Open Access Article
Open Access ArticleIn situ assembled ZIF superstructures via an emulsion-free soft-templating approach†
Namita
Singh
a,
Sana
Ahmed
a,
Aliyah
Fakim
a,
Somayah
Qutub
a,
Othman
Alahmed
a,
Omar
El Tall
b,
Osama
Shekhah
 c,
Mohamed
Eddaoudi
c,
Mohamed
Eddaoudi
 c and
Niveen M.
Khashab
c and
Niveen M.
Khashab
 *a
*a
aSmart Hybrid Materials (SHMs) Laboratory, Advanced Membranes and Porous Materials Center, King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Kingdom of Saudi Arabia. E-mail: Niveen.khashab@kaust.edu.sa
bKAUST Core Labs, King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Kingdom of Saudi Arabia
cProf. Mohamed Eddaoudi Functional Materials Design, Discovery & Development Research Group (FMD3) Advanced Membranes & Porous Materials Center, King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Kingdom of Saudi Arabia
First published on 22nd September 2020
Abstract
Assembling well-defined MOF superstructures remains challenging as it requires easily removable hard templates or readily available immiscible solutions for an emulsion-based soft-template approach. In this work, a single-step emulsion-free soft templating approach is reported to spontaneously prepare hollow ZIF-8 and ZIF-67 colloidosomes with no further purification. These superstructures can load different enzymes regardless of the size and charge with a high encapsulation efficiency of 99%. We envisage that this work will expand the repertoires of MOF superstructures by the judicious selection of precursors and the reaction medium.
Introduction
The fabrication of hollow metal–organic framework (MOF) superstructures enables superior guest encapsulation, complex microreactor preparation and highly selective molecular separation.1–6 An interesting class of superstructures7,8 are colloidosomes,9 which are microspheres with shells composed of self-assembled nanoparticles at the surface of emulsion droplets.10 To date, shaped metal oxides and orderly arranged polymeric/biomolecule templates have been successfully used as hard templates for the fabrication of complex MOF superstructures.11–13 On the other hand, emulsified droplets and micellar templates have emerged as soft templates14–24 where MOF colloidosomes are produced by mixing immiscible solutions or by controlling the flow of immiscible fluids in a microfluidic setup.1–5 The porosity of colloidosomes is defined by the size of the interstices between the colloidal particles which can be tuned by the corresponding particle size.25 MOF colloidosomes are especially attractive due to the intrinsic properties of MOFs such as porosity, high surface areas, high temperature stability and facile synthesis which impart different types of porosity and functionality to the colloidosomes and expand the scope of utilization of these higher order assemblies.26–30 Zeolitic imidazolate frameworks (ZIFs) are a well-known subclass of MOFs as they are topologically isomorphic to zeolites and have recently been extensively used in biomedical applications due to their biocompatibility.31–34 ZIF-8, in particular, has been extensively used for drug delivery, biomimetic mineralization and sheltering of biomacromolecules.35–41Hollow ZIF-8 and HKUST-1 colloidosomes were prepared via an emulsion-based soft interfacial formation method.1 A spray-drying technique has also been successfully employed to prepare a library of hollow MOF superstructures.2 More recently, a one-step emulsion-based strategy where MOFs are formed in solution has been employed to prepare such hollow assemblies.5 However, the use of emulsions can be greatly limited by the solubility compatibility between the MOF precursors and the constituent solvents of the emulsion.2
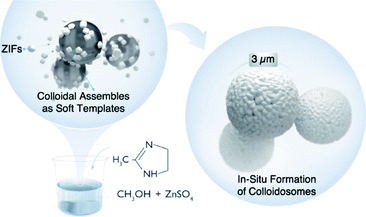
Here, we report an emulsion-free soft template approach for the preparation of hollow ZIF colloidosomes in a single step. The solubility of metal sulfate salts (Zn/Co) in methanol leads to the in situ formation of spherical colloidal assemblies that can act as soft templates for self-assembling nano ZIFs, formed in the reaction mixture, as stable colloidosomes (Scheme 1). Metal sulfates are insoluble in ethanol and longer chain alcohols; consequently they have been used as hard templates for the preparation of hollow inorganic nanoparticles due to their self-conglobation effect.42 In our case, the hydrated metal sulfate (ZnSO4·7H2O) is soluble in methanol. It forms a clear colloidal solution where the self-conglobation effect of the hydrated salt in alcohol leads to an aggregation of colloids that ultimately affords a soft-template for the colloidosome assembly. To the best of our knowledge, this is the first example of emulsion-free soft template preparation of ZIF colloidosomes via a facile single step approach. This synthesis does not require immiscible solvents, the use of surfactants or microfluidic set-ups. We believe that this approach will advance the design and synthesis of more complex MOF superstructures without the use of hard templating or emulsion techniques.
