Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Visualizing semipermeability of the cell membrane using a pH-responsive ratiometric AIEgen†
Yuan
Gu
ab,
Zheng
Zhao
 ab,
Guangle
Niu
ab,
Han
Zhang
ab,
Guangle
Niu
ab,
Han
Zhang
 c,
Yiming
Wang
ab,
Ryan T. K.
Kwok
ab,
Jacky W. Y.
Lam
ab and
Ben Zhong
Tang
c,
Yiming
Wang
ab,
Ryan T. K.
Kwok
ab,
Jacky W. Y.
Lam
ab and
Ben Zhong
Tang
 *abc
*abc
aDepartment of Chemical and Biological Engineering, Department of Chemistry, The Hong Kong Branch of Chinese National Engineering Research Center for Tissue Restoration and Reconstruction, Institute for Advanced Study, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong 999077, China. E-mail: tangbenz@ust.hk
bHKUST-Shenzhen Research Institute, No. 9 Yuexing 1st RD, South Area Hi-tech Park, Nanshan, Shenzhen 518057, China
cCenter for Aggregation-Induced Emission, SCUT-HKUST Joint Research Institute, State Key Laboratory of Luminescent Materials and Devices, South China University of Technology, Guangzhou 510640, China
First published on 15th May 2020
Abstract
In clinical chemotherapy, some basic drugs cannot enter the hydrophobic cell membrane because of ionization in the acidic tumor microenvironment, a phenomenon known as ion trapping. In this study, we developed a method to visualize this ion trapping phenomenon by utilizing a pH-responsive ratiometric AIEgen, dihydro berberine (dhBBR). By observing the intracellular fluorescence of dhBBR, we found that non-ionized dhBBR can enter cells more easily than ionized forms, which is in accordance with the concept of ion trapping. In addition, dhBBR shows superior anti-photobleaching ability to Curcumin thanks to its AIE properties. These results suggest that dhBBR can serve as a bioprobe for ion trapping.
Introduction
All living cells must exchange materials with their extracellular environment in order to be alive.1,2 The processes of substances entering and exiting cells are controlled by the cell membrane, a biological membrane consisting of a lipid bilayer with proteins embedded in, which separates the interior of cells from their external environment.3,4 Due to the semipermeable nature of the lipid bilayer, the cell membrane is permeable to non-ionized (fat-soluble) molecules, while the permeability to ionized (water-soluble) molecules is very limited, a phenomenon commonly known as ion trapping.5 Partial failure in cancer chemotherapy of some basic drugs, such as vinca alkaloids and anthracyclines, can be ascribed to ion trapping. The acidic tumor microenvironment prevents the ionized alkaline drugs from accumulating in cancer cells.5–9 Given this, the study of cell membrane's semipermeability, especially ion trapping, is not only fundamentally interesting, but also valuable in improving the clinical chemotherapeutic efficacy.Ion trapping is most widely studied by HPLC, a well-developed technology.5,9–12 Although HPLC enjoys the advantages of high sensitivity and selectivity, it usually brings about thorny issues like high equipment cost, complexity, complicated sample processing, and long runtime. More worse, the biological sample usually needs to be isolated and homogenized, which makes in situ monitoring of biological processes impossible.13,14
Fluorescence technology, with its charming merits of simplicity, high sensitivity, and low background noise, is becoming more and more popular in biomedical research.15 Thanks to the enthusiastic endeavors made by scientists, a lot of fluorescent bioprobes have been developed for various applications.16–19 Drugs with inherent fluorescence make real-time in situ tracking of drug molecules in vivo or in vitro possible, which is of critical importance in pharmaceutical research. However, so far, fluorescent drugs as probes for monitoring ion trapping have scarcely been reported in spite of their significance in studying drug delivery and drug distribution in the body. This is partly because it is still difficult to find a drug which shows pH-responsive fluorescence. And it is even more challenging to find a drug which has different emission behaviors between its non-ionized and ionized forms. What's more, fluorescent probes usually suffer from the aggregation-caused quenching (ACQ) effect.20 In 2001, Tang and co-workers discovered aggregation-induced emission (AIE), which is directly opposite to ACQ.21 Since then, a variety of AIEgens have been developed for many advanced applications.22,23 Recently, Tang et al. have put forward natural resources as a new source to explore AIEgens.24 AIEgens, usually obtained from herbal plants, animals, and other natural resources, have a lot of unique advantages, such as being synthesis-free, environmentally friendly, pharmaceutically active, etc.24–26 Previously, we have reported that berberine chloride, a natural isoquinoline isolated from Chinese herbal plants, is a rotor-free AIEgen.24 In this study, dihydro berberine (dhBBR) which is converted from berberine chloride by gut microorganisms was found to be an AIEgen with pH sensitivity. More importantly, the highly similar molecular structure of dhBBR to BBR enables it to work as a suitable drug-like probe for visualizing cell membrane's semipermeability, thanks to its ratiometric fluorescence.
Results and discussion
Synthesis and photophysical properties
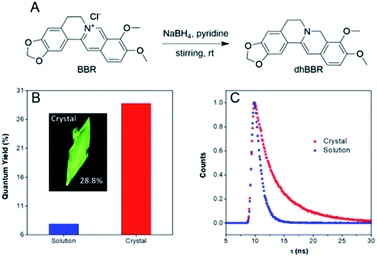
dhBBR was synthesized according to the route shown in Fig. 1A with 86% yield.27 The purity of the product was confirmed by 1H NMR and high resolution mass spectroscopy (HRMS) (Fig. S1 and S3 in the ESI†). The photoluminescence quantum yields (PLQYs) of dhBBR in DMSO solution and crystal states were measured to be 7.9% and 28.8%, respectively (Fig. 1B). The results indicate that dhBBR is a strong solid-state emitter. The fluorescence lifetime of dhBBR in the crystal state (4.68 ns) is longer than that in the solution state (1.1 ns) (Fig. 1C). In addition, from the solution to crystal state, the non-radiative decay rate decreases about 5.5 times (Knr,soln = 8.37 × 108 s−1, Knr,crystal = 1.52 × 108 s−1), which should be responsible for the AIE properties of dhBBR.Mechanism study
To have a clear picture of the photophysical properties of dhBBR, the single-crystal structure of dhBBR was analyzed.28 As shown in Fig. 2, the conformation of dhBBR in the crystal state is non-planar with a 27.94° twisted angle, which indicates that intramolecular vibration is possible. In addition, the intermolecular distance of the adjacent dhBBR molecule aligned in parallel was measured to be 3.771 Å, which exceeds the effective π–π stacking distance (3.5 Å) to quench the fluorescence.29 What's more, multiple intermolecular interactions (2.408–2.888 Å in distance) contribute to rigidification of the molecular conformation which makes dhBBR highly emissive in the crystal state. Thus, the brighter emission of dhBBR in the crystal state than that in solution can possibly be explained by the restriction of intramolecular vibration which suppresses the non-radiative decay pathway.30 To further clarify the intramolecular vibration for determining the photophysical properties of dhBBR, viscosity- and temperature-dependent fluorescence changes of dhBBR were investigated. As shown in Fig. S4(A and B),†dhBBR shows stronger emission in viscous solvents or at low temperature because the intramolecular vibration is suppressed.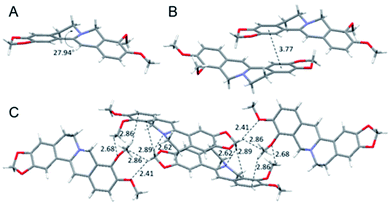 | ||
| Fig. 2 Single crystal packing of dhBBR. (A) Intramolecular torsional angle. (B) Intermolecular distance of adjacent molecules. (C) Intermolecular interactions. | ||
pH-dependent ratiometric fluorescent response
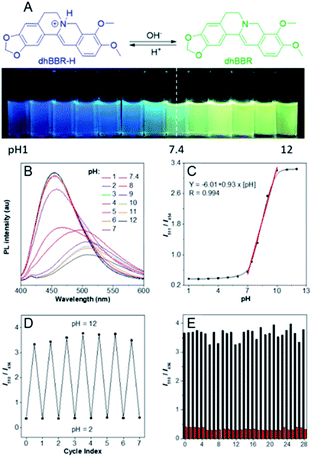
The nitrogen atom in the molecular backbone of dhBBR is speculated to be protonated in acid.31,32 As shown in Fig. 3A, dhBBR exhibits obvious pH-dependent fluorescence. Adding acid to dhBBR causes a gradual blue shift of dhBBR's emission from 511 nm to 454 nm as shown in Fig. 3A. To understand dhBBR's pH-dependent fluorescence, an absorption titration experiment is performed. As shown in Fig. S5,† there is a red shift of the maximum absorption band of dhBBR from 350 nm to 426 nm when the pH of the buffer was changed from 2 to 9. A 1H NMR titration study was performed by adding trifluoroacetic acid (TFA) to dhBBR. As shown in Fig. S2,† there are downfield shifts of