Open Access Article
Open Access ArticleRecruitment of receptors at supported lipid bilayers promoted by the multivalent binding of ligand-modified unilamellar vesicles†
Daniele
Di Iorio‡
,
Yao
Lu‡
,
Joris
Meulman
and
Jurriaan
Huskens
 *
*
Molecular Nanofabrication Group, MESA+ Institute for Nanotechnology, Faculty of Science and Technology, University of Twente, P.O. Box 217, Enschede, 7500 AE, The Netherlands. E-mail: j.huskens@utwente.nl
First published on 9th March 2020
Abstract
The development of model systems that mimic biological interactions and allow the control of both receptor and ligand densities, is essential for a better understanding of biomolecular processes, such as the recruitment of receptors at interfaces, at the molecular level. Here we report a model system based on supported lipid bilayers (SLBs) for the investigation of the clustering of receptors at their interface. Biotinylated SLBs, used as cell membrane mimics, were functionalized with streptavidin (SAv), used here as receptor. Subsequently, biotinylated small (SUVs) and giant (GUVs) unilamellar vesicles were bound to the SAv-functionalized SLBs by multivalent interactions and found to induce the recruitment of both SAv on the SLB surface and the biotin moieties in the vesicles. The recruitment of receptors was investigated with quartz crystal microbalance with dissipation monitoring (QCM-D), which allowed the identification of the biotin and SAv densities necessary to obtain receptor recruitment. At approx. 0.6% of biotin in the vesicles, a transition between dense and low vesicle packing was observed, which coincided with the transitions between recruitment in the vesicles vs. recruitment in the SLB and between full and partial use of the biotin moieties in the vesicle. Direct optical visualization of the clustering at the interface of individual GUVs with the SLB platform was achieved with fluorescence microscopy, showing recruitment of SAv at the contact area as well as the deformation of the vesicles upon binding. Different vesicle binding regimes were observed for lower and higher biotin densities in the vesicles and at the SLBs. A more quantitative analysis of the molecular parameters implied in the interaction, indicated that approx. 10% of the vesicle area constitutes the contact area. Moreover, the SUV binding and recruitment appeared to be fast on the analysis time scale, whereas the binding of GUVs is slower due to the larger SLB area over which SAv recruitment needs to occur. The mechanisms revealed in this study may provide insight in biological processes in which recruitment occurs.
Introduction
The clustering of receptors in cell membranes plays an important role in a large number of biological processes. In living cells, phenomena such as signal transduction, which are fundamental for immunological responses and neurotransmission, are often associated with the formation of domains and the clustering of receptors at a cell surface.1–3 Multivalent ligand–receptor interactions at the interface, in combination with the fluidity of the cell membrane, promote the rearrangement and co-localization of receptors on a surface, and thereby govern these biological responses.4The molecular processes of binding, diffusion and clustering occur at characteristic length and time scales, and these can be distinctly different from those associated with the biological processes that they induce. To deconvolute the various contributions to the biological processes, the molecular and biophysical aspects of receptor clustering need to be investigated. Molecular aspects, such as the number of receptors involved in the interactions, the local receptor density and the lateral diffusion on the cell membrane, are critical factors playing a role in these phenomena.5 To provide quantitative insight into these molecular parameters, it is necessary to build synthetic models that can mimic the biological interaction at the interface and in which parameters such as receptor density and surface fluidity can be controlled and quantified.
Supported lipid bilayers (SLBs) are used extensively as good cell-membrane mimics, in part for their excellent antifouling properties.6,7 Among their manifold advantages, SLBs present a two-dimensional fluidity, an important feature that allows the mimicking of mobility and rearrangement processes occurring at a cell membrane.8 The fluidity of an SLB can be tuned by changing the chemical composition of the lipids.9 Moreover, the possibility of modifying the composition of the SLB by incorporating a controllable fraction of a functionalized lipid, which, for example, allows the attachment of receptors, provides an exquisite method to control the surface density.10,11
The biotin–streptavidin interaction has been used as a workhorse for biomolecule immobilization since many years. The availability of biotinylated lipids has allowed extension to its use in SLBs.6 Several ligands have been anchored to SLBs by introducing biotinylated lipids in the lipid mixture, exploiting the strong non-covalent biotin–streptavidin interaction, in which density and mobility of the biotin influence the SAv coverage on the substrate.12 For example, the interactions of EphA2-expressing human breast cancer cells with SLBs displaying ephrin-A1 ligands has been reported.13 More recently, the use of SLBs for the study of interactions between membrane receptors in cellular membranes and of the cell mechanobiology has been reported.14 Arg–Gly–Asp (RGD) ligand have been introduced onto SLBs to study human MSC (hMSC) adhesion and differentiation and the role of ligand density and mobility in the interaction.15
SLBs have been used for the study and quantification of many more multivalent biological interactions at the interface. The multivalent interactions of the pentameric cholera toxin B subunits (CTB) with ganglioside GM1-modified lipid bilayers has been investigated by Cremer's group.16 Höök and coworkers mimicked the binding of virions to cell membranes by anchoring multivalent DNA-tethered vesicles to SLBs.17 With a different approach, Lin et al. used protein oligomer-coated NPs to investigate the multivalent adhesion of both p-CTB to GM1 and t-HA to GM3 glycans.18 These studies elucidated the important correlation between the mobile ligand densities on the surface and the overall affinity.