Results and discussion
A methanolic solution of 2-methyl-imidazole (MeIm) was added to an already prepared solution of ZnSO4·7H2O in methanol with constant stirring (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10/ZnSO4·7H2O
10/ZnSO4·7H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
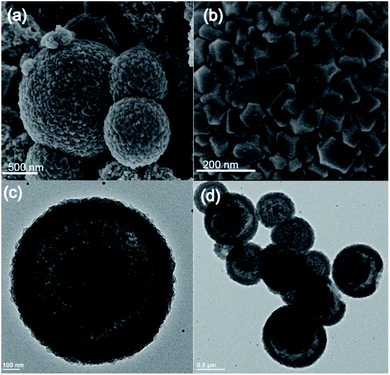
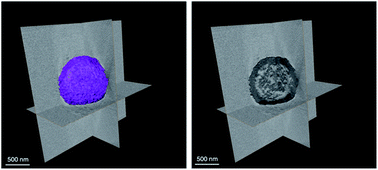
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeIm) at room temperature. The addition of the MeIm solution turned the clear methanolic ZnSO4·7H2O solution into a white turbid solution. After stirring for 1 h, the white solid was isolated by centrifugation, washed with water and methanol (2–3 times) and dried at 60 °C under vacuum. The powder X-ray diffraction (PXRD) pattern of the formed product matches perfectly the simulated XRD pattern and indicates the formation of phase pure ZIF-8 with the underlying sodalite (sod) topology (Fig. S1†). Furthermore, the scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images confirm the formation of well-defined spherical shaped colloidosomes (Fig. 1). Their size ranges between 600 nm and 2 μm with a shell thickness of 100 nm (Fig. 1c and d). The inner cavity of the colloidosomes is estimated to be between 500 nm and 1.9 μm depending on the overall size (Fig. 1d). A closer look at the external surface of these colloidosomes reveals that they are composed of small ZIF-8 nanocrystals, typically of 80–100 nm in size (Fig. 1b). The TEM images indicate that the colloidosome shell thickness corresponds to the presence of more than one layer of crystals (Fig. 1c and d). Tomography studies clearly reveal that the inner cavity of the colloidosome is not completely hollow and contains lumps of the ZIF nanocrystals in the cavity (Fig. 2 and ESI Video S1†).
MeIm) at room temperature. The addition of the MeIm solution turned the clear methanolic ZnSO4·7H2O solution into a white turbid solution. After stirring for 1 h, the white solid was isolated by centrifugation, washed with water and methanol (2–3 times) and dried at 60 °C under vacuum. The powder X-ray diffraction (PXRD) pattern of the formed product matches perfectly the simulated XRD pattern and indicates the formation of phase pure ZIF-8 with the underlying sodalite (sod) topology (Fig. S1†). Furthermore, the scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images confirm the formation of well-defined spherical shaped colloidosomes (Fig. 1). Their size ranges between 600 nm and 2 μm with a shell thickness of 100 nm (Fig. 1c and d). The inner cavity of the colloidosomes is estimated to be between 500 nm and 1.9 μm depending on the overall size (Fig. 1d). A closer look at the external surface of these colloidosomes reveals that they are composed of small ZIF-8 nanocrystals, typically of 80–100 nm in size (Fig. 1b). The TEM images indicate that the colloidosome shell thickness corresponds to the presence of more than one layer of crystals (Fig. 1c and d). Tomography studies clearly reveal that the inner cavity of the colloidosome is not completely hollow and contains lumps of the ZIF nanocrystals in the cavity (Fig. 2 and ESI Video S1†).
