality study (Fig. 1). The affinity-assisted photo-labeling capabilities of the aryl azide N-linked glycan polymer was demonstrated with aryl azide N-lactosyl polymer as a ligand for β-galactose-specific lectin from Arachis hypogaea (peanut agglutinin, PNA) and N-glucosyl polymer as a ligand for Glc-specific lectin from Canavalia ensiformis (Concanavalin A, ConA) after UV irradiation followed by SDS-PAGE confirmation with silver staining.
Cyanoxyl-mediated free radical polymerization (CMFRP) has been demonstrated for a straightforward synthesis of chain-end functionalized O-linked glycopolymers.17 In this report, aryl azide was employed as the initiator for the CMFRP with the mixture of N-acrylated glycomonomer and acrylamide to achieve N-linked glycan polymers with a chain-end photo-labelling functionality. First, free glycans were treated with ammonium bicarbonate in ammonium oxide to generate glycosylamines quantitatively,18,19 then the N-acryloyl group was introduced by treatment with acryloyl chloride in methanol–water while in the presence of sodium carbonate at 0 °C, followed by the removal of excess acryloyl chloride and sodium carbonate to yield N-acryloyl-glycosylamines as glycomonomers. Next, 4-azidoaniline was used as the initiator for CMFRP scheme in one-pot fashion to afford N-linked glycan polymers (Scheme 1). Overall, aryl azide chain-end functionalized N-glucosyl polymer, N-galactosyl polymer, and N-lactosyl polymer were synthesized in very good yields (over 60% yield).
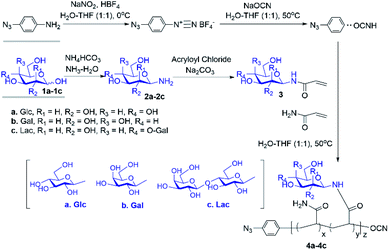 | ||
| Scheme 1 Straightforward synthesis of N-linked glycan polymers (4) from free glycans (1) via glycosylamine intermediates (2) followed by acrylation (3) and CMFRP. | ||
The progress of N-glycan polymer synthesis was monitored by 1H NMR spectra. As shown in Fig. 2 for N-lactosyl polymer, the successful amination of free lactose was confirmed by the signals of anomeric proton directly adjacent to the introduced amine (4.05 ppm), while the introduction of acrylamide was confirmed by the signals of protons on the alkene (5.82 and 6.25 ppm) as well as the downfield signal shift of the anomeric proton directly adjacent to amine (5.05 ppm) (Fig. 2). The saccharide density and average molecular weight of the resultant glycopolymers were also determined by 1H NMR spectrum. For example, for N-lactosyl polymer, the integration of protons on the terminal phenyl group (7.1 and 6.9 ppm), sugar anomeric carbons (4.4 and 4.9 ppm), sugar backbones (3.40 to 3.80 ppm) and polymer backbones (2.10 to 2.30 ppm and 1.80 to 1.30 ppm) were assigned and calculated for its saccharide density of 57 and an average molecular weight of 78,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 970 Mn (Fig. 2). In addition, the chain-end azido and O-cyanate functional groups of the glycopolymers were confirmed by FT-IR spectrum, in which azide absorption was observed at 2118.6 cm−1 while O-cyanate absorption was observed at 2217.6 cm−1 (Fig. 3B). In comparison, aryl chloride chain-end functionalized N-lactosyl polymer synthesized in our previous report9 showed only O-cyanate absorption at 2217.6 cm−1 (Fig. 3A).
970 Mn (Fig. 2). In addition, the chain-end azido and O-cyanate functional groups of the glycopolymers were confirmed by FT-IR spectrum, in which azide absorption was observed at 2118.6 cm−1 while O-cyanate absorption was observed at 2217.6 cm−1 (Fig. 3B). In comparison, aryl chloride chain-end functionalized N-lactosyl polymer synthesized in our previous report9 showed only O-cyanate absorption at 2217.6 cm−1 (Fig. 3A).