Here we report a well-defined SLB model system that allows the study and the microscopic visualization of the phenomenon of receptor clustering at a cell membrane that is the result of a biological multivalent interaction at the interface. Our goal is to investigate whether the interaction of multivalent ligand carriers, such as cells, vesicles and nanoparticles – here represented by vesicles with ligand binding sites anchored to fluidic membranes – can cause ligand clustering at the surface they bind to, and how this phenomenon is correlated to the densities of receptor and ligand sites exposed at the interface. Accordingly, SLBs and vesicles were chosen as mimics for the fluidic membrane and for the interacting multivalent biological system, respectively, owing to the possibility of accurately controlling both ligand and receptor densities at the interface in these systems. The strong, non-covalent biotin–streptavidin interaction was taken as a model for a biological interaction. The use of this, strong-binding, interaction pair enables a high level of control in the quantification of the number of interactions involved in the overall binding. Biotinylated SLBs were used as bio-mimetic platforms, which were functionalized with streptavidin (SAv), used here as receptor. Biotinylated small (SUVs; approx. 100 nm) and giant (GUVs; approx. 15–20 μm) unilamellar vesicles were employed as multivalent carriers to induce recruitment of SAv on the SLB surface. Specifically, SUVs were used in ensemble surface binding studies using quartz crystal microbalance with dissipation monitoring (QCM-D). Because of their size, GUVs were used for direct optical visualization of individual recruitment events by fluorescence microscopy. By controlling the densities of both biotin and SAv at the interface, we quantitatively analyzed the clustering of the receptor sites at the interface and investigated the role of both ligand and receptor density at the fluid surface. An analysis of the size of the contact area between vesicle and SLB is also provided in order to evaluate the number and local density of interacting sites involved during the receptor clustering.
Results
Ensemble binding of 100 nm unilamellar vesicles to supported lipid bilayers
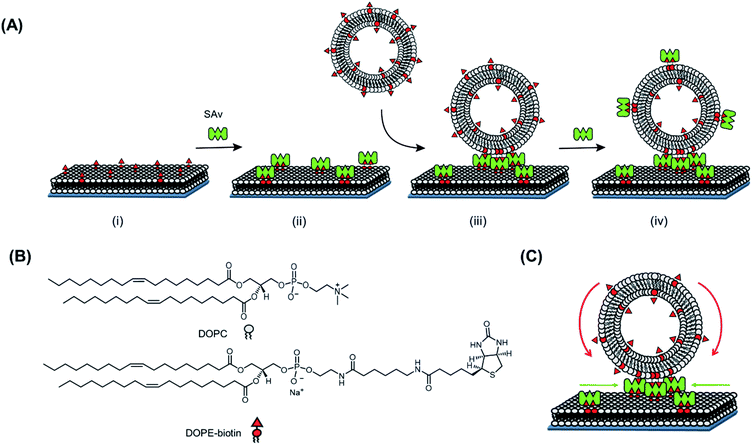
In order to investigate the recruitment of receptors at an SLB interface induced by a multivalent interaction, we first employed biotinylated small unilamellar vesicles (SUVs) with a diameter of 100 nm, able to bind multivalently to a SAv-functionalized SLB. Here, the strong non-covalent interaction between biotin and SAv (Kd = 10−14 M)19 was chosen as a model for a general ligand–receptor interaction occurring in a real biological system. The high affinity of this interaction is expected to provide the maximal possible recruitment.Fig. 1 reports a schematic representation of the formation of the platform used in this work and the interaction with SUVs. In a first step, a biotinylated SLB is formed on a (SiO2) substrate. For the formation of the SLB, unilamellar vesicles consisting of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) mixed with varying molar ratios of a 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine–N-(biotinyl) (DOPE–biotin) were formed (see ESI and Fig. S1†). These vesicles are known to rupture on activated surfaces forming stable and mobile bilayers.20 The presence of biotin in the SLB allows the binding of SAv in a following step. In particular, by mixing DOPC with DOPE–biotin in different molar ratios, it is possible to precisely tune the (average) density of biotin moieties displayed at the interface. Thereby, the average density of SAv absorbed on the surface during the second step, is determined during the vesicle preparation step.
In this work, three different biotin densities were chosen for the formation of SLB platforms (employing 0.1 mol%, 0.4 mol% and 2 mol% of DOPE–biotin in the lipid mixture). As we reported previously,10 an increasing molar fraction of DOPE–biotin in the SLB up to approximately 1% leads to a linear response of adsorbed SAv. Higher concentrations, however, cause a physical saturation of the surface with SAv. Taking these observations into account and assuming that a single SAv binds to two biotinylated lipids on the surface,12 the biotin molar ratios chosen for the formation of SLB lead to (average) SAv densities of 0.11, 0.46, and 1.57 pmol cm−2. Two remaining SAv binding pockets are therefore assumed to be available at SLB-bound SAv for further interactions with biotin ligands presented at the adsorbing vesicles.
In order to probe the lateral mobility of lipids within the SLB and of SAv bound onto the SLB, both the lateral diffusion of a dye-modified lipid added to the lipid membranes (at varying biotin mol%) and that of adsorbed and fluorescently labeled SAv were assessed through quantitative fluorescence recovery after photo bleaching (FRAP; see Fig. S2 and Table S1†).21 For all biotin densities used in this work, the SLB showed mobility, even when modified with a densely packed SAv layer (2% DOPE–biotin). A lower diffusion coefficient observed for lipids after the binding of SAv onto the surface (Table S1†) is in agreement with the previously made assumption that each SAv binds to two biotinylated lipids at an SLB. Additionally, the decreasing diffusion coefficient for higher biotin densities in the SLB after the adsorption of SAv, confirms the occurrence of steric hindrance between proteins due the high packing density of SAv, especially in the case of 2% DOPE–biotin.
After the formation of a SAv layer on the surface, biotinylated SUVs composed of DOPC mixed with DOPE–biotin were flowed over the surface. SUVs are ideal multivalent carriers, the size of which can be finely controlled in the extrusion step. This size control is important in our studies, because it allows proper quantification of the number of interactions and the interaction area at the interface. Like for the SLBs, varying the molar ratios of DOPE–biotin and DOPC in the lipid mixture allows control over the biotin density in the SUVs. In this work, SUVs were made containing DOPE–biotin concentrations in the range of 0.025% to 5% (Table S2†).
