Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
High salt compatible oxyanion receptors by dual ion imprinting†
Sudhirkumar
Shinde‡
 ,
Mona
Mansour§
,
Mona
Mansour§
 ,
Anil
Incel
,
Anil
Incel
 ,
Liliia
Mavliutova
,
Liliia
Mavliutova
 ,
Celina
Wierzbicka
,
Celina
Wierzbicka
 and
Börje
Sellergren
and
Börje
Sellergren
 *
*
Department of Biomedical Sciences, Faculty of Health and Society, Malmö University, 20506 Malmö, Sweden. E-mail: borje.sellergren@mau.se
First published on 14th April 2020
Abstract
The design of hosts for either cations or anions is complicated due to the competition for binding by the host or guest counterions. Imprinting relying on self-assembly offers the possibility to stabilize the guest and its counterion in a favorable geometry. We here report on a comprehensive supramolecular approach to anion receptor design relying on concurrent recognition of both anion and cation. This was achieved by high order complex imprinting of the disodium salt of phenyl-phosphonic acid in combination with neutral urea and sodium ion selective 18-crown-6 monomers. The polymers displayed enhanced affinity for the template or inorganic phosphate or sulfate in competitive aqueous buffers, with affinity and selectivity increasing with increasing ionic strength. The presence of engineered sites for both ionic species dramatically increases the salt uptake in strongly competitive media such as brine.
Introduction
Anion recognition drives a multitude of processes which are crucial for the living cell.1,2 The work horses in these processes are anion receptors refined by evolution to discriminatively bind anions in water. The performance of these receptors becomes particularly impressive when considering that they can exert their action in extreme environments in highly competitive media of high ionic strength. An often cited example in this regard refers to the proteins designed to specifically bind the pyramidal oxoanions phosphate, sulfate and arsenate.2–4 Discrimination between them occurs predominantly through multiple complementary hydrogen bonding involving main chain amides (nests) in a water poor microenvironment with less contribution by charged residues.5 The ability of phosphate binding protein to select phosphate over the isosteric arsenate with a selectivity factor of 4500 and sulfate binding protein to bind sulfate specifically with a binding constant Ka = 106 M−1 in water (pH 5–8) reflect the perfection of these receptors. Hence, this overturns the Hofmeister series of the salting out tendency for anions which otherwise falls in the order: SO42− > HPO42− > acetate.From the above perspective, efforts to design biomimetic anion hosts fall short. Anion recognition by neutral synthetic receptors can be highly selective but the receptors are challenging to construct since hydrogen bonding is considerably weaker in polar solvents.2,6 Moreover, ion recognition is complicated by the necessary presence of their counterions. For example, anion binding is electrostatically screened in high salt media, a situation commonly exploited in ion exchange chromatography for modulating ion retention.7 To avoid this screening effect, recognition at the air water interface8 or dual ion receptors9,10 comprising preorganized receptors for both anion and cation have been reported. The latter employ combinations of known cation or anion recognition motifs held together by appropriately designed spacers. In spite of the promising progress this approach suffers from significant synthetic challenges in correctly placing the anion and cation hosts to match the ion separation distance, hence requiring a priori knowledge about ion pair solvation i.e. contact or solvent separated.
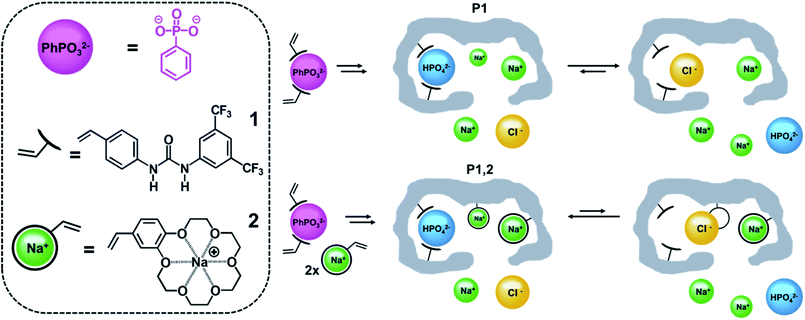
This dilemma we believe can be addressed by turning from bottom up design to top down strategies relying on self-assembly. Hence potent anion receptors can be prepared by polymerizing host monomers and a crosslinker in presence of the anion guest followed by guest removal.11–21 The anion preorganizes the host which is covalently fixed in a macromolecular scaffold with tunable local polarity. Permanent imprinted sites are achieved post template removal featuring enhanced affinity for the templated ion. This concept can be further exploited to incorporate sites for the counterion. Hence, combining anion and cation host monomers we anticipate will spontaneously lead to dual ion receptors¶ with optimally adjusted interhost distance for recognition in competitive aqueous media.22 To demonstrate this concept, we have here compared urea-based imprinted monoion phosphate receptors with corresponding dual ion receptors targeting sodium or potassium salts (Fig. 1).