The nitrogen adsorption analysis of the ZIF-8 colloidosomes shows a type IV isotherm with a small increase in the uptake at high relative pressure leading to a moderately hysteretic desorption profile (Fig. S2a†). This is in agreement with the surface mesoporosity arising from the interstices between the small ZIF-8 crystals comprising the sphere shell wall. The NLDFT pore size distribution analysis also supports the microporosity (<2 nm) and mesoporosity (2–20 nm) (Fig. S2b†). This was further verified by the ability of gold nanoparticles (5 nm) to diffuse through the colloidosome shell to the interior hollow pocket (Fig. S2c†). The apparent Brunauer–Emmet–Teller (BET) surface area and pore volume of ZIF-8 colloidosomes were calculated to be 1513 m2 g−1 and 0.76 cm3 g−1 respectively which are in good agreement with the reported ZIF-8 values.2 Thermogravimetric analysis (TGA) was performed under N2 and showed a thermal stability of up to 380 °C and a gradual weight-loss of 8.0% corresponding to the removal of unreacted species (e.g. 2-methylimidazole) (Fig. S3†). This is consistent with the reduced surface area of bulk ZIF-8 crystals.4
To further our understanding and gain more insight into the emulsion-free preparation of these superstructures, a time-based study was performed to investigate the step-by-step assembly. After mixing the reactants, the reaction mixture was stirred for 10, 20, 30, 40, 50 and 60 min, then washed and dried accordingly. The SEM results confirm the formation of ZIF-8 colloidosomes in the initial 10 min (Fig. S4†). No discrete small crystals were observed in SEM which supports the one step spontaneous fabrication of these colloidosomes. All the products obtained by varying the stirring duration show similar average outer/inner diameters.
As another control, the ratio of the MeIm was varied with respect to the metal salt to understand the influence of concentrations on the colloidosome assembly. No changes in the structure or size were observed when the metal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ligand was reduced from 1
ligand was reduced from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 1
10 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and then to 1
8 and then to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6. However, when the ratio was reduced to 1
6. However, when the ratio was reduced to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, only polyhedron crystals were identified in the SEM, which do not assemble into superstructures (Fig. S5†). On the other hand, the size of the colloidosomes decreased as the metal ligand ratio increased from 1
5, only polyhedron crystals were identified in the SEM, which do not assemble into superstructures (Fig. S5†). On the other hand, the size of the colloidosomes decreased as the metal ligand ratio increased from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 1
10 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 (Fig. S6†). The SEM images show the superstructure with an average size of 200–500 nm and consisting of one layer of the ZIF-8 crystal (20–25 nm in size) shell. Notably, this is much smaller than MOF colloidosomes synthesized by using surfactants or by the emulsion method. This could be claimed as the smallest MOF colloidosomes synthesized without using any templating reagents. The ZIF-8 superstructures with a 1
18 (Fig. S6†). The SEM images show the superstructure with an average size of 200–500 nm and consisting of one layer of the ZIF-8 crystal (20–25 nm in size) shell. Notably, this is much smaller than MOF colloidosomes synthesized by using surfactants or by the emulsion method. This could be claimed as the smallest MOF colloidosomes synthesized without using any templating reagents. The ZIF-8 superstructures with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18 metal ligand ratio show well defined structures with a uniform size of around 200 nm (Fig. S6d†). Replacing methanol with ethanol or other long chain alcohols did not yield any superstructure due to the insolubility of the metal salt in the solvent.
18 metal ligand ratio show well defined structures with a uniform size of around 200 nm (Fig. S6d†). Replacing methanol with ethanol or other long chain alcohols did not yield any superstructure due to the insolubility of the metal salt in the solvent.