Quartz crystal microbalance with dissipation monitoring (QCM-D) was used to monitor both the in situ formation of the interaction platform, consisting of the biotinylated SLB and the SAv layer attached onto it, and the subsequent interaction of SUVs with this platform in real time. Fig. 2A shows a typical QCM-D measurement in which every adsorption step described above is observed by monitoring the frequency shift (Δf5, blue line). Initially, after obtaining a stable baseline, an SLB (Δf5 = 24 ± 1 Hz and ΔD5 < 0.5 × 10−6) was formed on a SiO2-covered QCM sensor. Subsequently, the surface was rinsed with buffer, and a solution of SAv (1 μM) was flowed over the SLB. Adsorption of SAv onto the SLB was confirmed by a clear frequency shift, and surfaces were rinsed again with buffer in order to remove the non-bound SAv from the surface. As described before,10 the frequency shift of the SAv adsorption step depended on the fraction of DOPE–biotin used in the vesicles to make the SLBs. Consequently, the binding of SAv is specific, and occurs through biotin–SAv interaction pair formation.
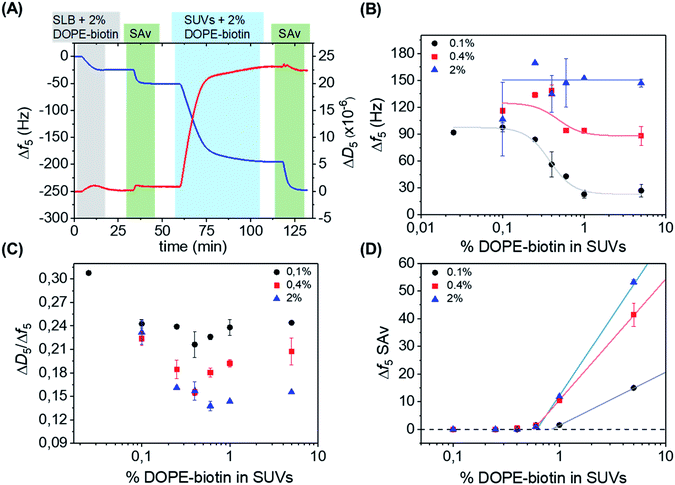 | ||
| Fig. 2 (A) QCM-D measurement showing stepwise: the formation of an SLB doped with 2 mol% of DOPE–biotin, adsorption of SAv, the interaction with SUVs containing 2 mol% of DOPE–biotin, and another adsorption step of SAv. The 5th overtone was used throughout the experiments. The frequency shift (Δf5, left axis) is shown in blue, and the change in dissipation (ΔD5, right axis) in red. The different shadings correspond to the different additions according to Fig. 1. (B) Limiting frequency shifts obtained for the SUV adsorption steps as a function of the DOPE–biotin fraction in the SUVs, in their interaction with SLBs containing different (0.1, 0.4 or 2%) biotin–DOPE fractions and saturated with SAv, after rinsing. Black, red and blue lines are guides to the eye. (C) Plot of ΔD/Δf values versus the molar fraction of DOPE–biotin in SUVs measured on SLBs with biotin fractions of 0.1% (black), 0.4% (red), and 2% (blue). (D) Frequency shifts of adsorption steps of SAv (final adsorption step shown in Fig. 1 and 2A) on SUVs bound to SAv-modified SLBs containing different percentages of biotin. | ||
In the following step, a frequency shift confirms the interaction of the vesicles at the SAv-covered platform. The addition of SAv or biotinylated SUVs on an SLB containing no DOPE–biotin did not show any frequency shift (Fig. S3†), thus confirming the specificity of the interactions observed. Additionally, the large change of frequency in the presence of SAv, accompanied by a large dissipation change, confirms that the vesicles remain intact upon binding, maintaining their vesicular structure while interacting with the platform.22 As a last step, SAv was again added onto the surface in order to detect any free biotin moieties in the SUVs remaining after the interaction with the SAv-coated SLB surface, thereby obtaining information about possible recruitment occurring in the adsorbed SUVs.
The effect of the biotin density, present in the vesicles ranging from 0.025% to 5%, on the interaction with the SAv-modified surfaces, was investigated by measuring the vesicle adsorption steps. In the first series of measurements, SLBs containing a low (0.1%) fraction of DOPE–biotin were chosen in order to provide a low-density SAv layer on the surface with a relatively large (average) spacing between each protein. Fig. 2B shows the values of the frequency shifts obtained for the binding of the SUVs as a function of their biotin. As expected, the vesicle adsorption onto the surface appeared to be correlated to the biotin content of the SUVs. Interestingly, both frequency and dissipation signals were found to stabilize at specific values depending on the biotin concentrations of the vesicles. In particular, a low (0.025% and 0.1%) content of biotin in the SUVs led to more vesicle adsorption, while higher biotin contents caused less vesicles to bind (Fig. 2B, black curve). A minimum adsorption was obtained for both the 1% and 5% molar fractions, and a clear transition between high and low vesicle binding was observed at approximately 0.4 mol% of biotin. The differences in vesicle adsorption are attributed exclusively to the differences in DOPE–biotin content in the SUVs as no other factors are expected to play a role (see ESI†).
Interestingly, when repeated on SLBs containing higher (0.4% and 2.0%) biotin densities, different trends were observed in comparison to the 0.1% biotin SLB (Fig. 2B, red and blue curves). In the case of an SLB with 0.4% biotin, a correlation between biotin content and vesicle adsorption, similar to the one found for the 0.1% platform, was observed. The values of the frequency shifts in this series of experiments were consistently higher than the ones measured for the 0.1% biotin SLB, suggesting that a higher amount of vesicles was adsorbed onto the surface. When the 2% biotin SLB was used, the biotin content in the SUVs appeared to have no influence at all on the adsorption of vesicles, and high frequency shifts were obtained for all of the SUV biotin densities used here. Most likely, the frequency shift (of approx. 150 Hz) obtained in this case corresponds to a dense packing of vesicles.