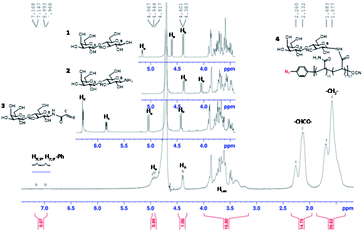 | ||
| Fig. 2 1H NMR spectrum of β-lactose (1), β-D-lactopyranosylamine (2), N-(prop-2-enoyl)-β-D-lactopyranosylamine (3), and p-azido-phenyl-β-D-Gal(1–4)-β-D-Glc-N-glycopolymer (4), (D2O, 400 MHz NMR). | ||
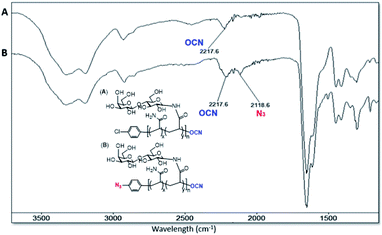 | ||
| Fig. 3 FT-IR spectrum of p-Cl-phenyl-β-D-Gal(1–4)-β-D-Glc-N-glycopolymer (A) and p-azido-phenyl-β-D-Gal(1–4)-β-D-Glc-N-glycopolymer (B). | ||
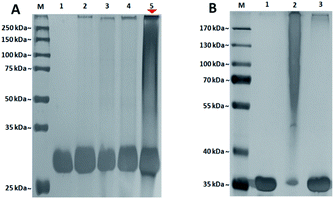
Once the aryl azide chain-end glycopolymers were in hands, their ability to photo-label a specific protein was evaluated with the aryl azide chain-end N-lactosyl polymer as ligand for the Galβ-1,4-Glc-specific Arachis hypogaea (PNA). Briefly, the aryl azide chain-end N-lactosyl polymer was incubated with PNA (lyophilized powder, Millipore-Sigma) in PBS buffer (pH 7.4) solution containing 0.2% Tween 20 for 4 hours at room temperature, then, while sitting on ice, exposed to a 366 nm UV source in the dark for 30 min. As controls, aryl azide chain-end acrylamide polymer, aryl azide chain-end N-glucosyl polymer and aryl azide chain-end N-galactosyl polymer were tested with PNA in the same manner, respectively. To evaluate which glycopolymer was accounted for selective ligand–protein binding with PNA, the UV-irradiated glycopolymer–PNA conjugates were subjected to SDS-PAGE and silver staining (Pierce™ Silver Stain Kit, ThermoFisher) (Fig. 4).
PNA is a homotetrameric protein with specificity for β-O-Gal such as Galβ-1,4-Glc in β-lactose, which in theory should display affinity only for the N-lactosyl polymer, but not acrylamide polymer, N-glucosyl polymer and N-galactosyl polymer. As shown in Fig. 4, only N-lactosyl polymer-PNA affinity-assisted photo-labeled complexes exhibited a definitive darkened smear staining (Fig. 4A, lane 5), which confirmed the formation of supramolecules via specific binding and photo-crosslinking. Like the N-lactosyl polymer, the other polymers all possessed the aryl azide chain-end groups but did not have β-O-Gal necessary for PNA affinity, which showed no noteworthy photo-crosslinking (Fig. 4A, lane 2, 3, and 4). Observations can be made that slight smearing exhibited in all lanes except for the negative control where only PNA was present, suggesting the expected functionality of the aryl azide groups, which may crosslink with proteins when they are close enough due to non-specific bindings. The SDS-PAGE results also indicated that it was, for the most part, unnecessary to wash away unbound glycopolymers before UV irradiation, as glycopolymers without assistance of protein affinity displayed drastically lower degree of supramolecule formation following photo-induced crosslinking, suggesting that specific affinity played a vital role in establishing proximity between the aryl azide functional groups and the amino acids of the proteins, where crosslinking and covalent bonding would take place. Compared to the single band of unlabeled protein, the smear staining of the photo-labeled proteins indicated variously-sized supramolecular formation due to multiple glycopolymers cross-linking to each lectin unit of homotetrameric PNA20 via the single aryl (azide) chain-end. Specifically, some lectin molecules have more glycopolymers conjugated while others have fewer glycopolymers conjugated, all together generating a mixture of lectin–glycopolymer conjugates as smear bands in SDS-PAGE. This phenomenon is often seen in protein–glycopolymer SDS-PAGE characterizations.21–23
In addition, an photocrosslinking experiment with aryl azide chain-end functionalized N-glycan polymers and ConA lectin, which specifically binds to α-Glc and α-Man,24 was conducted. As a result, aryl azide chain-end functionalized N-lactosyl polymer did not achieve any photo-crosslinking with ConA lectin, while aryl azide chain-end functionalized N-glucosyl polymer showed specific photo-crosslinking with ConA lectin (Fig. 4B). This result further confirmed the specific photocrosslinking capacity of the aryl azide chain-end functionalized N-lactosyl polymer for the target lectin PNA.