Results and discussion
Solution complex formation and polymer preparation
The dual ion imprinting receptor was constructed using 1,3-diarylurea monomer 1 as anion host and vinyl-benzo-18-crown-6 (monomer 2)23 as polymerizable macrocyclic cation host (Fig. 1). The former acts as twofold hydrogen bond donor to complex oxyanion guests and can be used to prepare high affinity anion receptors for highly diverse set of guests e.g. phosphopeptides, sulfopeptides, phospholipids, sugar acids, carboxylates.14–17,19 Complexation can be easily induced by proton transfer from the acid to bulky amine bases such as pentamethylpiperidine (PMP) or by the use of quaternary ammonium counterions (e.g. tetrabutylammonium, TBA) which moreover have been shown to produce pronounced counterion memory effects.14,24Crownethers on the other hand are macrocyclic hosts complexing size matched metal ions with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host guest stoichiometry.25–27 The 18-membered ring macrocycle 18-crown-6 (18C6) features a cavity size matching the abundant alkali cations sodium and potassium but with a strongly solvent dependent affinity (entries 3–5 in Table 1). Exposed to water its amphiphilic nature leads to cavity collapse and loss of binding but this may be counteracted by embedding it in scaffolds providing lower polarity microenvironments.
1 host guest stoichiometry.25–27 The 18-membered ring macrocycle 18-crown-6 (18C6) features a cavity size matching the abundant alkali cations sodium and potassium but with a strongly solvent dependent affinity (entries 3–5 in Table 1). Exposed to water its amphiphilic nature leads to cavity collapse and loss of binding but this may be counteracted by embedding it in scaffolds providing lower polarity microenvironments.
| Entry | Guest | Host | K a (M−1) | Solvent |
|---|---|---|---|---|
| a Determined by 1H-NMR titrations from the average of the individual complexation induced shifts of both urea protons. b Determined by conductometry of sodiumperchlorate solutions.28 c See ref. 15. | ||||
| 1 | PPA·TBA | 1 | 7005 ± 985a,c | DMSO-d6 |
| 2 | PPA·Na–18C6 | 1 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 181 ± 1655a 181 ± 1655a |
DMSO-d6 |
| 3 | Na+ | 18C6 | 25b | DMSO |
| 4 | Na+ | 18C6 | 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 622b 622b |
MeCN |
| 5 | Na+ | 18C6 | 6.3b | H2O |
To investigate whether monomer 2 could promote the association of host monomer 1 with the mono-sodium salt of phenylphosphonic acid (PPA·Na) we carried out 1H-NMR titrations in DMSO-d6 (Fig. S1 and S2†) and compared it with our previous records for the corresponding titration with PPA·TBA (Table 1). The titration of 1 with PPA·TBA in DMSO-d6 could previously be modelled using a one site host–guest model resulting in a binding constant Ka = 7005 M−1.
A different result was obtained using the crown-ether stabilized guest (Fig. S1 and S2†). Steep downfield shifts were observed for the urea protons Ha and Hb which inflected abruptly at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host guest ratio with only minor changes observed beyond this guest level. This isotherm was best fitted with the Hill equation resulting in a significantly higher association constant of Ka = 11
1 host guest ratio with only minor changes observed beyond this guest level. This isotherm was best fitted with the Hill equation resulting in a significantly higher association constant of Ka = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 181 M−1. A Hill coefficient of 1.7 indicates a strongly positive cooperativity, possibly caused by the multiple equilibria involved in forming the higher order complexes.
181 M−1. A Hill coefficient of 1.7 indicates a strongly positive cooperativity, possibly caused by the multiple equilibria involved in forming the higher order complexes.