In a QCM-D measurement, the monitoring of dissipation changes (ΔD5) concomitant to the frequency shifts provides useful information about changes of the viscoelastic properties of the system during a binding event. The ratio between ΔD5 and Δf5 can be therefore used as a tool to analyze such effects, as it allows one to examine the changes of energy dissipation of the system per adsorbed mass unit and thus provides insight into viscosity changes (i.e., changes in softness/rigidity) occurring at the interface upon binding. Here, the binding of the vesicles can intuitively be expected to provide smaller dissipation signals when a larger contact area is achieved (with more biotin–SAv bonds and larger vesicle deformation) and when the vesicles start touching each other (at high vesicle coverages). Therefore, the ratio between ΔD5 and Δf5 was determined for each SUV step (Fig. 2C). Interesting changes in the ratio were observed, indicating changes in viscoelastic properties as a function of biotin densities in SLB and SUVs (see ESI,† section on QCM-D for a more detailed explanation). All series showed a dip in the ΔD5/Δf5 ratio at 0.4–0.6% biotin fraction in the SUVs, coinciding with the biotin percentage in Fig. 2B at which the binding curves go from a relatively sparsely packed vesicle layer (at high biotin%) to a dense vesicle packing (at low biotin%). This clearly indicates the effect of dense vesicle packing on the viscoelastic properties of the layer.
After the adsorption of SUVs on the surface, as shown in Fig. 2A, a solution of SAv was added again. In this way, we aimed to determine the availability of free biotin moieties on the SUV surfaces after binding to the interacting platform and thus to investigate the recruitment of biotin moieties displayed at the SUVs. The SAv addition was performed for every combination of biotin densities in SUVs and SLBs. Fig. 2D reports the frequency changes observed upon SAv addition onto the SUVs pre-adsorbed on the various SLB surfaces. Notably, for SUVs containing biotin% below 0.6%, no SAv binding was observed for any of the SLB biotin densities. This observation implies that complete recruitment of all biotin moieties in the vesicles had occurred. It also confirms that competitive binding to already bound biotin moieties does not occur. Higher biotin contents in the SUVs led to increased adsorptions of SAv onto the vesicles. Notably, the onset of SAv binding to the vesicles (at approx. 0.6–0.9% of biotin in the SLB) coincides with the biotin density at which the transition occurs in the vesicle binding density (Fig. 2B) and in the ΔD5/Δf5 ratio (Fig. 2C).
Single-vesicle binding of giant unilamellar vesicles to supported lipid bilayers
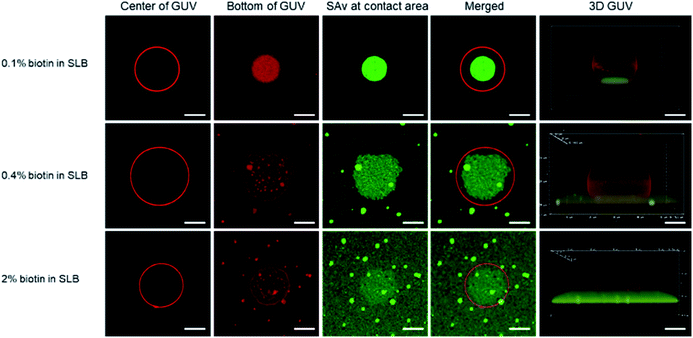
In order to visualize the clustering of receptors at the SLB interface with imaging techniques such as fluorescence microscopy, giant unilamellar vesicles (GUVs) were used as a model for immobilization onto the SLB. Although the biotin density of SUVs can be precisely controlled, their nanoscale size prevents the direct visualization of receptor clustering and contact area, hence necessitating the use of larger vesicles (here, GUVs). Both the GUVs and the SLBs were made from lipid mixtures of DOPC, DOPE–biotin, and Texas Red-functionalized 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Texas Red-DHPE, Fig. S4†). After the deposition of the biotinylated SLB, Alexa Fluor 488-labeled SAv was anchored onto the membrane surface to allow direct visualization of recruited SAv molecules in the contact area between the GUV and the SLB, which occurs upon adhesion of biotinylated GUVs by the formation of biotin–SAv affinity pairs.Fluorescence microscopy images of typical individual, bound vesicles, obtained at different focal planes and at different biotin fractions (0.1, 0.4, and 2%) in the SLB, are shown in Fig. 3. The red color of the Texas Red label implemented in the GUVs appears to be evenly spread in the GUV membrane, and allows visualization of the size and shape of the vesicles. Cross section views of the center section of the GUVs are shown in the 1st column, and the bottom section that is in contact with the SLB surface is shown in the 2nd column. The data shows that the GUVs (with diameters of 15 to 20 μm) stayed intact upon adhesion to the SLB. By gradually lowering the focal plane closer to the binding interface at the SLB, the contact area between the adhered GUVs and the SLB was made visible. Most notably, this area overlapped with the area of green fluorescence emitted by the labeled SAv (3rd column in Fig. 3). Whereas the green SAv fluorescence was homogeneously distributed over the surface before vesicle adhesion (Fig. S5†), binding of the vesicles caused an increase of the local concentration of SAv at the vesicle-SLB contact area. Some bright green specks of higher fluorescence intensity were observed on the SLBs with 0.4 and 2 mol% biotin receptors, which is likely due to some aggregation of SAv at higher biotin concentrations. Qualitatively, the co-localized fluorescence of the vesicles and SAv in the contact area supports the interpretation of recruitment of SAv into the contact area upon adsorption of the vesicles.