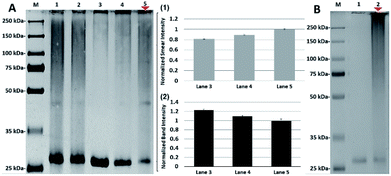
To further investigate the specificity of N-lactosyl polymer for PNA labelling, selected control conditions focusing on competitivity and concentration were performed (Fig. 5). For example, free β-lactose disaccharide was introduced to PNA either competitively alongside N-lactosyl polymer or sequentially prior to the introduction of N-lactosyl polymer followed by UV irradiation. Both conditions displayed smears in a similar manner (Fig. 5A, lane 1 and 2) and intensity as determined using ImageJ with the former possibly being slightly more intense than the latter (Fig. 5A, lane 1 and 2). This observation can contribute to the consensus that the multivalent nature of carbohydrates presented on polymer backbones is much more preferred by lectins as multivalent ligands compared to monosaccharide single-unit ligands. In addition, a gradient can be achieved in the intensity of the smears when the reaction molar ratio between N-lactosyl polymer and PNA is manipulated. As a result, when the molar ratio between N-lactosyl polymer and PNA are equal, fewer supramolecules were formed following affinity-assisted photo-labelling (Fig. 5A, lane 3), however, proportionally more supramolecules were formed when more N-lactosyl polymer was used as observed using ImageJ measurements (Fig. 5A, lane 4 and 5). Roughly, 20% increase was measured in smear staining intensity when the molar ratio of N-lactosyl polymer to PNA was increased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 6
1 to 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 5A(1)), which is coherent with a 20% decrease measured in band intensity in respective lanes (Fig. 5A(2)). These results suggest that the level of crosslinking is directly correlated to the molar ratio between the ligand and protein, indicating the existence of a specific interaction between N-lactosyl polymer and PNA.
1 (Fig. 5A(1)), which is coherent with a 20% decrease measured in band intensity in respective lanes (Fig. 5A(2)). These results suggest that the level of crosslinking is directly correlated to the molar ratio between the ligand and protein, indicating the existence of a specific interaction between N-lactosyl polymer and PNA.
Finally, to investigate the specific reactivity of aryl azide of the N-lactosyl polymer for PNA labeling, aryl chloride chain-end functionalized N-lactosyl polymer synthesized in our previous report9 was investigated for the same photo-crosslinking procedure for aryl azide N-lactosyl polymer with PNA (Fig. 5B). As a result, aryl chloride chain-end functionalized N-lactosyl polymer did not achieve any photo-crosslinking with PNA (Fig. 5B, lane 1) compared with aryl azide of the N-lactosyl polymer, which showed apparent PNA labelling (Fig. 5B, lane 2). This observation confirmed that the chain-end aryl azide generates highly reactive species that reacts with adjacent molecules, resulting in the direct covalent modification of proteins.
In summary, we have developed aryl azide chain-end functionalized N-linked glycan polymers as photoactivatable glyco-ligands and characterized their application for affinity-assisted photo-labelling of the specific protein. CMFRP is an attractive method for the synthesis of chain-end functionalized N-linked glycan polymers, as it possesses several advantages, such as direct polymerization, reaction in aqueous solution, exclusion of tedious protection and deprotection steps, and compatibility with a broad range of functional groups. The present synthetic method can be applied to synthesizing polymers with any kind of glycan and alternative chain-end functionalizable groups, such as benzophenone and diazirine. On the other hand, the reported aryl azide chain-end functionalized N-linked glycan polymers have demonstrated undeniable capabilities for specifically labelling proteins and will be useful multivalent ligands for specific protein labelling and functionality study both in vitro and in vivo. In addition, the present polymers can undergo further modification to its O-cyanate chain-end group to yield an additional chain-end functional group, such as a biotin or fluorophore, thus achieving dual functionalities on both ends of the polymers for protein imaging and proteomics applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Faculty Research Fund from the Center for Gene Regulation in Health and Disease (GRHD) and Faculty Research Development (FRD) Fund (X.-L. Sun) and Graduate Student Research Award (K. K. Chan) at Cleveland State University.Notes and references
- A. Varki, Glycobiology, 2017, 27, 3–49 CrossRef CAS
.