After verifying the influence of the MeIm concentration and solvents on the colloidosome assembly, we ventured to understand the effect of the metal salt. ZIF-67 colloidosomes were successfully prepared by replacing the ZnSO4 salt with CoSO4, keeping the reaction conditions constant. The formation of the ZIF-67 colloidosomes was confirmed by PXRD and SEM (Fig. S7 and S8†). Employing non-sulfate-based metal salts (Cl− and NO3−) did not yield any colloidosome. Literature reports support the effective templating role of metal sulphate salts MSO4·xH2O (M = Zn, Fe, Co, etc.).42 More recently, tripodal N-methylated(1,3,5-benzene-tricarboxamide)-tris(phenylurea) BTA ligands were able to form different self-assemblies including nanospheres via hydrogen bonding templated by sulfate anions.43
As ZIF-8 has been used as the best candidate for protein encapsulation due to its ease of assembly, biocompatibility, and eventual biodegradation, we tested the applicability of our hollow ZIF-8 colloidosomes for enzyme encapsulation.29,34 ZIF-8 colloidosomes are excellent candidates due to their hollow interior and microporous shell where different size and charged enzymes can diffuse.44 Moreover, ZIF-8 colloidosomes are extremely stable in water with no disassembly over 1 month in solution. Fluorescein isothiocynate (FITC) tagged BSA (FBSA) was added to water dispersed ZIF-8 colloidosomes followed by sonication for 10 min and further stirring at 100 rpm for 48 h. Following a reported procedure,45 the sample was then washed with trypsin and SDS to eliminate any surface bound protein followed by several washings with water. Confocal laser scanning microscopy (CLSM) analysis indicates that FBSA molecules are incorporated into the colloidosomes where green spheres and green rings were obtained (Fig. 3a). This can be interpreted as corresponding to the intercalated proteins in the hollow interior (spheres) and in the outside shell (rings).
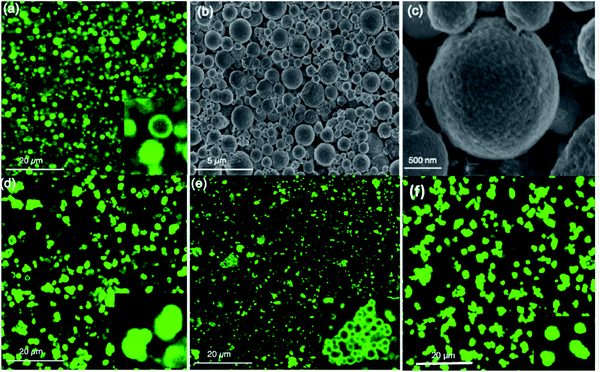 | ||
| Fig. 3 (a) CLSM images (b and c) SEM images of FITC-BSA@ZIF-8 colloidosome. CLSM images of (d) FITC-Catalase (e) FITC-HRP and (f) FITC-Lysozyme@ZIF-8 colloidosomes. | ||
The SEM images of FBSA@ZIF-8 colloidosomes confirm the structural integrity of the colloidosomes after encapsulation (Fig. 3b and c). The PXRD pattern of the BSA@ZIF-8 colloidosome is in agreement with that of the ZIF-8 colloidosome (Figure S9a†). The IR spectrum for the BSA@ZIF-8 colloidosome post incubation shows significant characteristic BSA peaks between 1640 and 1660 and 1510–1560 cm−1, corresponding to amide I and II bands for the adsorbed BSA at the surface of the colloidosome (Fig. S9b and c†).36 After washing, BSA@ZIF-8 colloidosomes show no significant IR peak for BSA, which is consistent with reported literature on enzyme encapsulation.46 The ZIF-8 colloidosomes show a positive ζ-potential/surface charge (29 mV) but after encapsulation of BSA, the ζ-potential is measured at +15 mV (Fig. S10†). Encapsulation of other enzymes namely FITC tagged catalase (negatively charged), HRP (negatively charged) and lysozyme (positively charged) was also studied (Fig. S11†). Encapsulation of positively charged enzymes such as lysozymes is challenging using the infiltration approach however, loading readily occurs using the ZIF-8 colloidosomes. We hypothize that the mesopores facilitate the loading of the positively charged enzymes that usually face charge repulsion that hinders their encapsulation. The encapsulation efficiency and loading capacity were measured for BSA, albumin and HRP proteins (Table S1†). The encapsulation efficiency (%EE) was calculated to be 99% in all cases using the Bradford assay.47 The loading capacity (%LC) was measured to be 10% with 0.5 mg protein (Table S1†). When we varied the protein to colloidosome ratio, we concluded that the best loading ratio (protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) colloidosome) is 1
colloidosome) is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (Table S2†). SEM images confirm the integrity of the colloidosomes after loading (Fig. S12†). CLSM analysis shows catalase and lysozyme loaded colloidosomes mainly as green spheres while HRP loaded colloidosomes appeared as green rings (Fig. 3d–f). Moreover, the coordination environment of the colloidosome prevents the leaking of the enzymes as fluorescence experiments showed a slow release of the proteins over 48 h (Fig. S13†). To verify that the enzyme activity was maintained after encapsulation, HRP was tested at pH 7 and 5 and showed constant activities of 81% and 76% respectively after 48 h (Fig. S14†).