To study the effect of the receptor density on the clustering process, we first fixed the biotin density in the GUVs (0.1 mol%) but varied the biotin concentration in the SLB, similar to the QCM-D experiments with SUVs described above. At this low biotin% in the GUVs, all or most biotin moieties of the GUVs are expected to be used in binding to the SLB. As shown in Fig. 3 (top row), the binding area, visualized both in red and green (2nd and 3rd columns), was much smaller (4th column) than the cross section area (i.e., the center, 1st column) of the GUV. The green fluorescence was bright and with high contrast (Fig. S6A†), indicating an efficient, high density clustering, with no or only few SAv molecules remaining unbound outside the contact area of the vesicle.
When the biotin concentration in the SLB was increased to 0.4 mol%, the contact area, relative to the center (4th column in Fig. 3, second row), became larger than for the SLB with 0.1 mol% biotin. At the same time, a weak but clearly visible green fluorescence remained outside of the binding site, yielding a lower contrast (Fig. S6B†). Both trends continued for the SLB with 2 mol% biotin (Fig. 3, 3rd row, and Fig. S6C†), which can be attributed to the increased biotin density at the SLB interface that led to a higher density of fluorescent SAv. Most likely, as described for the low-biotin% SUVs discussed above, all or most biotin moieties of the GUVs are being used in binding to the SLB.
During the receptor clustering process, deformable GUVs may undergo morphological changes upon binding to the surface of the SLB. In order to visualize the deformation of GUVs, confocal microscopy was used to provide 3D structures of the GUVs. As the 5th column in Fig. 3 shows, a GUV bound to the surface of the SLB with 0.1 mol% biotin exhibited a clear deformation leading to a flat contact area between the GUV and the SLB. As the biotin density in the SLB was increased to 0.4 and 2 mol%, a larger deformation of the GUVs became apparent from the sideview of their 3D structures. It is likely that the increased biotin density at the SLB facilitated more biotin receptors in the GUV to move into the contact area and interact with SAv, thus providing more biotin–SAv interaction pairs that induce larger morphological changes with concomitantly larger contact areas. A more quantitative analysis is presented in the ESI (Fig. S7–S9† and accompanying text).
In the case of GUVs with 2 mol% biotin, the stability was not as high as in the other cases, and most vesicles ruptured (Fig. S10†). It is likely that the high biotin% in the GUVs drives the system towards an increased contact area between the GUV and the SLB that promotes vesicle rupture. This is also an indirect confirmation that, at 0.1% biotin in the GUVs, all biotins are being used in the binding to the SLB, whereby an increase of the biotin% in the SLB cannot lead to further increase of the contact area.
Discussion
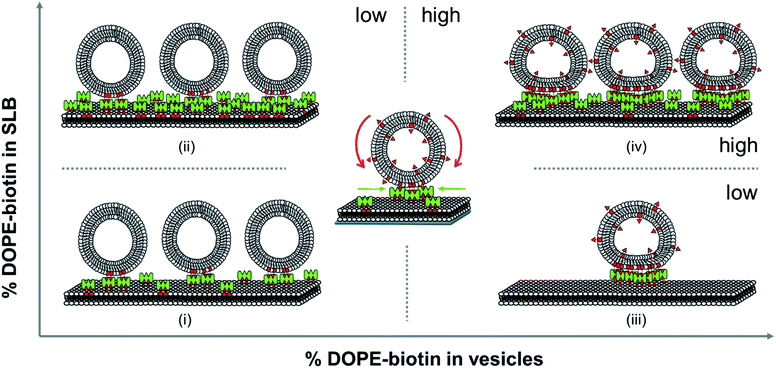
All the distinctive trends occurring in the vesicles binding to the SLBs as observed above show that the biotin densities both in the vesicles and in the SLB play an important role in the recruitment of biotin/SAv interaction pairs into the contact area. Here we attempt to analyze what effects play at the molecular level during the binding of the vesicles to the SLB and how this affects the recruitment and the deformation of the vesicles at the interface. When viewing the biotin% in the SLBs and the vesicles as ranging from low to high, various regimes can be distinguished. The different regimes in these vesicles binding processes are depicted in Fig. 4.First we consider the effect of a low biotin density in the vesicles (0.025 to 0.1 mol%) on their binding at SLB surfaces functionalized with a low density of SAv. As shown in Fig. 2B, low biotin densities in the SUVs cause a high coverage of vesicles on surfaces functionalized with low SAv densities (0.1 and 0.4 mol% biotin), leading to large frequency shifts (case (i) in Fig. 4). Similarly, when surfaces are functionalized with a higher density of SAv on the SLB, a similar coverage of vesicles is observed (Fig. 2B and case (ii) in Fig. 4), implying that the density of adsorbed vesicles is not influenced by the SAv density at the SLBs. This suggests that the adsorption of vesicles is determined by the amount of biotin in the vesicles. In particular, all the biotin moieties on the vesicle are employed in the binding with SAv on the SLB surface, as demonstrated by the absence of SAv adsorption on bound vesicles for such biotin density (Fig. 2D). Apparently, the low biotin density in the vesicles in cases (i) and (ii) promotes the formation of only a limited number of biotin–SAv interaction pairs, resulting in recruitment of SAv molecules from a limited area surrounding the vesicle, thus promoting a relatively high coverage of vesicles at the SLB surface. This was confirmed by the ΔD5/Δf5 ratios calculated at low biotin contents in the SUVs. In fact, a similar ratio was obtained for all three SAv densities (Fig. 2C), suggesting that no difference in the density of the vesicles on the surface occurred. The measured stiffness can therefore be attributed to a densely packed vesicle layer formed on the SLB. Moreover, the absence of SAv adsorption after vesicle deposition (Fig. 2D), demonstrates that the biotin moieties displayed on the SUVs are all bound to SAv at the surface. Therefore, in this regime, recruitment occurs exclusively for the biotins present in the SUVs.