the isoquinoline protons induced by TFA. The new peak at 8.0 suggested that the nitrogen atom in the isoquinoline is protonated. A pH titration experiment was then conducted. The fluorescence maximum of dhBBR exhibited a gradual red shift when the pH increased from 1 to 12 (Fig. 3B). More importantly, dhBBR showed an excellent linearity of I511/I454 in the pH range of 7–10 with a pKa value of 8.4 (Fig. 3C), which indicated that dhBBR can serve as a ratiometric pH probe. Furthermore, dhBBR showed a good reversibility between pH 2 and pH 12 as shown in Fig. 3D. In addition, the intensity ratios (I511/I454) of dhBBR in pH 3 and pH 10 buffer remain nearly unchanged after 10 min (Fig. S6†). The fluorescence response of dhBBR towards different interfering species was evaluated (Fig. 3E). The intensity ratio (I511/I454) was negligibly affected by common metal ions (Na+, K+, Ca2+, Mg2+, Fe2+, Cu2+, Zn2+, Mn2+, Al3+, and Pb2+; 0.1 mM for Na+, K+, Ca2+, and Mg2+ and 0.01 mM for Fe2+, Cu2+, Zn2+, Mn2+, Al3+, and Pb2+), negatively charged species (Ac−, SO42−, NO3−, PO42−, S2O32−, and CO32−; 0.01 mM for SO42−, PO42−, S2O32−, and CO32− and 0.02 mM for Ac− and NO3−), amino acids (Phe, His, Met, Pro, Arg, Asp, Cys, and Hcy; 0.01 mM for each), GSH (0.01 mM), glucose (0.01 mM), and reactive oxygen species (H2O2, 0.01 mM; NaClO, 0.01 mM).Visualizing semipermeability of the cell membrane
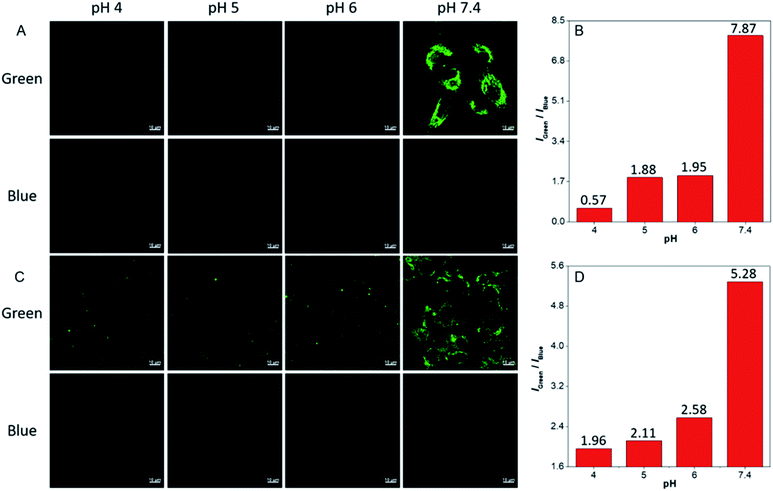
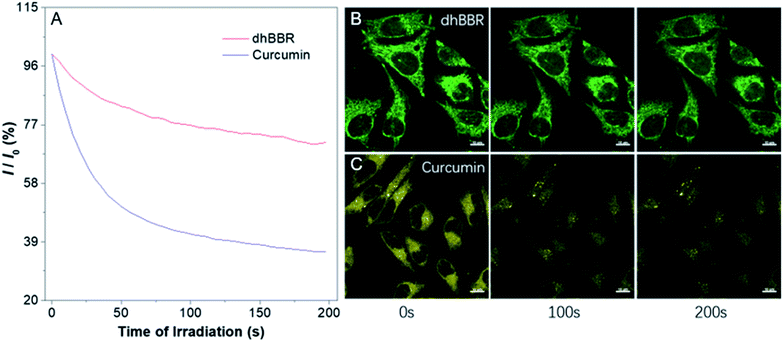
The ratiometric fluorescent response of dhBBR towards pH and the pharmacological properties of dhBBR33,34 inspired us to further explore the potential of dhBBR to be utilized as a probe for ion trapping. Before such an exploration, the biocompatibility of dhBBR with various cell lines was investigated first. As shown in Fig. S7,†dhBBR shows little toxicity towards different cell types at a concentration of 1 μM (with more than 80% cells being alive), indicating the feasibility of a cell imaging study. For cell imaging, different from the reported pH-responsive probes for which nigericin and monensin are usually used for intracellular pH calibration,35–38 we didn't use any chemicals to calibrate intracellular pH because we wanted intracellular pH to be constant. Instead, we incubated cells with dhBBR-containing PBS buffer of varied pH (pH from 4 to 7.4) for a period. Cells were then imaged under confocal microscopy. It was intriguing to notice that A549 cells incubated with dhBBR-containing PBS with a pH range of 4–6 show almost no emission from both blue and green channels (blue channel: 400–460 nm; green channel: 500–600 nm) (Fig. 4A). In contrast, obvious green fluorescence was observed inside cells when A549 cells were incubated with dhBBR-containing PBS buffer of pH 7.4 (Fig. 4A). Also, increasing the pH of the PBS buffer from 4 to 7.4 can also induce a gradual increase of the emission ratio (IGreen/IBlue) (Fig. 4B). Similar results were also observed in HEK 293T cells (Fig. 4C and D). Additionally, different concentrations of dhBBR were used and similar results were obtained in HeLa cells (Fig. S8†). This “acid out, base in” phenomenon can be ascribed to the semipermeability of the cell membrane as mentioned above. Due to the hydrophobic nature of the cell membrane, it is easier for non-ionized dhBBR to enter cells than for ionized forms, a phenomenon known as “ion trapping”, which well explains why incubating cells with dhBBR-containing PBS of higher pH value causes stronger intracellular fluorescence than in the case of dhBBR-containing PBS with lower pH value. Photostability is an important parameter in evaluating a bio-probe's anti-photobleaching ability. As shown in Fig. 5, more than 70% of dhBBR's fluorescence is retained even after 200 seconds of irradiation, while less than 40% of Curcumin's fluorescence is retained after the same irradiation time, suggesting that dhBBR has superior anti-photobleaching ability compared to Curcumin thanks to its AIE properties.Conclusions
In summary, dihydro berberine (dhBBR) was found to be an AIEgen. The single crystal analysis, viscosity effect, and low-temperature effect revealed that the AIE phenomenon of dhBBR originates from the restriction of intramolecular vibration (RIV). Moreover, dhBBR can serve as a fluorescent probe for visualizing ion trapping thanks to its ratiometric fluorescent response to pH and superior anti-photobleaching ability.Experimental procedures
Materials and instrumentation
Chemicals for synthesis were purchased from Sigma-Aldrich or Meryer and used as received without any further purification. Dihydro berberine was prepared according to the reported literature.39 The 1H-NMR spectrum was obtained on a Bruker AV 400 spectrometer. High resolution mass spectra (HRMS) were recorded on a GCT premier CAB048 mass spectrometer operating in MALDI-TOF mode. Ultraviolet-visible (UV-vis) absorption spectra were taken on a PerkinElmer Lambda 25 UV-vis absorption spectrophotometer. Photoluminescence spectra were recorded on a PerkinElmer LS 55 fluorescence spectrometer. The absolute fluorescence quantum yields were measured on a Hamamatsu Absolute Quantum Yield Spectrometer C11347.Synthesis of dihydro berberine (dhBBR)
Anhydrous berberine hydrochloride (1 mmol, 371.8 mg) and sodium borohydride (1 mmol, 37.8 mg) were dissolved in pyridine, and then the mixture was stirred slowly at room temperature. After that, ice water was added to the system. The obtained light yellow powdery solid was filtered, dried in vacuo to afford pure dhBBR (290.1 mg, 86%). 1H NMR (400 MHz, CDCl3, 25 °C), δ (ppm): 7.14 (1H), 6.71 (2H), 6.55 (1H), 5.92 (3H), 4.29 (2H), 3.82 (6H), 3.12–3.09 (2H), 2.87–2.84 (2H); HRMS (MALDI-TOF, m/z): [M]+ calcd. for C20H19NO4, 337.1314; found, 337.1320.Spectral measurement
The absorption and emission spectra were measured in PBS buffer solutions (10 mM). A stock solution of dhBBR (1 mM) was prepared in DMSO and was subsequently diluted to prepare 10 μM solutions of dhBBR in PBS buffer with various pH (1, 2, 3, 4, 5, 6, 7, 7.4, 8, 9, 10, 11, and 12). PBS buffer solutions (10 mM) with varied pH were prepared by using NaOH (1.0 M) or HCl (1.0 M) to adjust the pH. For the calibration curve, 20 μL of stock solutions of dhBBR (1 mM) were mixed with 980 μL of PBS buffer with different pH in a quartz optical cell with a 1.0 cm optical path length at 25 °C, and spectral data were recorded immediately. Excitation was at 365 nm and emission was detected at 454 nm and 511 nm.Cell culturing
HeLa cells, A549 cells, and HEK 293 T cells were purchased from ATCC. All cells were cultured in Dulbecco's Modified Eagle's Medium with 1% penicillin–streptomycin and 10% FBS, at 37 °C in a humidified incubator with 5% CO2. The culture medium was replaced every second day. By treating with 0.25% trypsin–EDTA solution, the cells were collected after they reached confluence.Cytotoxicity assay
HeLa cells, A549 cells, and HEK 293T cells were seeded in 96-well plates at a density of 5000 cells per well, respectively. After 24 h of cell culture, various concentrations of dhBBR were added to the 96-well plates. After another 24 h of cell culture, the medium was removed and freshly prepared MTT medium solution (0.5 mg mL−1, 100 μL) was added to the 96-well plates. After incubation at 37 °C, with 5% CO2, for 6 h, the MTT medium solution was removed carefully. After that, 100 μL of DMSO was added to each well and the plate was gently shaken at room temperature to dissolve all the formed precipitates. A microplate reader was utilized to measure the absorbance at 570 nm from which the cell viability could be determined. Cell viability was expressed as the ratio of absorbance of the cells incubated with dhBBR solution to that of the cells incubated with culture medium only.Cell imaging
Cells were grown in a 35 mm Petri dish with a cover slip at 37 °C, with 5% CO2. First, cells were incubated with different-pH buffer solutions containing dhBBR (0.5, 1, and 10 μM) or Curcumin (10 μM) for 30 min at 37 °C, with 5% CO2. Then, the staining solution was removed and the cells were washed with PBS of the same pH as the staining solution three times. After that, the cells were imaged using confocal microscopy (Zeiss laser scanning confocal microscope LSM7 DUO). For dhBBR, the excitation wavelength was 405 nm, and the emission filter was 400–460 nm and 500–600 nm, respectively; for Curcumin, the excitation wavelength was 405 nm, and the emission filter was 500–600 nm.40Photobleaching assay
HeLa cells stained with dhBBR were irradiated using a 405 nm laser for 200 s continuously using confocal microscopy to evaluate dhBBR's photostability. For comparison, HeLa cells stained with Curcumin were irradiated using a 405 nm laser under the same conditions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (21788102), the Research Grants Council of Hong Kong (16305518, N-HKUST609/19, A-HKUST605/16, and C6009-17G), the Innovation and Technology Commission (ITC-CNERC14SC01) and the Shenzhen Science and Technology Program (JCYJ20180507183832774). We are also grateful for the technical support from AIEgen Biotech Co., LtdNotes and references
- P. Marrack, D. Lo, R. Brinster, R. Palmiter, L. Burkly, R. H. Flavell and J. Kappler, Cell, 1988, 53, 627–634 CrossRef CAS PubMed.