- L. L. Kiessling and R. A. Splain, Annu. Rev. Biochem., 2010, 79, 619–653 CrossRef CAS
.
- H. Wu and J. Kohler, Curr. Opin. Chem. Biol., 2019, 53, 173–182 CrossRef CAS
.
- A. Fujita and J. J. Kohler, Trends Glycosci. Glycotechnol., 2015, 156, E1–E7 Search PubMed
.
- A. Wibowo, E. C. Peters and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2014, 136, 9528–9531 CrossRef CAS
.
- G. Mattson, E. Conklin, S. Desai, G. Nielander, M. D. Savage and S. Morgensen, Mol. Biol. Rep., 1993, 17, 167–183 CrossRef CAS
.
- P. Yella Reddy, S. Kondo, T. Toru and Y. Ueno, Chemistry of Protein Conjugation and Cross-linking, 1996, vol. 36, pp. 2652–2654 Search PubMed
.
- Y. Miura, Polym. J., 2012, 44, 679–689 CrossRef CAS
.
- S. N. Narla and X.-L. Sun, Lab Chip, 2012, 12, 1656–1663 RSC
.
- M. L. Huang, R. A. Smith, G. W. Trieger and K. Godula, J. Am. Chem. Soc., 2014, 136, 10565–10568 CrossRef CAS
.
- H. Zhang, J. Weingart, V. Gruzdys and X.-L. Sun, ACS Macro Lett., 2016, 5, 73–77 CrossRef CAS
.
- W. Dennis, I. R. Nabi and M. Demetriou, Cell, 2009, 139, 1229–1241 CrossRef
.
- R. J. Solá and K. Griebenow, BioDrugs, 2010, 24, 9–21 CrossRef
.
- M. Dalziel, M. Crispin, C. N. Scanlan, N. Zitzmann and R. A. Dwek, Science, 2014, 343, 1235681 CrossRef
.
- M. Sinclair and S. Elliott, J. Pharm. Sci., 2005, 94, 1626–1635 CrossRef
.
- A. M. Sinclair and S. Elliott, J. Pharm. Sci., 2005, 94, 1626–1635 CrossRef CAS
.
- S. Hou, X.-L. Sun, C.-H. Dong and E. L. Chaikof, Bioconjugate Chem., 2004, 15, 954–959 CrossRef CAS
.
- J. Tang, E. Ozhegov, Y. Liu, D. Wang, X. Yao and X.-L. Sun, ACS Macro Lett., 2017, 6, 107–111 CrossRef CAS
.
- L. M. Likhosherstov, O. S. Novikova, V. A. Derevitskaja and N. K. Kochetkov, Carbohydr. Res., 1986, 146, C1–C5 CrossRef CAS
.
- S. Mukhopadhyay, P. K. Panda, B. Behera, C. K. Das, M. K. Hassan, D. N. Das, N. Sinha, A. Bissoyi, K. Pramanik, T. K. Maiti and S. K. Bhutia, Food Chem. Toxicol., 2014, 64, 369–377 CrossRef CAS
.
- T. H. Nguyen, S. H. Kim, C. G. Decker, D. Y. Wong, J. A. Loo and H. D. Maynard, Nat. Chem., 2013, 5, 221–227 CrossRef CAS
.
- K. L. Heredia, G. N. Grover, L. Tao and H. D. Maynard, Macromolecules, 2009, 42, 2360–2367 CrossRef CAS
.
- B. A. Killinger and A. Moszczynska, Anal. Chem., 2016, 88, 4071–4084 CrossRef CAS
.
- I. J. Goldstein and R. D. Poretz, Lectins, 1986, 2, 33–247 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra08400j |
| This journal is © The Royal Society of Chemistry 2020 |