10 (Table S2†). SEM images confirm the integrity of the colloidosomes after loading (Fig. S12†). CLSM analysis shows catalase and lysozyme loaded colloidosomes mainly as green spheres while HRP loaded colloidosomes appeared as green rings (Fig. 3d–f). Moreover, the coordination environment of the colloidosome prevents the leaking of the enzymes as fluorescence experiments showed a slow release of the proteins over 48 h (Fig. S13†). To verify that the enzyme activity was maintained after encapsulation, HRP was tested at pH 7 and 5 and showed constant activities of 81% and 76% respectively after 48 h (Fig. S14†).
As multistep enzymatic reactions in living organisms commonly function without the separation of intermediates, this ultimately provides high selectivity and limits by-product formation.48 Mimicking natural design, the application of cascade reactions in organic synthesis offers far more advantages over the classical step-by-step approach mainly because there is no need for further purification with reduced operation time and costs. Interestingly, nano MOFs have been used as nanoreactors for evaluating the bio-catalytical properties of enzyme cascades.48–51 The microporosity of the ZIF-8 nanoparticles coupled with the mesoporosity of the colloidosome shell implies that the diffusion of two different size proteins into the colloidosomes can lead to distribution or localization preferences. As a proof of concept, equal molar ratios of FITC (green) tagged catalase (≈9 nm) and rhodamine B isothiocyanate (red) tagged HRP (≈2 nm) were left to diffuse into the colloidosomes over 24 h followed by a trypsin wash and successive washings with water. The CLSM images indicate the presence of both FITC–catalase and RhB–HRP in the ZIF-8 colloidosomes (Fig. 4). We hypothesize that the bigger size catalase occupies the interior, diffusing through the mesopores, while the smaller HRP concentrated in the microporous ZIF-8 shell (Fig. 4). However, CLSM images alone are not conclusive enough to verify the compartmentalization of different enzymes. Further studies are underway to understand how enzyme compartmentalization can be fully exploited in these superstructures to be implanted in enzyme cascade reactions in the future.
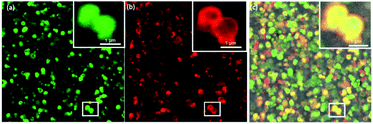 | ||
| Fig. 4 CLSM images of (a) FITC-Catalase@ZIF-8 colloidosome (b) RhB-HRP@ZIF-8 colloidosome and (c) merged image. | ||
Conclusion
In conclusion, we report a facile and effective approach for the synthesis of hollow ZIF colloidosomes by exploiting the soft templating effect of sulfate salts in methanol. To the best of our knowledge, this is the first demonstration of an emulsion free methodology to prepare 200–500 nm size MOF colloidosomes in situ. Furthermore, the mesoporosity of ZIF-8 colloidosomes facilitates the high loading of different enzymes regardless of the size or charge. We envision that this approach will promote the synthesis of more complex MOF superstructures without the need for surfactants, hard templating or emulsion techniques.Conflicts of interest
There are no conflicts to declare.References
- R. Ameloot, F. Vermoortele, W. Vanhove, M. B. J. Roeffaers, B. F. Sels and D. E. De Vos, Nat. Chem., 2011, 3, 382–387 Search PubMed.
- A. Carné-Sánchez, I. Imaz, M. Cano-Sarabia and D. Maspoch, Nat. Chem., 2013, 5, 203–211 Search PubMed.
- M. Pang, A. J. Cairns, Y. Liu, Y. Belmabkhout, H. C. Zeng and M. Eddaoudi, J. Am. Chem. Soc., 2013, 135, 10234–10237 Search PubMed.
- L. Lupica-Spagnolo, D. J. Ward, J.-J. Marie, S. Lymperopoulou and D. Bradshaw, Chem. Commun., 2018, 54, 8506–8509 Search PubMed.
- G. Zhu, M. Zhang, Y. Bu, L. Lu, X. Lou and L. Zhu, Chem.–Asian J., 2018, 13, 2891–2896 Search PubMed.
- F. Zhang, Y. Wei, X. Wu, H. Jiang, W. Wang and H. Li, J. Am. Chem. Soc., 2014, 136, 13963–13966 Search PubMed.
- L. Feng, K.-Y. Wang, T.-H. Yan and H.-C. Zhou, Chem. Sci., 2020, 11, 1643–1648 Search PubMed.
- K. Sumida, N. Moitra, J. Reboul, S. Fukumoto, K. Nakanishi, K. Kanamori, S. Furukawa and S. Kitagawa, Chem. Sci., 2015, 6, 5938–5946 Search PubMed.
- M. Li, D. C. Green, J. L. R. Anderson, B. P. Binks and S. Mann, Chem. Sci., 2011, 2, 1739–1745 Search PubMed.
- S. Li, B. A. Moosa, J. G. Croissant and N. M. Khashab, Angew. Chem., Int. Ed., 2015, 54, 6804–6808 Search PubMed.
- H. J. Lee, W. Cho and M. Oh, Journal, 2012, 48, 221–223 Search PubMed.
- S. El-Hankari, J. Aguilera-Sigalat and D. Bradshaw, J. Mater. Chem. A, 2016, 4, 13509–13518 Search PubMed.
- L. Lin, T. Zhang, H. Liu, J. Qiu and X. Zhang, Nanoscale, 2015, 7, 7615–7623 Search PubMed.
- S. Jiang, Q. Chen, M. Tripathy, E. Luijten, K. S. Schweizer and S. Granick, Adv. Mater., 2010, 22, 1060–1071 Search PubMed.
- M. Fujiwara, K. Shiokawa, Y. Tanaka and Y. Nakahara, Chem. Mater., 2004, 16, 5420–5426 Search PubMed.
- Y.-S. Cho, S.-H. Kim, G.-R. Yi and S.-M. Yang, Colloids Surf., A, 2009, 345, 237–245 Search PubMed.
- I. Akartuna, E. Tervoort, A. R. Studart and L. J. Gauckler, Langmuir, 2009, 25, 12419–12424 Search PubMed.
- M. F. Hsu, M. G. Nikolaides, A. D. Dinsmore, A. R. Bausch, V. D. Gordon, X. Chen, J. W. Hutchinson, D. A. Weitz and M. Marquez, Langmuir, 2005, 21, 2963–2970 Search PubMed.
- S. Jiang and S. Granick, Langmuir, 2008, 24, 2438–2445 Search PubMed.
- O. D. Velev, K. Furusawa and K. Nagayama, Langmuir, 1996, 12, 2374–2384 Search PubMed.
- Q. Wu, Z. Wang, X. Kong, X. Gu and G. Xue, Langmuir, 2008, 24, 7778–7784 Search PubMed.
- R. McGorty, J. Fung, D. Kaz and V. N. Manoharan, Mater. Today, 2010, 13, 34–42 Search PubMed.
- A. D. Dinsmore, M. F. Hsu, M. G. Nikolaides, M. Marquez, A. R. Bausch and D. A. Weitz, Science, 2002, 298, 1006 Search PubMed.
- O. D. Velev, A. M. Lenhoff and E. W. Kaler, Science, 2000, 287, 2240 Search PubMed.
- H. C. Zeng, J. Mater. Chem., 2011, 21, 7511–7526 Search PubMed.
- B. Chen, C. Liang, J. Yang, D. S. Contreras, Y. L. Clancy, E. B. Lobkovsky, O. M. Yaghi and S. Dai, Angew. Chem., Int. Ed., 2006, 45, 1390–1393 Search PubMed.
- O. K. Farha, A. Özgür Yazaydın, I. Eryazici, C. D. Malliakas, B. G. Hauser, M. G. Kanatzidis, S. T. Nguyen, R. Q. Snurr and J. T. Hupp, Nat. Chem., 2010, 2, 944–948 Search PubMed.
- J. Huo, J. Aguilera-Sigalat, S. El-Hankari and D. Bradshaw, Chem. Sci., 2015, 6, 1938–1943 Search PubMed.
- S. Wang, C. M. McGuirk, A. d'Aquino, J. A. Mason and C. A. Mirkin, Adv. Mater., 2018, 30, 1800202 Search PubMed.