Imprinted and nonimprinted polymers were prepared and characterised using the urea host monomers 1 and 2 as listed in Table S1† (Fig. 1). Nonimprinted polymers (PN) were prepared identically to the imprinted polymers but omitting the template. Characterisation of the polymers by scanning electron microscopy (SEM) (Fig. S3†), elemental analysis and transmission FTIR (Fig. S4†) gave data supporting the formation of polymers with a macroporous morphology, a stoichiometric monomer incorporation reflecting the feed ratio and a successful template removal.
pH Dependence of ion recognition in buffer
To evaluate the anion recognition properties of the materials we assessed their ability to bind both organic (PPA and phenylsulphonic acid PSA) and inorganic (HPO42− and SO42−) oxyanions. The polymers were incubated in buffers adjusted to different pH values in presence of the sodium salts of the anions. Fig. 2A and S5† show the uptake of PPA and PSA by the different polymers in the pH range 1–9.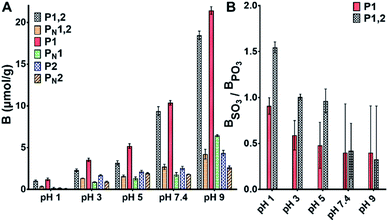 | ||
| Fig. 2 (A) Binding of PPA (0.6 mM) on PPA imprinted polymers in buffers of different pH. (B) Sulfo-selectivity expressed as the ratio of bound PSA to PPA based on the binding data in Fig. 2A and S5.† | ||
First, we note that binding increases with increasing pH, the trend being more pronounced for PPA compared to PSA. This is most likely related to the different protonation states of the two anions and the stronger hydration tendency of sulphate. The steepest increase in anion uptake is observed for the imprinted materials P1 and P1,2 containing anion host monomer 1 whereas the polymers prepared using host monomer 2 alone as in P2 did not display strong imprinting. In agreement with our previous observations,15 acidic conditions favored selectivity for sulphonate. This is reflected in the selectivity factor shown in Fig. 2B. Interestingly, only P1,2 featured a PhSO3/PhPO3 selectivity factor exceeding 1 under such conditions.
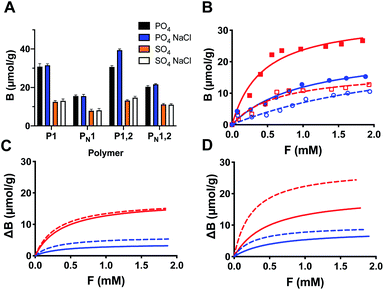
We then investigated the ion selectivity of all polymers in buffer pH 9. Binding of the inorganic anions was measured by conductometry with both anions carrying a net 2-fold negative charge (Fig. 3A). As expected, the polymers showed a preference for phosphate over sulphate and imprinted polymers showed a significantly larger uptake than the nonimprinted reference polymers. To prove the presence of dual ion receptors we then performed the same experiment in presence of 1 M NaCl, corresponding to ca. 1/5th of a saturated salt solution (brine). As seen in Fig. 3A the salt had a positive effect on the ion binding to P1,2 whereas binding to P1 appeared less affected. These results are contrary to salt induced ionic screening effects seen in charge driven molecular recognition. We attribute this effect to the colocalization of both cation and anion hosts caused by the imprinting process.
Adsorption isotherms and binding parameters in low and high salt media
The binding-energy distributions of the polymers P1 and P1,2 and their corresponding nonimprinted polymers are given by single-component adsorption isotherms determined by batch equilibration in 0.1 M sodium bicarbonate buffer at pH 9 (Fig. 3B and S6†). The imprinted polymers consistently displayed a higher uptake of both PPA and PSA compared to their nonimprinted counterparts with the isotherms showing clear saturation behaviour. This contrasts with the results for the nonimprinted polymers were only the PPA isotherms displayed curvature whereas those of PSA appeared linear (Fig. S6†). As expected from the initial batch binding experiments in Fig. 3A, the saturation capacity for PPA is overall higher than that for PSA. Fitting these data with the one site host–guest model resulted in the curves shown in Fig. S6† with the fitting parameters Ka and Bmax given in Table S2.† The preference for PPA is reflected in the higher association constants recorded for this anion again with the highest values obtained for the imprinted polymers.The same experiment was then performed in presence of 1 M NaCl. Overall, this resulted in little or no change in binding affinity for P1 whereas P1,2 now bound the oxyanions more tightly. This agrees with the results reported in Fig. 3A and shows a clear positive effect of salt on the anion binding affinity of P1,2. Hence, P1,2 showed the highest affinity with a Ka = 3700 M−1 and a Bmax = 45 μmol g−1 for PPA (Table S2†).