- R. Sutherland, Science, 1988, 240, 177–184 CrossRef CAS PubMed.
- K. Simons and E. Ikonen, Nature, 1997, 387, 569–572 CrossRef CAS PubMed.
- S. J. Singer and G. L. Nicolson, Science, 1972, 175, 720–731 CrossRef CAS PubMed.
- N. Raghunand, B. P. Mahoney and R. J. Gillies, Biochem. Pharmacol., 2003, 66, 1219–1229 CrossRef CAS PubMed.
- H. Arnold, F. Bourseaux and N. Brock, Nature, 1958, 181, 931 CrossRef CAS PubMed.
- B. A. Webb, M. Chimenti, M. P. Jacobson and D. L. Barber, Nat. Rev. Cancer, 2011, 11, 671–677 CrossRef CAS PubMed.
- F. Lucien, P.-P. Pelletier, R. R. Lavoie, J.-M. Lacroix, S. Roy, J.-L. Parent, D. Arsenault, K. Harper and C. M. Dubois, Nat. Commun., 2017, 8, 15884 CrossRef CAS PubMed.
- N. Raghunand and R. J. Gillies, Drug Resist. Updates, 2000, 3, 39–47 CrossRef CAS PubMed.
- M. K. Nair, D. J. Chetty, Y. W. Chien and H. Ho, J. Pharm. Sci., 1997, 86, 257–262 CrossRef CAS PubMed.
- L.-L. H. Chen, D. J. Chetty and Y. W. Chien, Int. J. Pharm., 1999, 184, 63–72 CrossRef CAS PubMed.
- H. M. Nielsen and M. R. Rassing, Eur. J. Pharm. Sci., 2002, 16, 151–157 CrossRef CAS PubMed.
- C. Yin, F. Huo, J. Zhang, R. Martínez-Máñez, Y. Yang, H. Lv and S. Li, Chem. Soc. Rev., 2013, 42, 6032–6059 RSC.
- S. Wang, H. Yin, Y. Huang and X. Guan, Anal. Chem., 2018, 90, 8170–8177 CrossRef CAS PubMed.
- C. Caskey, Science, 1987, 236, 1223–1229 CrossRef CAS PubMed.
- R. Wirth, P. Gao, G. U. Nienhaus, M. Sunbul and A. Jäschke, J. Am. Chem. Soc., 2019, 141, 7562–7571 CrossRef CAS PubMed.
- C. J. Macnevin, T. Watanabe, M. Weitzman, A. Gulyani, S. Fuehrer, N. K. Pinkin, X. Tian, F. Liu, J. Jin and K. M. Hahn, J. Am. Chem. Soc., 2019, 141, 7275–7282 CrossRef CAS PubMed.
- Q. Qi, W. Chi, Y. Li, Q. Qiao, J. Chen, L. Miao, Y. Zhang, J. Li, W. Ji, T. Xu, X. Liu, J. Yoon and Z. Xu, Chem. Sci., 2019, 10, 4914–4922 RSC.
- H. Y. Kwon, X. Liu, E. G. Choi, J. Y. Lee, S. Y. Choi, J. Y. Kim, L. Wang, S. J. Park, B. Kim, Y. A. Lee, J. J. Kim, N. Y. Kang and Y. T. Chang, Angew. Chem., Int. Ed., 2019, 58, 8426–8431 CrossRef CAS PubMed.
- M. Yamaguchi, S. Ito, A. Hirose, K. Tanaka and Y. Chujo, Mater. Chem. Front., 2017, 1, 1573–1579 RSC.
- C. Zhu, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, ACS Appl. Bio Mater., 2018, 1, 1768–1786 CrossRef CAS.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718–11940 CrossRef CAS PubMed.
- R. T. K. Kwok, C. W. T. Leung, J. W. Y. Lam and B. Z. Tang, Chem. Soc. Rev., 2015, 44, 4228–4238 RSC.
- Y. Gu, Z. Zhao, H. Su, P. Zhang, J. Liu, G. Niu, S. Li, Z. Wang, R. T. K. Kwok, X.-L. Ni, J. Sun, A. Qin, J. W. Y. Lam and B. Z. Tang, Chem. Sci., 2018, 9, 6497–6502 RSC.
- B. Li, X. Xie, Z. Chen, C. Zhan, F. Zeng and S. Wu, Adv. Funct. Mater., 2018, 28, 1800692 CrossRef.
- T. He, N. Niu, Z. Chen, S. Li, S. Liu and J. Li, Adv. Funct. Mater., 2018, 28, 1870068 CrossRef.
- S. Fu, Y. Xie, J. Tuo, Y. Wang, W. Zhu, S. Wu, G. Yan and H. Hu, MedChemComm, 2015, 6, 164–173 RSC.
- K. Biradha, CrystEngComm, 2003, 5, 388–391 RSC.
- M. D. Curtis, J. Cao and J. W. Kampf, J. Am. Chem. Soc., 2004, 126, 4318–4328 CrossRef CAS PubMed.
- N. L. C. Leung, N. Xie, W. Yuan, Y. Liu, Q. Wu, Q. Peng, Q. Miao, J. W. Y. Lam and B. Z. Tang, Chem.–Eur. J., 2014, 20, 15349–15353 CrossRef CAS PubMed.
- W. Adam, A. Grimison and G. Rodríguez, Tetrahedron, 1967, 23, 2513–2521 CrossRef CAS.
- G. A. Olah, A. M. White and D. H. Obrien, Chem. Rev., 1970, 70, 561–591 CrossRef CAS.
- R. Feng, J.-W. Shou, Z.-X. Zhao, C.-Y. He, C. Ma, M. Huang, J. Fu, X.-S. Tan, X.-Y. Li, B.-Y. Wen, X. Chen, X.-Y. Yang, G. Ren, Y. Lin, Y. Chen, X.-F. You, Y. Wang and J.-D. Jiang, Sci. Rep., 2015, 5, 12155 CrossRef CAS PubMed.
- D. Yu, L. Lv, L. Fang, B. Zhang, J. Wang, G. Zhan, L. Zhao, X. Zhao and B. Li, Plos One, 2017, 12(8), e0181823 CrossRef PubMed.
- X. Liu, Y. Su, H. Tian, L. Yang, H. Zhang, X. Song and J. W. Foley, Anal. Chem., 2017, 89, 7038–7045 CrossRef CAS PubMed.
- K. Li, Q. Feng, G. Niu, W. Zhang, Y. Li, M. Kang, K. Xu, J. He, H. Hou and B. Z. Tang, ACS Sens, 2018, 3, 920–928 CrossRef CAS PubMed.
- G. Niu, P. Zhang, W. Liu, M. Wang, H. Zhang, J. Wu, L. Zhang and P. Wang, Anal. Chem., 2017, 89, 1922–1929 CrossRef CAS PubMed.
- J. Zhang, M. Yang, W. Mazi, K. Adhikari, M. Fang, F. Xie, L. Valenzano, A. Tiwari, F.-T. Luo and H. Liu, ACS Sens, 2015, 1, 158–165 CrossRef PubMed.
- S. Fu, Y. Xie, J. Tuo, Y. Wang, W. Zhu, S. Wu, G. Yan and H. Hu, MedChemComm, 2015, 6, 164–173 RSC.
- A. Kunwar, A. Barik, B. Mishra, K. Rathinasamy, R. Pandey and K. Priyadarsini, Biochim. Biophys. Acta, 2008, 1780, 673–679 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods; 1H NMR and HRMS spectra of compounds; photophysical data and imaging data. See DOI: 10.1039/d0sc02097d |
| This journal is © The Royal Society of Chemistry 2020 |