Subsequently, we consider the effect of a high biotin density in the vesicles, i.e. in the range of 1 to 5 mol%, on the vesicle binding at the SLB interface functionalized with a low density of SAv (case (iii) in Fig. 4). As shown in Fig. 2B, high biotin densities in the vesicles cause a lower coverage of the vesicles on the surface functionalized with low SAv densities (0.1 and 0.4 mol% biotin), leading to small frequency shifts. This phenomenon can be attributed to the enhanced recruitment of SAv on the surface promoted by the binding of the vesicles. Due to the mobility of the SLB, SAv can diffuse at the interface and as a consequence, it is likely that as soon as a single SAv–biotin interaction occurs between a vesicle and the SLB, more proteins at the SLB interface bind to the bottom of the vesicle. The anchoring of a large number of SAv with vesicles thus causes a depletion of free SAv on the SLB interface surrounding the vesicle. The recruitment of SAv was confirmed by the observation of the contact area in GUVs adsorbed to SLBs and of depletion of SAv outside the contact area, which provided a high contrast (Fig. 3). Overall, compared to case (i), the higher biotin% in the vesicles in case (iii) promotes stronger recruitment and the formation of a larger number of interaction pairs involving a larger number of SAv molecules, leading to a concomitantly larger contact area.
The higher coverage of vesicles with a high biotin% observed at larger biotin densities on the SLB (2 mol%), demonstrated that the depletion of SAv on the surface is prevented in this case. In other words, when the surface can supply more SAv, it is possible to accommodate a larger density of vesicles (case (iv) in Fig. 4). When surfaces are densely covered with SAv, the depletion is fully suppressed and the binding of vesicles is purely determined by the packing density of SUVs on the surface. The adsorption of SUVs on SLBs containing increasing SAv densities, leads to an increase in stiffness of the surface, as also demonstrated by the different ΔD5/Δf5 ratios obtained for SLBs containing 0.1, 0.4 and 2% of biotinylated lipids (Fig. 2C). However, the similar vesicle coverage obtained with 1% to 5% of biotin in the vesicles, suggests that no further depletion of SAv on the SLB occurs, meaning that the maximal density of SAv at the interface with a vesicle is reached with 1% of biotin density in the SUVs. Beyond this value, the contact area of the SUVs does not further increase. As a consequence, a higher content of biotin in the SUVs implies a higher amount of free biotin in the SUVs remaining after adsorption. Conclusively, in both cases (iii) and (iv), the biotin density at the vesicle is higher than that is used for bond formation at the contact area, leaving free unused biotin moieties available upon adsorption, as witnessed by the SAv attachment to these moieties (Fig. 2D).
The cut-off values obtained from Fig. 2D allow a quantitative estimation of the contact area of a vesicle with the surface. Assuming an average diameter of an SUV of approx. 100 nm and the footprint of a single lipid molecule of 0.725 nm (ref. 23) (leading to approx. 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lipids in the outer leaflet at an area of 31
000 lipids in the outer leaflet at an area of 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm2), we estimate that approximately 260 biotins can be involved in the interaction at the interface at the observed cut-off of 0.6 mol% biotin in the SUVs. As a consequence, a maximum of approx. 130 SAv can be recruited in the contact area. By setting a single SAv area to 25 nm2, the area occupied by densely packed SAv is approximately 3250 nm2, corresponding to 10% of the SUV surface area. At the same time, this cut-off represents roughly the boundary at which the biotin moieties in SLB and vesicle are balanced: at lower biotin% in the SUVs, the biotin density in the SUV is limiting, and recruitment occurs primarily within the SUV, consuming all of its biotin moieties in its interaction with the SLB. Above the 0.6% boundary, the biotin (and SAv) density in the SLB is limiting, and recruitment occurs primarily within the SLB, thus recruiting SAv molecules from larger areas, leading to concomitantly lower vesicle coverages. Yet, small deviations in the estimation of the contact area can occur due to additional factors such as a different residual valency of the SAv adsorbed on the SLB or the flip-flop of biotinylated lipids in the SLBs. In particular, when not every SAv binds to the biotinylated vesicles in a 1
000 nm2), we estimate that approximately 260 biotins can be involved in the interaction at the interface at the observed cut-off of 0.6 mol% biotin in the SUVs. As a consequence, a maximum of approx. 130 SAv can be recruited in the contact area. By setting a single SAv area to 25 nm2, the area occupied by densely packed SAv is approximately 3250 nm2, corresponding to 10% of the SUV surface area. At the same time, this cut-off represents roughly the boundary at which the biotin moieties in SLB and vesicle are balanced: at lower biotin% in the SUVs, the biotin density in the SUV is limiting, and recruitment occurs primarily within the SUV, consuming all of its biotin moieties in its interaction with the SLB. Above the 0.6% boundary, the biotin (and SAv) density in the SLB is limiting, and recruitment occurs primarily within the SLB, thus recruiting SAv molecules from larger areas, leading to concomitantly lower vesicle coverages. Yet, small deviations in the estimation of the contact area can occur due to additional factors such as a different residual valency of the SAv adsorbed on the SLB or the flip-flop of biotinylated lipids in the SLBs. In particular, when not every SAv binds to the biotinylated vesicles in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, a larger number of SAv can be involved in overall binding, thus causing a relatively larger contact area than the one estimated above. Additional uncertainty is also given by the flip-flop of biotinylated lipids from one leaflet to another, which could cause alterations in number of SAv anchored onto the surface. However, because of the low flip-flop rate,24 this effect is here expected to be of only limited influence.
2 ratio, a larger number of SAv can be involved in overall binding, thus causing a relatively larger contact area than the one estimated above. Additional uncertainty is also given by the flip-flop of biotinylated lipids from one leaflet to another, which could cause alterations in number of SAv anchored onto the surface. However, because of the low flip-flop rate,24 this effect is here expected to be of only limited influence.