To highlight the contribution to binding caused by the template effect we subtracted binding to the nonimprinted from the imprinted polymer (assuming the former to reflect the nonspecific binding contribution) (Fig. 4) and compared the resulting binding parameters in graphic format (Fig. 3C and D). The graphs offer a convincing evidence for the synergistic effect of the dual ion host on the binding affinity and capacity. Whereas this host (P1,2) displayed a concomitant increase in both binding constant (Ka = 2000 to 4000 M−1 for PPA) and saturation capacity (Bmax = 20 to 28 μmol g−1) in the presence of 1 M NaCl, the corresponding mono-ion host (P1) showed a decreased PPA affinity (Ka = 3100 to 2600 M−1) and no change in capacity (Bmax = 18 μmol g−1).
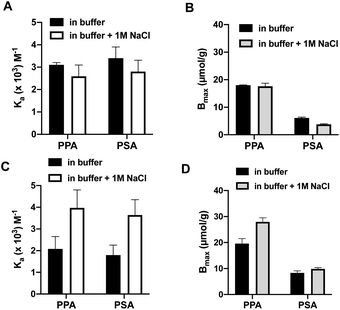 | ||
| Fig. 4 Association constants (Ka) (A and C) and binding capacities (Bmax) (B and D) for PPA and PSA interacting with P1 (A and B) and P1,2 (C and D) in 0.1 M sodium bicarbonate buffer pH 9 with or without addition of extra salt (1 M NaCl). The binding parameters were derived from the corrected curves in Fig. 3C and D. | ||
Conclusions
Charge neutral receptors interacting with ligands via hydrogen bonding have for long been associated with poor compatibility with aqueous media. Presence of water and other protic solvents effectively disrupt the host–guest interactions in these systems. Recently however some exceptions to this rule have been reported although neutral receptors displaying affinity for phosphate and sulfate in pure water or buffer or as here investigated, high levels of salt, are rare.2 The concept of dual ion hosts suggests a way forward.9 These display high affinity and selectivity under physiological conditions by providing recognitive sites for both cation and anion in a geometry defined by the covalent chemistry used to link the two hosts. As shown in this work, template driven self assembly of the two hosts offers a straightforward alternative to construct these receptors. Combining macrocyclic cation hosts with neutral urea-based anion receptors can thus significantly boost ion affinity and capacity making these receptors a possible alternative for ion scavenging applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support from the Marie Skłodowska-Curie Actions (H2020-MSCA-ITN-2016, 722171), Vetenskapsrådet (VR) under grant number 2014-3794 and the Sweden Knowledge-Foundation (KKS) for research under project number 20150086. CW acknowledges support from KKS-Prospekt program under project number 20170040.Notes and references
- Supramolecular chemistry in water, ed. S. Kubik, Wiley-VCH, 2019 Search PubMed.
- M. J. Langton, C. J. Serpell and P. D. Beer, Angew. Chem., Int. Ed. Engl., 2016, 55, 1974–1987 CrossRef CAS PubMed.
- P. Chakrabarti, J. Mol. Biol., 1993, 234, 463–482 CrossRef CAS PubMed.
- A. E. Hargrove, S. Nieto, T. Zhang, J. L. Sessler and E. V. Anslyn, Chem. Rev., 2011, 111, 6603–6782 CrossRef CAS PubMed.
- J. D. Watson and E. J. Milner-White, J. Mol. Biol., 2002, 315, 171–182 CrossRef CAS PubMed.
- I. Ravikumar and P. Ghosh, Chem. Soc. Rev., 2012, 41, 3077–3098 RSC.
- P. M. Cummins, K. D. Rochfort and B. F. O'Connor, in Protein Chromatography: Methods and Protocols, ed. D. Walls and S. T. Loughran, Springer New York, New York, NY, 2017, pp. 209–223, DOI:10.1007/978-1-4939-6412-3_11.
- (a) D. Y. Sasaki, K. Kurihara and T. Kunitake, J. Am. Chem. Soc., 1992, 114, 10994–10995 CrossRef CAS; (b) J. F. Neal, W. Zhao, A. J. Grooms, A. H. Flood and H. C. Allen, J. Phys. Chem. C, 2018, 122, 26362–26371 CrossRef CAS.
- (a) S. K. Kim and J. L. Sessler, Chem. Soc. Rev., 2010, 39, 3784–3809 RSC; (b) D. Smith, in Macrocyclic Chemistry: Current Trends and Future Perspectives, ed. K. Gloe, Springer Netherlands, Dordrecht, 2005, pp. 137–151, DOI:10.1007/1-4020-3687-6_9.
- M. M. G. Antonisse and D. N. Reinhoudt, Chem. Commun., 1998, 443–448, 10.1039/A707529D.
- G. Wulff, T. Gross and R. Schönfeld, Angew. Chem., Int. Ed. Engl., 1997, 36, 1962–9164 CrossRef CAS.
- G. Wulff and K. Knorr, Bioseparation, 2002, 10, 257 CrossRef PubMed.
- C. Lübke, M. Lübke, M. J. Whitcombe and E. N. Vulfson, Macromolecules, 2000, 33, 5098–5105 CrossRef.
- M. Emgenbroich, C. Borrelli, S. Shinde, I. Lazraq, F. Vilela, A. J. Hall, J. Oxelbark, E. De Lorenzi, J. Courtois, A. Simanova, J. Verhage, K. Irgum, K. Karim and B. Sellergren, Chem.–Eur. J., 2008, 14, 9516–9529 CrossRef CAS PubMed.
- S. Shinde, A. Bunschoten, J. A. W. Kruijtzer, R. M. J. Liskamp and B. Sellergren, Angew. Chem., Int. Ed., 2012, 51, 8326–8329 CrossRef CAS PubMed.
- W. Wan, M. Biyikal, R. Wagner, B. Sellergren and K. Rurack, Angew. Chem., Int. Ed., 2013, 52, 7023–7027 CrossRef CAS PubMed.
- S. Shinde, Z. El-Schich, A. Malakpour, W. Wan, N. Dizeyi, R. Mohammadi, K. Rurack, A. Gjörloff Wingren and B. Sellergren, J. Am. Chem. Soc., 2015, 137, 13908–13912 CrossRef CAS PubMed.
- A. Cutivet, C. Schembri, J. Kovensky and K. Haupt, J. Am. Chem. Soc., 2009, 131, 14699–14702 CrossRef CAS PubMed.
- R. Sulc, G. Szekely, S. Shinde, C. Wierzbicka, F. Vilela, D. Bauer and B. Sellergren, Sci. Rep., 2017, 7, 44299 CrossRef CAS PubMed.
- M. Liu, S. B. Torsetnes, C. Wierzbicka, O. N. Jensen, B. Sellergren and K. Irgum, Anal. Chem., 2019, 91, 10188–10196 CrossRef CAS PubMed.
- P. Manesiotis, A. Riley and B. Bollen, J. Mater. Chem. C, 2014, 2, 8990–8995 RSC.
- For examples of molecular recognition in water using MIPs see: (a) Y. Hoshino, T. Kodama, Y. Okahata and K. J. Shea, J. Am. Chem. Soc., 2008, 130, 15242–15243 CrossRef CAS PubMed; (b) G. Pan, Y. Zhang, Y. Ma, C. Li and H. Zhang, Angew. Chem., Int. Ed., 2011, 50, 1–14 CrossRef; (c) F. Canfarotta, A. Poma, A. Guerreiro and S. Piletsky, Nat. Protoc., 2016, 11, 443 CrossRef CAS PubMed.
- J. Smid, S. C. Shah, R. Sinta, A. J. Varma and L. Wong, Pure Appl. Chem., 1979, 51, 111 CAS.
- S. Wagner, C. Zapata, W. Wan, K. Gawlitza, M. Weber and K. Rurack, Langmuir, 2018, 34, 6963–6975 CrossRef CAS PubMed.
- C. J. Pedersen, J. Am. Chem. Soc., 1967, 89, 2495–2496 CrossRef CAS.
- D. J. Cram, Angew. Chem., Int. Ed. Engl., 1988, 27, 1009 CrossRef.
- J. S. Bradshaw and R. M. Izatt, Acc. Chem. Res., 1997, 30, 338–345 CrossRef CAS.
- Y. Takeda, Bull. Chem. Soc. Jpn., 1981, 54, 3133–3136 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section and supporting data. See DOI: 10.1039/c9sc06508c |
| ‡ Current address: School of Chemistry and Chemical Engineering, Queens University Belfast, Northern Ireland, UK. |
| § Current address: Analysis and evaluation department, Egyptian petroleum research institute, 1 Ahmed el zomor street, Nasr city, Cairo, Egypt. |
| ¶ This ion-pair imprinting approach contrasts with previously reported dual-ion imprinting of two or more cations. See: Prasad B., Jauhari D. and Verma A., Talanta, 2014, 120, 398–407. |
| This journal is © The Royal Society of Chemistry 2020 |