- J. Navarro-Sánchez, N. Almora-Barrios, B. Lerma-Berlanga, J. J. Ruiz-Pernía, V. A. Lorenz-Fonfria, I. Tuñón and C. Martí-Gastaldo, Chem. Sci., 2019, 10, 4082–4088 Search PubMed.
- G. Chen, S. Huang, X. Kou, S. Wei, S. Huang, S. Jiang, J. Shen, F. Zhu and G. Ouyang, Angew. Chem., Int. Ed., 2019, 58, 1463–1467 Search PubMed.
- F. Carraro, M. d. J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Chem. Sci., 2020, 11, 3397–3404 Search PubMed.
- K. S. Park, Z. Ni, A. P. Côté, J. Y. Choi, R. Huang, F. J. Uribe-Romo, H. K. Chae, M. O'Keeffe and O. M. Yaghi, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10186 Search PubMed.
- R. Banerjee, A. Phan, B. Wang, C. Knobler, H. Furukawa, M. Keeffe and O. M. Yaghi, Science, 2008, 319, 939 Search PubMed.
- C. Doonan, R. Riccò, K. Liang, D. Bradshaw and P. Falcaro, Acc. Chem. Res., 2017, 50, 1423–1432 Search PubMed.
- M. A. Luzuriaga, R. P. Welch, M. Dharmarwardana, C. E. Benjamin, S. Li, A. Shahrivarkevishahi, S. Popal, L. H. Tuong, C. T. Creswell and J. J. Gassensmith, ACS Appl. Mater. Interfaces, 2019, 11, 9740–9746 Search PubMed.
- G. Chen, X. Kou, S. Huang, L. Tong, Y. Shen, W. Zhu, F. Zhu and G. Ouyang, Angew. Chem., Int. Ed., 2020, 59, 2867–2874 Search PubMed.
- A. C. McKinlay, R. E. Morris, P. Horcajada, G. Férey, R. Gref, P. Couvreur and C. Serre, Angew. Chem., Int. Ed., 2010, 49, 6260–6266 Search PubMed.
- M. B. Majewski, A. J. Howarth, P. Li, M. R. Wasielewski, J. T. Hupp and O. K. Farha, CrystEngComm, 2017, 19, 4082–4091 Search PubMed.
- S. K. Alsaiari, S. Patil, M. Alyami, K. O. Alamoudi, F. A. Aleisa, J. S. Merzaban, M. Li and N. M. Khashab, J. Am. Chem. Soc., 2018, 140, 143–146 Search PubMed.
- M. Z. Alyami, S. K. Alsaiari, Y. Li, S. S. Qutub, F. A. Aleisa, R. Sougrat, J. S. Merzaban and N. M. Khashab, J. Am. Chem. Soc., 2020, 142, 1715–1720 Search PubMed.
- X. Lü, F. Huang, X. Mou, Y. Wang and F. Xu, Adv. Mater., 2010, 22, 3719–3722 Search PubMed.
- A. B. Aletti, S. Blasco, S. J. Aramballi, P. E. Kruger and T. Gunnlaugsson, Chem, 2019, 5, 2617–2629 Search PubMed.
- L. Zhang, W. Baslyman, P. Yang and N. M. Khashab, Chem. Commun., 2019, 55, 620–623 Search PubMed.
- P. Li, J. A. Modica, A. J. Howarth, E. Vargas L, P. Z. Moghadam, R. Q. Snurr, M. Mrksich, J. T. Hupp and O. K. Farha, Chem, 2016, 1, 154–169 Search PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Nat. Commun., 2015, 6, 7240 Search PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 Search PubMed.
- X. Wu, J. Ge, C. Yang, M. Hou and Z. Liu, Chem. Commun., 2015, 51, 13408–13411 Search PubMed.
- W.-H. Chen, M. Vázquez-González, A. Zoabi, R. Abu-Reziq and I. Willner, Nat. Catal., 2018, 1, 689–695 Search PubMed.
- X. Lian, Y.-P. Chen, T.-F. Liu and H.-C. Zhou, Chem. Sci., 2016, 7, 6969–6973 Search PubMed.
- J. Liang, F. Mazur, C. Tang, X. Ning, R. Chandrawati and K. Liang, Chem. Sci., 2019, 10, 7852–7858 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04513f |
| This journal is © The Royal Society of Chemistry 2020 |