Also the SLB areas from which recruitment occurs can be estimated. A maximal dense packing of equally sized spheres on a surface is 90.6%, while the random parking limit is approx. 55% of the underlying surface area.25 The QCM data for the additional SAv bound to SUVs on an SLB (Fig. 2D) can provide additional information. Of all values shown in this graph, the one with the highest value, corresponding to 5% biotin-modified SUVs at a 2% biotin SLB, is the most reliable, as it was measured at a dense SUV layer (150 Hz for the vesicle adsorption step) and the 5% vesicles have the highest fraction of unused biotins. The observed frequency in this case (53 Hz) is approximately twice that of a densely packed SAv layer on a 2% biotin SLB (27 Hz). When we assume that the QCM sensitivity for the SAv adsorption is approximately the same on an SLB vs. a vesicle-coated SLB, the SAv density can be calculated as 0.08 SAv per nm2 of SLB area. This SAv density can be reached when the vesicle packing density is approx. 66%, which is well within the range of the random parking limit and the densest possible packing. At vesicle frequency changes below 150 Hz, the vesicle density is correspondingly lower, and the SLB area per vesicle increases. As shown in Fig. 2B, this effect is most pronounced for high biotin% in the SUVs and low biotin% in the SLB. But also at low biotin% in the SUVs (left side of Fig. 2B), the small differences in frequency change may indicate that the coverage at lower biotin% is not completely dense (approx. 60% of the densest packing at 0.1% biotin in the SLB). However, some more considerations should be taken into account when estimating the contact area of a SUV and the surface coverage. In fact, the change in frequency upon formation of a SAv layer on a planar surface, such as a SLB, is not directly comparable with the binding of SAv to adsorbed vesicles. The latter value is, in fact, expected to be underestimated, since a considerable, yet not quantifiable, part of the proteins replaces water molecules trapped between the adsorbed vesicles before SAv binding and sensed by the QCM. For comparison with the SUV data, the recruited numbers of SAv inside the contact area of bound GUVs were analyzed as well. It is, however, important to note that the GUV assembly is generally done for a relatively short time at much lower vesicle concentrations than in the case of the SUVs. Therefore, high vesicle densities are never observed, and in principle every vesicle can recruit SAv molecules from a sufficiently large SLB area. Because of their size, the contact area of a bound GUV can actually be measured (Fig. 3). The projected area ratios shown in Fig. S8† can be recalculated in a straightforward manner to the contact area per vesicle area. The high area ratios of approx. 0.8 correspond to a relative contact area of 21% of the vesicle surface area (Table S3†). This is somewhat higher than the 10% estimated from the SUV data described above. Only when the biotin% in both the SLB and the GUV is 0.1%, the area ratio is lower (approx. 0.3), which corresponds to a contact area of 7%. When we take the number of biotin moieties in the 0.1% GUVs into account, we note that the measured contact areas are quite a lot (a factor 4–15) larger than needed for a dense SAv packing that would saturate all these biotin groups. Most likely, the energy penalty required to deform the GUVs is a lot lower than for the SUVs, thus allowing a larger contact area and a relatively low-density SAv packing within it. The lower contact areas observed at the 0.1% biotin in both GUV and SLB is attributed to insufficient recruitment time, see below.
To estimate the timescale at which recruitment and depletion may occur, we can consider the diffusion time. In a homogeneous and two-dimensional system, the diffusion coefficient D can be defined through the relation:26t ≈ x2/4D. Here x indicates the displacement of molecules (here biotin-lipid-anchored SAv) in a clustering process and t the time. For SUVs of 100 nm with 0.1 mol% biotin, and assuming that all biotins of the SUV are involved in binding, all SAv of a 200 nm diameter area of an SLB with 0.1 mol% biotin are expected to be recruited. When the SUV has a 5 mol% biotin density, the recruited area increases to a diameter of 1.4 μm. Assuming a diffusion coefficient on the order of 1 μm2 s−1 (Table S1†), these areas correspond to recruitment times of 0.01–0.5 s, which is way faster than the QCM times used here. This means that recruitment is expected to be complete and equilibrium to be reached during QCM observation. For GUVs, however, the larger vesicle size leads to larger recruitment areas and therefore longer recruitment times, which can for a 10 μm vesicle range from 100–5000 s for recruitment within the SLB and on the order of 100 s for recruitment within the GUV itself. Typically, images of the GUVs were taken approx. 30 min after adsorption, which is an adequate time to achieve practically complete recruitment for the SLBs with 0.4 and 2% of biotin. The longest recruitment time is expected for the 0.1% of biotin in both GUV and SLB, and therefore the observed smaller contact areas, combined with the complete absence of SAv surrounding the bound vesicles in this case, may be a sign of incomplete recruitment within the contact area to achieve saturation of all biotins of the GUV within the given time.
Conclusions
In this work, the clustering of SAv molecules on biotin-modified SLBs induced by the strong multivalent binding of biotinylated SUVs and GUVs was investigated in both comparative and quantitative manners. Because both the SLB platform and the biotin–streptavidin interaction are workhorses in the studies and applications of (bio)molecular recognition, cell adhesion and biosensing, the recruitment observed here can provide an insight into possibly unexpected behavior as well as provide new functionalities of such platforms.In particular, both recruitment of SAv on modified SLBs and of biotin moieties in the vesicles were observed upon adsorption of SUVs and GUVs on the surface. This study provides an insight in the relationships between surface densities of both the ligands and receptors and the packing density. Four different regimes of the binding of SUVs were discerned in which the modulation of clustering is attributed to the densities of biotin moieties both in the vesicles and the SLBs. This led to the observation that a transition occurs between dense and low vesicle packing at 0.6% of biotinylated lipids. A quantitative analysis of the number of receptors recruited at the interacting area allowed a correlation between the receptor density at the interface and the contact area, and the contact area was estimated to be 10% of the total SUV area. The dependence of the contact area between the vesicles and the SLB was measured also with a GUV model system to understand how the receptor density affected the formation of receptor clustering. The deformation of individual GUVs was quantitatively analyzed to show the induced morphological changes during the binding process. Both SUVs and GUVs show that the clustering response upon vesicle binding is dependent on the receptor density and recruitment in the SLB and vesicle membranes.
In order to further understand the recruitment process in model systems and in biological interactions, it is useful to investigate the time scale of these phenomena. However, these phenomena appear to be very fast and, as a consequence, do not allow easy measurements of their rates. Further work will also focus on the development of different systems based on weaker multivalent interactions. In fact, the weak affinity is of great relevance for the understanding of biological systems, in which the rate of recruitment is also a critical issue for quantitative analysis. At the same time, we believe that with such systems, the weaker binding affinity may prevent full recruitment, which might lead to much more complex analysis. Moreover, for such weak multivalent systems, competitive binding studies might provide useful information and insights to effectively interfere with multivalent biological interactions. Yet, therefore, the data presented in this work may serve as a benchmark for such weaker multivalent systems. These results may offer a useful tool for assisting the analysis of biological examples of ligand/receptor clustering, such as for example the one of virus binding on cell membranes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the European Commission, Marie Curie Innovative Training Network MULTI-APP (No. 642793) and the Netherlands Organization for Scientific Research (NWO-NSFC grant no. 729001032 to J. H.).Notes and references
- P. Tolar, J. Hanna, P. D. Krueger and S. K. Pierce, Immunity, 2009, 30, 44–55 CrossRef CAS PubMed.
- A. Grochmal, E. Ferrero, L. Milanesi and S. Tomas, J. Am. Chem. Soc., 2013, 135, 10172–10177 CrossRef CAS PubMed.
- M. Renner, C. G. Specht and A. Triller, Curr. Opin. Neurobiol., 2008, 18, 532–540 CrossRef CAS PubMed.
- J. R. Cochran, D. Aivazian, T. O. Cameron and L. J. Stern, Trends Biochem. Sci., 2001, 26, 304–310 CrossRef CAS PubMed.
- G. V. Dubacheva, T. Curk, D. Frenkel and R. P. Richter, J. Am. Chem. Soc., 2019, 141, 2577–2588 CrossRef CAS PubMed.
- J. van Weerd, M. Karperien and P. Jonkheijm, Adv. Healthcare Mater., 2015, 4, 2743–2779 CrossRef CAS PubMed.
- K. Glasmästar, C. Larsson, F. Höök and B. Kasemo, J. Colloid Interface Sci., 2002, 246, 40–47 CrossRef PubMed.
- H. Jung, A. D. Robison and P. S. Cremer, J. Struct. Biol., 2009, 168, 90–94 CrossRef CAS PubMed.
- K. J. Seu, L. R. Cambrea, R. M. Everly and J. S. Hovis, Biophys. J., 2006, 91, 3727–3735 CrossRef CAS PubMed.
- D. Di Iorio, M. L. Verheijden, E. van der Vries, P. Jonkheijm and J. Huskens, ACS Nano, 2019, 13, 3413–3423 CrossRef CAS PubMed.
- D. Di Iorio and J. Huskens, ChemistryOpen, 2020, 9, 53–66 CrossRef CAS PubMed.
- G. V. Dubacheva, C. Araya-Callis, A. Geert Volbeda, M. Fairhead, J. Codée, M. Howarth and R. P. Richter, J. Am. Chem. Soc., 2017, 139, 4157–4167 CrossRef CAS PubMed.
- K. Salaita, P. M. Nair, R. S. Petit, R. M. Neve, D. Das, J. W. Gray and J. T. Groves, Science, 2010, 327, 1380–1385 CrossRef CAS PubMed.
- R. Glazier and K. Salaita, Biochim. Biophys. Acta Biomembr., 2017, 1859, 1465–1482 CrossRef CAS PubMed.
- G. Koçer and P. Jonkheijm, Adv. Healthcare Mater., 2017, 6, 1600862 CrossRef PubMed.
- J. Shi, T. Yang, S. Kataoka, Y. Zhang, A. J. Diaz and P. S. Cremer, J. Am. Chem. Soc., 2007, 129, 5954–5961 CrossRef CAS PubMed.
- S. Block, V. P. Zhdanov and F. Höök, Nano Lett., 2016, 16, 4382–4390 CrossRef CAS PubMed.
- J. Lin, K. Wang, X. Xia and L. Shen, Langmuir, 2018, 34, 8415–8421 CrossRef CAS PubMed.
- N. M. Green, in Advances in Protein Chemistry, Academic Press, 1975, vol. 29, pp. 85–133 Search PubMed.
- H. Schönherr, J. M. Johnson, P. Lenz, C. W. Frank and S. G. Boxer, Langmuir, 2004, 20, 11600–11606 CrossRef PubMed.
- D. M. Soumpasis, Biophys. J., 1983, 41, 95–97 CrossRef CAS PubMed.
- I. Reviakine, F. F. Rossetti, A. N. Morozov and M. Textor, J. Chem. Phys., 2005, 122, 204711 CrossRef PubMed.
- J. F. Nagle and S. Tristram-Nagle, Biochim. Biophys. Acta Rev. Biomembr., 2000, 1469, 159–195 CrossRef CAS.
- M. M. Sperotto and A. Ferrarini, in The Biophysics of Cell Membranes: Biological Consequences, Springer Singapore, Singapore, 2017 Search PubMed.
- G. Y. Onoda and E. G. Liniger, Phys. Rev. A, 1986, 33, 715–716 CrossRef CAS PubMed.
- P. W. Atkins, Physical Chemistry, Oxford University Press, Oxford, 5th edn, 1994 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: DLS, FRAP and QCM measurements, fluorescence microscopy images and calculation details. See DOI: 10.1039/d0sc00518e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |