How does DNA ‘meet’ capillary-based microsystems?
Tingting
Hong†
 a,
Lin
Qiu†
ab,
Shuwen
Zhou
a,
Zhiqiang
Cai
a,
Lin
Qiu†
ab,
Shuwen
Zhou
a,
Zhiqiang
Cai
 ac,
Pengfei
Cui
a,
Ronghui
Zheng
a,
Jianhao
Wang
*ad,
Songwen
Tan
*e and
Pengju
Jiang
*a
ac,
Pengfei
Cui
a,
Ronghui
Zheng
a,
Jianhao
Wang
*ad,
Songwen
Tan
*e and
Pengju
Jiang
*a
aSchool of Pharmacy, Changzhou University, Changzhou, Jiangsu 213164, China. E-mail: minuswan@cczu.edu.cn; pengju.jiang@gmail.com
bJiangsu Science Standard Medical Testing Co., Ltd, Changzhou, Jiangsu 213164, China
cJiangsu Dawning Pharmaceutical Co., Ltd, Changzhou, Jiangsu 213100, China
dChangzhou Le Sun Pharmaceuticals Co., Ltd, Changzhou, Jiangsu 213125, China
eXiangya School of Pharmaceutical Sciences, Central South University, 172 Tongzipo Road, Changsha, Hunan 410013, China. E-mail: songwen.tan@csu.edu.cn
First published on 2nd November 2020
Abstract
DNA possesses various chemical and physical properties which make it important in biological analysis. The opportunity for DNA to ‘meet’ capillary-based microsystems is rapidly increasing owing to the expanding development of miniaturization. Novel capillary-based methods can provide favourable platforms for DNA–ligand interaction assay, DNA translocation study, DNA separation, DNA aptamer selection, DNA amplification assay, and DNA digestion. Meanwhile, DNA exhibits great potential in the fabrication of new capillary-based biosensors and enzymatic bioreactors. Moreover, DNA has received significant research interest in improving capillary electrophoresis (CE) performance. We focus on highlighting the advantages of combining DNA and capillary-based microsystems. The general trend presented in this review suggests that the ‘meeting’ has offered a stepping stone for the application of DNA and capillary-based microsystems in the field of analytical chemistry.
1. Introduction
Nowadays, capillary-based microsystems have rapidly gained popularity due to their prominent advantages, including low reagent consumption, fast analysis time, and desirable portability.1–6 DNA have attracted considerable attention in the analysis domain due to its high affinity towards various molecules.7–10 Efforts have been made on developing novel CE-based methods for DNA assay. Meanwhile, DNA is a desirable alternative for fabricating state-of-the-art capillary-based analytical microsystems (Fig. 1).Since DNA chains possess remarkable charge density and predictable charge-to-size ratio, CE is regarded an efficient technique for DNA analysis, such as DNA–ligand interaction assay, DNA translocation study, DNA separation, DNA aptamer selection, DNA amplification assay, and DNA digestion.11–15 Multiple CE modes including kinetic CE (KCE), capillary isoelectric focusing (cIEF), micellar electrokinetic chromatography (MEKC) and affinity CE (ACE) have emerged as promising approaches to investigate the thermodynamic and kinetic interactions between DNA and various binding ligands. Moreover, CE-mass spectrometry (MS) and CE-laser-induced fluorescence (LIF) offer satisfactory strategies for the investigation of the DNA-binding reactivity of metal-based anticancer drugs, facilitating the successful selection of metallodrug candidates.16,17 Online real-time CE-LIF has also been developed for detecting DNA base pair mutation in the cancer diagnosis process.18 In addition to CE, a novel method based on open microcapillary hydrodynamic chromatography (HDC) is able to explore DNA–ligand interactions with improved sensitivity and quantification capacity.19 For the DNA translocation study, glass capillary nanopores present specific merits depending on their simple manufacturing method with adjustable size and shape.20 Exploring effective capillary-based technologies for DNA separation is another essential domain in DNA assay. During the separation process, DNA molecules are able to be isolated according to the molecular size difference. Gelatinous materials and artificial sieves have been successfully used for the separation of biomolecules containing DNA.21,22 In addition to the separation by length, DNA separation by sequence is important for genome sequencing, genotyping, and phenotyping of microbial communities.23 As we know, DNA aptamers exhibit high affinity and specificity toward various targets.24 However, methods applied for selecting aptamers have an essential effect on the application of aptamers. Compared with the conventional systematic evolution of ligands via the exponential enrichment (SELEX) process, CE-SELEX can save selection time and greatly enhance the efficiency. For whole-cell aptamer selection, the CE-based strategy also presents tremendous advantages. Although CE-SELEX is widely utilized in DNA aptamer selection based on its less nonspecific binding and fast selection speed, only limited amounts of target aptamers are collected at each round because of the small injection volume of CE. To avoid this shortcoming, the combination of CE-SELEX and magnetic nanoparticle-involved selection provides an innovative alternative for enriching aptamers.25,26 Since the traditional polymerase chain reaction (PCR) is time-consuming, different modes of CE have been coupled with the PCR to decrease reagent consumption, improve efficiency, and simplify procedure.27,28 Moreover, novel capillary-based equipment also presents specific merits for performing digital droplet recombinase polymerase amplification (ddRPA) and other types of DNA amplification-based assays.29 We have summarized the application of capillary-based enzymatic reactors for protein digestion.30 In the present review, we will further discuss the advantages of using capillary-based bioreactors for DNA digestion.
Attributing to its specific affinity to various molecules, DNA presents a promising potential in fabricating biosensors and enzymatic reactors. The studies of capillary-based biosensors have been growing for decades.31–33 DNA hydrogels have been utilized as sensors by transducing the analyte-dependent hydrogel changes into an optical or electrical signal.34 Compared with conventional DNA-based sensors, these novel biosensors can avoid the use of assistant techniques and a large volume of DNA hydrogels. For the fabrication of capillary-based enzymatic reactors, immobilization methods of enzymes have an important effect on reactor performance. The DNA-directed immobilization (DDI) strategy has attracted tremendous research interest.35 Different from covalent interactions, this mild and biocompatible immobilization process can protect biological enzyme activity. Moreover, DDI enables the regeneration of enzymatic microreactors by removing the enzyme–DNA conjugates and then rehybridizing with a new DNA linker solution. In addition to fabricating capillary-based biosensors and enzymatic reactors, DNA has been used in CE separation and detection with improved efficiency. DNA can be added in the CE running buffer for analyzing chiral drugs and biomolecules to enhance resolution and sensitivity. DNA aptamers are regarded as efficient ligands for capillary-based affinity analysis.36 Different types of aptamer/NP probes have been employed to improve CE detection sensitivity. As the essential element of fabricating affinity open tubular (OT) and monolithic capillaries, aptamer immobilization techniques have also attracted tremendous attention.37,38
In this manuscript, we want to review developments in the combination of DNA and capillary-based microsystems, focusing on the recent four years. Various capillary-based methods utilized for DNA–ligand interaction assay, DNA translocation study, DNA separation, DNA aptamer selection, DNA amplification assay, and DNA digestion will be discussed. The advantages of applying the CE-based technique for DNA assay will be highlighted. In the section of introducing DNA-based methods for microsystem fabrication, innovative capillary-based biosensors and bioreactors are described. In addition, DNA employed for improving the performance of capillary electrophoresis analysis will be discussed.
2. Capillary for DNA assay
Capillary-based microsystems provide an efficient platform for DNA assay due to their low reagent consumption. Multiple modes of CE have attracted much attention for use in the investigation of DNA–biomolecule and DNA–metallodrug interactions. In addition to microcapillaries, glass capillary nanopores are a desirable choice for DNA translocation study. The on-line capillary-based methods have been applied for DNA separation according to the differences in DNA molecular size or sequence. To achieve DNA aptamer selection, CE-SELEX can greatly improve the efficiency of standard SELEX separation. Moreover, various capillary-based methods utilized for DNA amplification assay and DNA digestion are also reviewed.2.1. Capillary-based methods for DNA–ligand interaction assay
Attributing to the high affinity towards targets and diverse three-dimensional structures, DNA molecules can be used as effective probes for assaying DNA–biomolecule interactions. CE offers a desirable way for characterizing binding properties and interaction types. For instance, ACE can be used to assess DNA–DNA, DNA–protein, and DNA–small molecule interaction. KCE is able to offer the electrophoretograms for extracting the on- and off-rates of DNA–protein interaction. Electrophoresis-based strategies are usually limited in the type of compatible run buffer. Traditional KCE experiments are performed in low-conductivity run buffers. Since the ionic strength of physiological fluids in biological systems can influence DNA–ligand interactions, it is meaningful to construct a physiological environment for the studied biomolecules. Phosphate buffered saline (PBS), as a type of physiological buffer, was used in the KCE method for analyzing a platelet-derived growth factor (PDGF) protein–DNA aptamer interaction.39 A pressure-assisted modification of KCE was applied to solve the DNA detectability problem under standard KCE conditions. The analysis time could be shortened by adding a pressure-driven hydrodynamic flow, and the velocity of DNA could be improved. Moreover, the desirable reproducibility of the peak area for analytes was achieved with a relative error of less than 2.4%. To investigate the interactions between peptide nucleic acids (PNAs) and proteins, multiple modes of CE including cIEF and MEKC were used.40 We discussed KCE utilized for exploring DNA aptamer–protein interactions. Since PNAs possess desirable biological stability in vivo, they are regarded as better aptamers than DNA-based aptamers. A study about PNAs could provide an efficient molecular tool for nucleic acid research.41,42 The results suggested that the cIEF mode enabled the determination of the PNA isoelectric point (pI), and MECK mode could be applied for the direct identification of PNA, PNA–ssDNA and PNA–thrombin complexes.For the DNA–ligand interaction assay, a strategy based on online CE hyphenated to MS has attracted much attention in recent years. MS can discriminate a variety of complexes which cannot be completely resolved by CE alone. In addition to DNA–protein interactions, research was also focused on investigating DNA-vector complexation, which is important for designing gene delivery vectors.43 This integrated ACE-MS system offered a satisfactory method for analyzing DNA-β-cyclodextrin (CD) complexation in less than 6 min. Information including the stoichiometry, the micro and macroscopic dissociation constants and the cooperativity and Gibbs free energy data could be obtained. Compared with conventional DNA–CD complex characterization strategies such as dynamic light scattering and isothermal titration calorimetry, the proposed ACE-MS method could be performed by using 20 fold lower amount of β-CD and DNA oligomers.
Metal-based anticancer agents are necessary in the treatment of cancer. Since traditional MS could not detect the binding sites between metallodrugs and biomolecules, Keppler et al. coupled CZE to electrospray ionization (ESI)- and inductively coupled plasma (ICP)-MS.16 The CE-MS strategy allowed the simultaneous quantification and characterization of the binding of metallodrugs to biomolecules. Before analysis by MS, the initial metabolites of metal-based agents were able to be separated by CZE in aqueous solutions. As seen in Fig. 2, cisplatin and RM175 were chosen as model anticancer drugs to explore the binding preference of metal-based agents to DNA and proteins. For the analysis process, ESI-MS could be used for monitoring specific mass shifts that were induced by covalent binding of the drug to either DNA or protein. CZE-ESI-MS2 enabled the identification of the binding site of metallodrugs on the biomolecules. Moreover, CZE-ICP-MS was applied for the quantification by detecting the isotopes 31P for DNA and 195Pt or 102Ru for the drugs. Matczuk et al. also utilized the CE-ICP-MS method to investigate the DNA-binding reactivity of indazolium trans-[tetrachloridobis(1H-indazole)ruthenate(III).44 In addition to CE-MS, CE-LIF was applied to determine the DNA sequence specificity of the anti-tumor drugs.17 The results indicated that CE-LIF fragment analysis technology could achieve the detection of single-stranded DNA (ssDNA) cleavage induced by bleomycin. Furthermore, the obtained bleomycin sequence specificity data were compared with the mitochondrial genome-wide data, and the overall bleomycin DNA sequence specificity was very similar in the two environments.
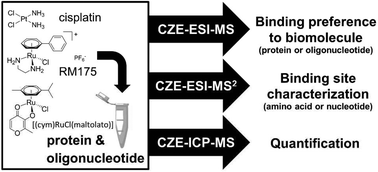 | ||
| Fig. 2 A CZE-MS approach for elucidating the binding selectivity of metallodrugs on the levels of intact biomolecules and binding sites (Reproduced from ref. 16 with permission from [The Royal Society of Chemistry], copyright [2017].). | ||
Developing an efficient method for detecting DNA base pair mutation is important in cancer diagnosis. Traditional approaches usually involve high consumption of reagents and need expensive instrumental investment. Two types of CE-based micro-scale techniques including restriction fragment length polymorphism analysis and single-stranded conformation polymorphism analysis have been applied for mutation detection. Wang et al. constructed a rapid and online real-time detection system on DNA hybridization and base pair mutations.18 The CE-LIF method was used to evaluate the influence of injection time, interval time, and molar ratio of oligos on DNA hybridization. As seen in Fig. 3, in-capillary assay could be achieved by performing different steps of mixing, reaction, separation and hybridization within one capillary. First, the thrombin-binding ssDNA oligonucleotide (ON) labeled with 6-carboxyfluorescein (DNA1-FAM) and its complementary DNA containing different number of mismatch bases (cDNAms) were injected into the capillary sequentially. After mixing in the capillary, the hybridization peaks were monitored based on the recorded electrophoretograms. This in-capillary assay presented comparable results with the post-capillary assay. The results suggested that the proposed CE-LIF method was suitable for detecting DNA point mutations.
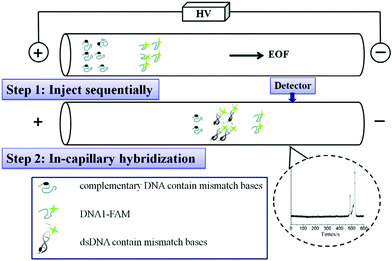 | ||
| Fig. 3 Schematic illustration on hybridization analysis of DNA1-FAM and cDNA1 and cDNAms (Reproduced from ref. 18 with permission from [Elsevier], copyright [2016].). | ||
As mentioned above, CE is a common approach employed for analyzing DNA–ligand interactions. However, the electrophoretic mobility shift utilized in CE-based strategies is not desirable for detecting the size or molecular weight of an interspecies complex. The utilized gel matrix during separation can also influence the stability of intermolecular interactions. The open microcapillary HDC offers an alternative method for the separation of biomacromolecules. To enhance the sensitivity and quantification ability of HDC, we developed a single-molecule free solution hydrodynamic separation (SML-FSHS) platform.19 In the referenced work, bound DNA–protein (E. coli single stranded binding protein) complexes can be separated from unbound DNA oligos depending on the differences in hydrodynamic mobility, and single molecule detection enabled the characterization of binding behavior, such as affinity, cooperativity, and stoichiometry. Moreover, single molecule burst analysis could achieve decoupled analysis of quantity, size, and conformation within a single measurement. This technology provided a satisfactory approach for the quantitative and sensitive assessment of various types of DNA interactions and binding properties in free solution. In addition to microcapillaries, nanocapillaries were used in single molecule assay of DNA–protein complexes.22 Since the electroosmotic flow of nanocapillaries could be controlled, nanocapillaries were tailored to be either sensitive to the charge or size of proteins. The glass nanocapillaries were combined with optical tweezers to localize and discriminate DNA–protein complexes by controlled translocation. This single molecule force spectroscopy-based technology could recognize high affinity sites and characterize their charge and size, enabling the detection of DNA–protein interaction dynamics.
A variety of capillary-based methods including KCE, ACE, cIEF, MEKC, CE-MS, CE-LIF, and SML-FSHS and nanocapillaries used to explore DNA–ligand interactions are summarized. Furthermore, the advantages of each approach are described (Table 1).
| Type of capillary-based method | Advantages and disadvantages | Ref. |
|---|---|---|
| Affinity CE | * Available for assessing DNA–biomolecule/small molecule interaction | 5 |
| Limited in the type of running buffers | ||
| Kinetic CE | * Desirable reproducibility | 39 |
| Limited in the type of running buffers | ||
| Capillary isoelectric focusing | * Available in the determination of the DNA isoelectric point | 41 and 42 |
| Unable to separate neutral substrates | ||
| Micellar electrokinetic chromatography | * Available for identifying PNA, PNA–ssDNA and PNA–thrombin complex | 40 |
| Stable micelle needs to be formed | ||
| CE-mass spectrometry | * Promising in the simultaneous quantification and characterization of DNA–ligand interactions | 16, 43 and 44 |
| Complex sample preparation process | ||
| CE-laser-induced fluorescence | * Available for detecting DNA point mutations and DNA hybridization | 17 and 18 |
| Unable to detect DNA without fluorescence labeling | ||
| Microcapillary hydrodynamic chromatography | * Enhanced sensitivity and quantification ability | 19 |
| Nanocapillary | * Promising in single molecule assay of DNA–protein complexes | 22 |
| The capillary fabrication process is complicated |
2.2. Glass capillary nanopores for the DNA translocation study
In section 2.1, we discussed the application of nanocapillaries for the controlled translocation of DNA–protein complexes.22 For the DNA translocation study, nanopores are another desirable choice. Glass capillary nanopores are solid-state nanopores. Compared with biological nanopores, glass capillary nanopores possess a stable structure and can be easily modified.45,46The properties of translocating molecules including their charge, structure, and length can be monitored by nanopore sensing. However, it is difficult to reduce the nanopore diameter down to sub-10 nm. A novel wet-chemical approach was explored to shrink the nanopore diameter by disodium silicate hydrolysis.20 By this method, DNA translocation could be slowed down and the signal-to-noise ratio was enhanced. Research focused on DNA nanostructures has attracted tremendous interest due to their promising potential for use in the area of drug delivery, cell imaging, and information storage.47–49 Jin et al. developed a small orifice glass nanopore for multiarm DNA concatemer sensing.50 The relationship between the number of DNA arms and translocation frequency was investigated. The experimental principle and setup are shown in Fig. 4. The three-arm DNA concatemers translocated through the nanopore at a voltage of 1 V, and three types of responses could be obtained. The results indicated that the DNA concatemers exhibited more complicated states in buffer solution when compared with the one-dimensional structure of ds-DNA. Gel electrophoresis was utilized to confirm the successful preparation of DNA concatemers with different arms (Fig. 4b). The transmission electron microscopy (TEM) image of the glass capillary nanopore is presented in Fig. 4c. It was found that the use of glass capillary nanopores was a desirable strategy that could sensitively characterize the single molecule conformational information on artificial DNA nanostructures and concatemers in solution. Moreover, we foresee the promising potential of glass capillary nanopores for biomolecule sensing applications, such as DNA–protein interactions.
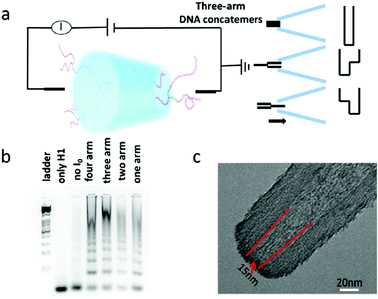 | ||
| Fig. 4 (a) Schematic diagram of the experimental principle and setup. (b) Agarose electrophoresis image of multiarm DNA concatemers. (c) Typical TEM images of the glass nanopore used in this study (Reproduced from ref. 50 with permission from [American Chemical Society], copyright [2019].). | ||
2.3. Capillary-based methods for DNA separation
Different types of CE methods including KCE, ACE, cIEF and MEKC were mentioned in section 2.1. Capillary gel electrophoresis (CGE) is another efficient CE mode used for biomolecule separation. Polyacrylamide (PAA) gels and agarose gels are considered common separation media in GE assay. However, agarose gel cannot be utilized for the separation of small size differences based on its large pores. PAA gels also present limitations because of the toxicity caused by the acrylamide monomer and the non-specific interactions by amide groups toward biomolecules. Compared with these two gels, poly(ethylene glycol) (PEG) and cellulosic polymers possess specific advantages such as higher biocompatibility, lower toxicity of the monomers and non-specific interaction.51,52 For instance, to effectively separate biomolecules containing DNA and sugar, a variety of PEG-based hydrogels were employed for performing CGE.21 The results presented that DNA ladder in the range of 10–1100 base pair could be separated depending on the molecular size effect. During the separation process, the separation ranges against the molecular size were controllable by optimizing the ethylene oxide concentration in the PEG-based crosslinker. As we discussed above, gelatinous materials were suitable to be used as the sieving medium based on the cross-linked network of pores. In addition, artificial sieves are micro/nanostructures containing a regular pattern of pores and present enhanced performance for the electrophoresis of macromolecules. Different mixtures of DNA and proteins were able to be separated into sharp peaks by using an entropic trap array.53,54 To improve the amount of sample that could be analyzed at a time, Yobas et al. further developed a 2D capillary-well array that relied on the previous fabricated sieve.22 In this structure, the self-enclosed capillaries were aligned orthogonally to the deep wells. For the continuous-flow electrophoresis of DNA and proteins, the proposed sieve possessed comparable performance to the 2D slit-well array. An on-line capillary isotachophoresis (CITP)-CZE system with a UV detector was constructed for separating ten DNA fragments.55 This technique could combine the high concentrating ability of the CITP and high efficiency of CZE. To improve the separation performance, native α-, β- and γ-CDs were chosen as buffer additives. DNA ladder fragments with sizes ranging from 100 to 1000 bp were able to be separated within 10 minutes. Satisfactory repeatability of migration time and peak area was observed, with RSDs in the range of 1–3 and 3–9%, respectively.In section 2.1, we described the online real-time CE-LIF system utilized for exploring DNA hybridization and base pair mutations.18 Conventional methods applied for sequencing the same-length strands usually change the stable secondary structure of single DNA strand or involve a target to hybridize with the single strand. Therefore, it is necessary to develop a simple and robust strategy. Separation of a set of ten 15-mers including poly-dT and nine variants was performed using CZE with LIF detection.23 The results suggested that migration times increased with increasing content of purine, and adenosine (A) having a much larger effect than guanosine (G). Moreover, A was sensitive to the chemical microenvironment during the separation process. This work provided a basic platform for further investigating the sequence-dependence and nucleotide-dependence of the electrophoretic mobility of ssDNA. A simple CE approach was applied to the separation of single-base sequential isomers of DNA based on charge differences of DNA bases.56 An acidic aqueous urea solution was employed as the electrophoretic buffer solution to enhance the resolution. A mixture of 13 single-base isomers of 12-mer ssDNA could be separated under identical electrophoretic conditions. Moreover, this method was also able to detect the polymerase chain reaction products of a 68-mer gene fragment and its single-base isomers by combining with the genomic DNA extraction technology.
Generally, CGE is an efficient CE mode used for DNA separation. In addition to gelatinous materials, artificial sieves containing a regular pattern of pores exhibit improved performance for the electrophoresis of DNA. The combination of high concentrating ability of the CITP and high efficiency of CZE facilitates DNA fragment separation. Moreover, CE-based methods utilized for DNA separation depending on sequence differences have been discussed as well.
2.4. Capillary electrophoresis for DNA aptamer selection
ssDNA aptamers are usually selected from a random prepared oligonucleotide (ON) library using the SELEX process. Due to their advantages including high affinity to various targets, high chemical stability and easy modification, aptamers are considered desirable ‘chemical antibodies’.57 The SELEX process used for aptamer selection is time consuming, and it often takes four to six weeks or more. In comparison, CE has attracted tremendous attention in the selection of aptamers based on its high efficiency and fast separation of unbound ssDNA and target-ssDNA complex.58,59 CE-SELEX can greatly enhance the efficiency of standard SELEX separation. For instance, the combination of CE-SELEX and six rounds of amplification was used to successfully select DNA aptamer S1 against swine anaphylatoxin C5a.60 The procedure of CE-SELEX for selecting DNA-aptamers against activated protein C (APC) was investigated.61 Qu et al. developed an online reaction based single-step CE-SELEX (ssCE-SELEX) mode by using human thrombin as a model target.25 As seen in Fig. 5, human thrombin (H-Thr), selection buffer, and 82 nt ssDNA library were orderly injected into the modified capillary. Since the mobility of ssDNA library was larger than H-Thr, the ssDNA could mix and react with H-Thr when they moved towards the anode. Subsequently, unbound ssDNA and the H-Thr/ssDNA complex were able to be monitored by LIF detection. The obtained H-Thr/ssDNA complex fraction was collected and subjected to symmetric PCR amplification, and then a portion of the products was sequenced. For the next selection round, a new ssDNA pool could be generated by the asymmetric PCR of the remaining products. Different procedures including mixture, incubation, reaction, separation, detection, and collection were performed in a single step by using ssCE-SELEX mode. The results indicated that the selected sequences presented higher specificities towards H-Thr than bovine thrombin, IgG, lysozyme, and lactoferrin. Binding affinity and specificity of the selected sequences were evaluated by AuNP based colorimetric analysis and molecular dynamics study. Compared with conventional selection, this ssCE-SELEX method was time-saving with only two rounds of selection, and only 59.83 nL of sample was needed for one injection. Another easily-adoptable method for one-step selection of aptamers was reported by Krylov et al.62 Ideal-filter capillary electrophoresis (IFCE)-based partitioning was desirable for effectively selecting high-affinity aptamers against the protein target. During the separation process, binders and nonbinders moved in the opposite directions, facilitating fast and robust selection of binders from a random-sequence ON library.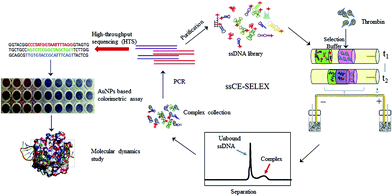 | ||
| Fig. 5 Schematic illustration of the ssCE-SELEX process (Reproduced from ref. 25 with permission from [Elsevier], copyright [2019].). | ||
Aptamers against whole cells are considered promising tumor cell detection probes in early diagnosis since they are able to be utilized for cell recognition with high specificity and affinity. However, the selection of whole-cell aptamers was much more complicated than purified molecules. For instance, factors such as the complexity of cell surface ligands and molecular-level minor difference between batches of cells can influence aptamer selection. It is difficult to detect the distribution of all ssDNA sequences in the SELEX process. Therefore, Qu et al. employed a CE-based strategy to characterize complete ssDNA quantitative distribution of the cell–ssDNA complex, unbound ssDNA, and dissociated ssDNA.63 The distribution percentage of FAM labeled Sgc8c and 41 mer random ssDNA library against different types of cancer cell lines including PC3, U251 and HeLa was investigated. The results presented that the CE-based method could be utilized to distinguish the binding difference among cells under the same condition. In one CE run, this strategy was able to get complete ssDNA distribution and cell–ssDNA complex separation. Therefore, CE is promising for high efficiency whole-cell aptamer selection.
Magnetic bead-based SELEX is another commonly used method for aptamer selection, and it often suffers from some disadvantages, such as time-consuming procedure and nonspecific binding toward ONs. To avoid these drawbacks, Liu et al. investigated a hybrid method that combined CE-SELEX and boronate affinity magnetic nanoparticle (BA-MNP)-based selection for selecting glycoprotein-binding aptamers.26 The selection process is shown in Fig. 6. First, an alkaline pH was chosen for the immobilization of target glycoprotein onto BA-MNPs, and then the BA-MNPs were incubated with a 5′-FAM-labeled ssDNA library. Subsequently, the resulting target-ssDNA complexes could be collected after eluting with an acidic solution. After PCR amplification of the complexes, CE-SELEX was applied to analyze the PCR products. When the binding affinity met the requirement, the PCR products were used for a new round CE-SELEX. Finally, the obtained ssDNA with optimum affinity was sent to cloning and sequencing. In this hybrid strategy, BA-MNP-based selection facilitated the effective enrichment of ssDNA in the nucleic acid library before CE-SELEX. In addition, steric hindrance effects involved in traditional magnetic bead-based selection could be eliminated. As a result, this work provided a novel tool for efficiently selecting high-performance aptamers.
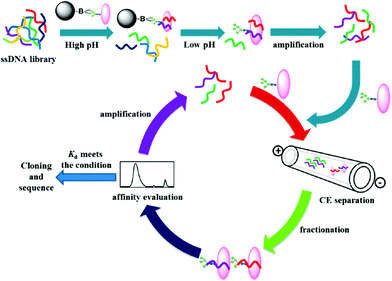 | ||
| Fig. 6 Schematic illustration of the hybrid SELEX approach (Reproduced from ref. 26 with permission from [American Chemical Society], copyright [2016].). | ||
Various aptamers can be efficiently selected by using CE-SELEX. The novel CE-based strategy presents substantial benefits in the selection of whole-cell aptamers. To avoid the disadvantages of magnetic bead-based SELEX, a hybrid approach that combines CE-SELEX and BA-MNP-based selection has attracted much research interest. The merits of these CE-based methods over the traditional method used for aptamer selection are summarized in Table 2.
| Type of capillary-based method | Advantages and disadvantages | Type of aptamer | Ref. |
|---|---|---|---|
| CE-SELEX | * Enhanced selection efficiency, time-saving | * DNA aptamer S1 against swine anaphylatoxin C5a | 25 and 57–61 |
| Buffer types can influence the complex formation in ssCE | DNA-aptamers against activated protein C | ||
| DNA-aptamers against human thrombin | |||
| Ideal-filter CE | * Promising in fast and robust selection of aptamers | * DNA aptamers against the protein target | 62 |
| CE-magnetic nanoparticles | * Improved performance, time-saving | * Glycoprotein-binding aptamers | 26 |
| Nonspecific binding toward ssDNA | |||
| CE-LIF | * Low detection limit | * DNA-aptamers against thrombin | 93 |
| Sample pretreatment is necessary | |||
| CE | * High sensitivity and accuracy | * Organophosphorus pesticide-binding aptamers DNA aptamers against adenosine triphosphate | 36 |
| Sample pretreatment is necessary | |||
| Nanomaterial-modified capillaries | * High target loadings, low cost | * Adenosine-binding aptamers | 94–97 and 37 |
| Metal ions influence the separation | |||
| GNP-modified monoliths | * Improved sensitivity, high coverage density of aptamers | * DNA-aptamers against thrombin | 38 |
| POSS-modified monoliths | * Low detection limit | * Ochratoxin A-binding aptamers | 105–107 |
| Polymerization reaction affects nonspecific adsorption |
2.5. Capillary-based methods for DNA amplification assay
PCR can help templates of nucleic acid to be amplified at the rate of exponential growth. PCR thermal cycling processes usually involve different temperature-dependent reaction stages, including denaturation, annealing, and extension. To solve the time-consuming problem of conventional PCR, some research studies focused on developing microfluidic PCR with decreased reagent volume, improved efficiency and simplified procedure.64 However, the relatively low amount of nucleic acid templates could cause low detection sensitivity, and a complex temperature control system was also needed for repeatedly switching between heating and cooling. In comparison, convective PCR is able to achieve efficient thermal cycling depending on an easier setup. Convective PCR introduces stable thermal convection within a capillary reactor with a desirable steady-stable temperature gradient between two ends of the tube. The real-time horizontal convective PCR was reported to perform efficient heating with a fixed temperature and fluorescence detection with a smartphone.28 Compared with vertical convective PCR, the horizontal PCR presented less convective heat transfer. Therefore, the spatial temperature distribution in the horizontal position was more suitable for DNA amplification.Research on multiplex pathogens is important in the fields of pathogen diagnosis and therapy. For enumerating pathogens, the quantitative polymer chain reaction (qPCR) is able to monitor the number of DNAs during the reaction process and determine the number of pathogens depending on the cycle number. Whereas the application of multiplex qPCR is limited due to its low capacity and high cost. To avoid these drawbacks, end-point multiplex PCR (mPCR) is considered a type of stepwise PCR strategy that can separate and detect PCR products via off-line techniques. Three pathogens including Porphyromonas gingivalis (P.g.), Treponema denticola (T.d.) and Tannerella forsythia (T.f.) were quantified by using the mPCR-quantitative CE (qCE) assay.27 To improve the precision of qCE, a time modified internal standard (TMIS) method was applied. The amplification efficiency of mPCR could be evaluated by qCE. This proposed mPCR-qCE protocol could be performed with lower reagent consumption and decreased turn-around time. Therefore, CE with low reagent consumption and rapid speed was a satisfactory technique in the application of DNA analysis. Different modes of CE have been coupled with PCR. For instance, Jian et al. developed a system combining random amplified polymorphic DNA (RAPD)-PCR and capillary gel electrophoresis (CGE) for the epidemiological typing of E. anophelis.65 Multiplex PCR was combined with non-gel sieving CE for the identification of four serotypes of V. cholerae in marine products within 8 min66 Satisfactory intraday repeatability values in the ranges of 1.60–2.56% and 1.60–6.29% for determining DNA marker and PCR products were obtained, respectively. As we mentioned above, the quantitative analysis of nucleic acids was important for molecular diagnostics and gene expression. Compared with traditional qPCR, digital PCR (dPCR) exhibits better repeatability and linearity by transferring the exponential nature of PCR amplification into a digital format. Manz et al. developed a capillary-based setup to perform ddRPA.29 Since RPA is able to start amplification at room temperature in the presence of a chemical initiator, the number of false positives will increase if all reagents are mixed before the compartmentalization. To solve this problem, a capillary-based equipment was designed to control the start of RPA reactions by encapsulating the chemical initiator to independent reaction compartments. Moreover, the performance of ddRPA was evaluated via the counting of positive application results of Avian virus DNA confined in the partitions. This proposed ddRPA capillary-based technique facilitated the digital quantification of nucleic acids with desirable simplicity and reliability.
We discussed the application of PCR-based technology for pathogen diagnosis. As an alternative, multiplexed loop mediated isothermal amplification (LAMP) based assay can also be used for point-of-care diagnostics with high specificity and sensitivity. To facilitate point-of-care (POC) diagnostics in resource-limited settings, capillary-based platforms have attracted tremendous attention because of their usefulness for multiplexing and in line sample processing. For the rapid diagnosis of C. trachomatis and N. gonorrhoea, three capabilities were integrated into the capillary-based system.67 During the analysis process, magnetic beads were able to move through the plugs to extract DNA from complex samples, and heated water could be used to drive the reaction. Compared with the complicated DNA amplification method such as PCR, precise thermal control was not necessary for LAMP employed in this study. After amplification, the fluorescence signal of the amplified product was detected by using a hand-held UV flashlight. As a result, capillary-based platforms were available to perform the LAMP-based assay due to their low cost and simplicity. The multiplexed microcapillary-based LAMP system (cLAMP) could efficiently avoid contamination during the process of nucleic acid amplification in microchips.68 Furthermore, Jiang et al. improved the performance of cLAMP by designing an integrated microcapillary-based LAMP (icLAMP).69 Without using advanced instruments, the proposed icLAMP system containing prefixed reagents and DNA extraction cards was able to achieve on-site pretreatment, extraction, amplification, and detection of nucleic acids within 150 min. Since the device production process was complex and expensive, a novel mini-disk capillary array combined with LAMP (mDC-LAMP) was explored to simplify sample loading and operation steps.70 This mDC-LAMP system provided a promising platform for analyzing a wide range of multiple nucleic acids with enhanced specificity and sensitivity.
Capillary-based methods provide a desirable choice for DNA amplification assay. Convective PCR can achieve more efficient thermal cycling with an easier setup when compared with conventional PCR. Different modes of CE including GCE and non-gel sieving CE have been coupled with PCR. An innovative mPCR-qCE strategy presents specific advantages such as lower reagent consumption and faster analysis speed. Moreover, the capillary-based ddRPA and LAMP also possess better performance than traditional PCR.
2.6. Capillary-based bioreactors for DNA digestion
As we know, the modification of nucleosides and phosphate moieties in DNA chains can cause DNA damage. Since it is difficult to directly monitor different modifications in genomic DNA, DNA samples need to be digested into single nucleosides or mononucleotides. Capillary-based enzymatic reactors have attracted much attention in applications such as protein digestion and peptide identification.71,72 Different from proteomic digestion, genomic DNA strands cannot be digested by one enzyme because of their larger molecular weight. A DNA digestion system was fabricated by immobilizing three types of enzymes including alkaline phosphatase (ALPase), snake venom phosphodiesterase (SVP) and deoxyribonuclease (DNase I) on the inner surface of the monolithic capillary.73 For the preparation of immobilized enzymatic reactors, different enzymes were immobilized onto silica monoliths by using the glutaraldehyde-based approach. Subsequently, DNase I, SVP, and ALPase bioreactors were connected in sequence for assembling the cascade bioreactors (Fig. 7). Since the silica monolith possessed a macro-porous structure, genomic DNA was able to pass through the reactor via a low-pressure microinjection pump. The obtained three-enzyme cascade capillary bioreactor could be used to digest genomic DNA into single nucleosides and mononucleotides. Finally, liquid chromatography-tandem MS (LC-MS/MS) was coupled with the bioreactor to analyze the collected products. The results indicated that the proposed cascade bioreactor exhibited high enzymatic activity and capacity, resulting in satisfactory digestion efficiency. This capillary-based platform is promising in the application of DNA digestion. Table 3 concludes the advantages of using capillary-based methods for constructing biosensors and bioreactors.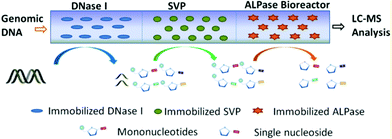 | ||
| Fig. 7 Schematic diagram of the three-enzyme cascade capillary monolithic bioreactor (Reproduced from ref. 73 with permission from [American Chemical Society], copyright [2016].). | ||
| Type of capillary-based microsystem | Applications | Advantages and disadvantages | Ref. |
|---|---|---|---|
| Glass capillary nanopore-based sensors | * Detecting properties of DNA translocation, multiarm DNA concatemer sensing | * Enhanced signal-to-noise ratio, available for sensitively characterizing the single molecule conformational information | 20 and 50 |
| Monolithic capillary-based enzymatic reactors | * DNA digestion | * High enzymatic activity and capacity, satisfactory digestion efficiency | 73 |
| Connection sequence influences digestion efficiency | |||
| Capillary self-driven regulator sensors | * Determining the concentration of Pb2+ and cocaine | * Improved sensitivity, reduced volume of DNA hydrogels | 34 and 76 |
| Concentration of crosslinker DNA influences hydrogel formation | |||
| Trypsin capillary microreactors | * Protein digestion | * Satisfactory reproducibility | 35 |
| Multienzyme immobilized capillaries | * Detecting glucose | * Improved reusability and stability, high performance | 81 |
| The enzyme ratio needs to be optimized |
3. DNA for capillary-based assay
DNA exhibits promising potential in capillary-based assay due to its specific affinity to various molecules. The integration of DNA with capillary-based microsystems provides a desirable strategy for fabricating biosensors and enzymatic reactors. Additionally, DNA has attracted tremendous attention for use in CE separation and detection with improved efficiency and sensitivity. Some of the prominent advantages of applying DNA for capillary-based assay are reviewed in this article.3.1. DNA hydrogel for the fabrication of capillary-based biosensors
Since DNA can selectively respond to various types of molecules, DNA hydrogels have been used as biosensors by transducing the hydrogel changes into an optical or electrical signal. During the analysis process, functional nucleic acids facilitated hydrogel response to external stimuli such as pH, metal ions and molecules.74,75 The conventional DNA sensors usually need additional equipment to achieve precise measurement, and utilizing a large volume of DNA hydrogels can also increase the cost of the detection.To avoid the disadvantages of applying DNA hydrogels in visual quantitative sensors, Yang et al. developed capillary self-driven regulator sensors (CSDR-Sensors) via the combination of the properties of the DNA hydrogel and the principle of capillary action.34,76 For the fabrication of CSDR-Sensors, hot capillary was put in the DNA hydrogel, and the gel could be turned into a liquid solution and injected into the capillary. Subsequently, the DNA hydrogel film was formed in the capillary tube at room temperature. In the detection process, the interaction between target analytes and DNA linkers could cause the structural changes in the gel, and the increased permeability of the hydrogel film could further affect the flow velocity of the solvent in the capillary. The concentration of sample solutions was able to be determined by characterizing the duration time of the target analytes flowing through the capillary tube with a specified length. For instance, a DNAzyme-based hydrogel capillary sensor was designed for detecting Pb2+. The GR-5 DNAzyme was activated by Pb2+ and cleaved the substrate strand, causing the transition of the crosslinked polymer from the hydrogel into solution (Fig. 8). Therefore, the concentration of Pb2+ could be determined by recording the flow behavior of the solution in the capillary tube. Another CSDR-Sensor utilized for detecting cocaine was fabricated based on an aptamer-functionalized DNA hydrogel film. Compared with traditional DNA hydrogel-based sensors, this CSDR-Sensor provided a sensitive way to measure cocaine by using ultra-trace hydrogel (0.01 μL).
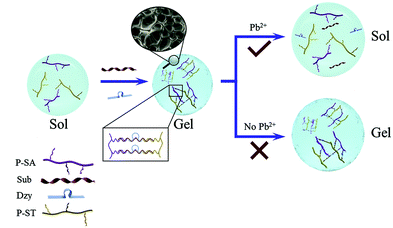 | ||
| Fig. 8 Schematic illustration of the DNA-based hydrogel for the detection of Pb2+ (‘Sub’ is the abbreviation of ‘substrate’. ‘Dzy’ is the abbreviation of ‘GR-5 DNAzyme’.) (Reproduced from ref. 34 with permission from [Elsevier], copyright [2020].). | ||
3.2. DNA linker for the fabrication of capillary-based enzymatic reactors
As presented in section 2.6, enzymatic microreactors can be efficiently used for DNA digestion. CE-integrated IMERs have attracted considerable interest due to their advantages including high separation efficiency, rapid analysis, and low reagent consumption. The enzyme immobilization method is considered an important factor that influences the performance of capillary-based enzymatic reactors. In this section, the DDI method for enzymatic microreactor fabrication is discussed.Since microreactors constructed by conventional strategies are usually difficult to regenerate and exhibit low immobilization capacities, the DDI approach has attracted increasing attention. During the preparation process, enzymes are able to bind to the carrier by Watson–Crick base pairing between two complementary DNA strands.77,78 An immobilized trypsin capillary microreactor was constructed based on the DDI technique.35 A polyamidoamine (PAMAM) dendrimer-coated capillary was developed for further modification of 5′-aminated ssDNA (NH2-ssDNA). And then, the NH2-ssDNA-PAMAM functionalized capillary was incubated with trypsin-SH-ssDNA conjugates to prepare the trypsin immobilized microreactor. The performance of this microreactor was characterized by detecting the enzymatic hydrolysis of Nα-benzoyl-L-arginine ethyl ester (BAEE). In addition, the renewability of the trypsin-functionalized reactor was achieved via DNA double-strand hybridization and dehybridization. Satisfactory reproducibility of the capillary enzyme microreactor was observed, with an RSD of 2.91%. Since the application of single enzyme-immobilized microreactors is limited, some research studies have focused on the fabrication of multienzyme immobilized reactors.79,80 Two types of enzymes including glucose oxidase (GOx) and horseradish peroxidase (HRP) were coated onto the PAMAM-functionalized capillary.81 The enzyme ratio could be optimized by adjusting the ratio of DNA strands α and β. Compared with IMER constructed by nonspecific adsorption, the proposed reactors exhibited better reusability and stability. Therefore, the DNA linker was suitable to fabricate capillary-based enzymatic reactors with high performance.
3.3. DNA additive for capillary electrophoresis analysis
We concluded the application of DNA for the fabrication of capillary-based biosensors and enzymatic reactors. DNA can be added in the CE running buffer for analyzing enantiomers, microRNAs (miRNAs), and proteins as well.The importance of chiral discrimination has generated a tremendous demand for developing efficient enantioseparation methods. CE is considered a high-resolution separation technique used for chiral recognition.82–84 Nucleic acids present specific chirality due to their backbone sugar units and secondary/tertiary topologies. Some research studies have been focused on the exploration of DNA molecules in the enantioseparation field.85 For instance, double-stranded (ds) DNA molecules and G-quadruplexes were employed either as surface coated selectors or as background electrolyte additives in the CE approach.86,87 Various DNA ONs were utilized as chiral selectors in partial-filling CE for enantioresolution of low-affinity DNA binders.88 Peyrin et al. investigated the chiral separation ability of homopolymeric sequences (Poly-dT) with different lengths (from 5 to 60-mer). The results presented that two hairpins of similar composition but with different sizes possessed different chiral separation ability, indicating that the sequence length played an important role in the separation process. Moreover, ONs with an unpaired-base displayed increased enantiomer selectivity range than those exhibiting secondary structure motifs. For exploring the impact of chemical diversity on the enantioresolution ability of the ONs, four based were incorporated into a 30-mer sequence. It was found that heteropolymers possessed better chiral resolution capability than homopolymers. This work provided an efficient ON chiral selector for the CE separation of weak DNA binder enantiomers.
CE coupled with fluorescence detection has many applications in separating and detecting biomolecules such as DNA, peptides and proteins, since it provides high resolution and utilizes small amounts of samples. As short single-strand RNA molecules, miRNAs are important for cell differentiation, proliferation, and apoptosis. Conventional methods used for miRNA detection usually need to separate RNA from the sample before the measurement. In comparison, gel-free CE is regarded an efficient technique for the direct quantitative analysis of multiple miRNAs (DQAMmiR). During the analysis process, ssDNA binding protein (SSB) was added to the running buffer to bind with the ssDNA probes, enabling the separation of the excess probes from the miRNA-probe hybrids.89 The probes were modified with different drag tags, and hybrids with different size to charge ratios could be separated from each other. However, the application of SSB can influence the robustness of the DQAMmiR assay. The second-generation DQAMmiR was developed based on peptide nucleic acid (PNA)–miRNA hybrids.90 PNA was an electrically neutral analogue of DNA containing repeating N-(2-amino-ethyl)-glycine units. Three types of miRNAs (miR-21, miR147a, and miR-378g) were chosen as model analytes and incubated with their respective complementary PNA probes (Fig. 9). The negatively charged PNA–miRNA hybrids could be separated from PNA probes without adding SSB. Moreover, the three hybrids were able to be baseline separated from each other. High accuracy (recovery >90%) and precision (RSD ≈10%) suggested that DQAMmiR with PNA hybridization probes may potentially serve as a tool for practical miRNA analyses in clinical samples. Moreover, protein detection and quantification are important in cell biology and medicine. cIEF combined with antibody-based detection is a useful technique for resolving protein isoforms based on their different pIs. The DNA-assisted technique can improve the sensitivity of detection of proteins and their isoforms, whereas insufficient sensitivity prevented the detection of proteins present at low concentrations. Compared with the standard assay utilizing the same primary antibodies, a great signal increase was observed via rolling circle amplification (RCA)-assisted reactions.91 The results suggested that the concentration of the probe and the duration of RCA were both positively related to the signal intensity. In addition to biomolecules, DNA could also improve the detection sensitivity of TiO2 nanoparticles in the CE assay.92
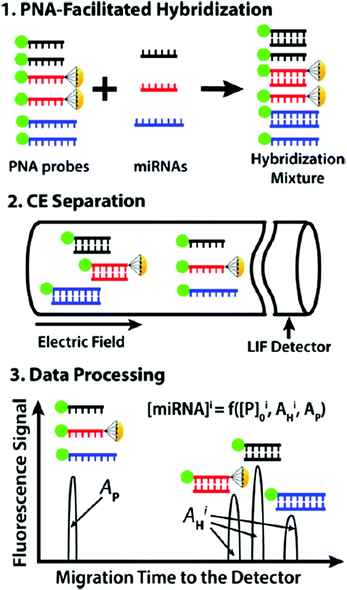 | ||
| Fig. 9 Schematic depiction of DQAMmiR with PNA hybridization probes (Reproduced from ref. 90 with permission from [American Chemical Society], copyright [2018].). | ||
DNA is a desirable chiral selector used in partial-filling CE enantioseparation with improved resolution. Moreover, the DNA-assisted technique also exhibits specific merits to enhance the sensitivity of quantifying and detecting biomolecules. The application of DNA aptamers for CE analysis will be further discussed in detail in section 3.4.
3.4. DNA aptamers for capillary-based affinity analysis
DNA aptamers possess specific affinity to various targets, such as small organic molecules, proteins, and inorganic ions. Aptamers are suitable for affinity CE-LIF analysis of aptamer-protein and small molecule–aptamer interactions.93 To improve the binding affinity of the 15-mer DNA aptamer (Apt15), a polyT tail was attached on the 3′-end of Apt15.36 For α-thrombin detection, polyT tail could also interact with human α-thrombin when the Apt15 section bound to α-thrombin. By using Apt15 containing a T25 tail, sensitive detection of thrombin was achieved with a detection limit of 0.1 nM. Different types of nanomaterials can be combined with aptamers to prepare novel probes. For instance, Zhou et al. combined the high sensitivity of CdTe/CdS core–shell quantum dots (QDs) with the broad-specificity of aptamers for simultaneously detecting organophosphorus pesticides.94 Graphene oxide (GO) was able to adsorb and quench a fluorescent aptamer with a sequence of 5′-carboxyfluorescein (FAM)-TCCTCCTCCTTCACACGGAAC-3′.95 Subsequently, aptamer/GO was incubated with the target adenosine triphosphate (ATP). The desorption of aptamers occurred due to the stronger affinity between ATP and the aptamer. Finally, a mixture containing the ATP–aptamer complex and excess aptamer/GO and GO was separated and quantified by CE-LIF detection. During the CE analysis process, the isolation of the fluorophore from GO could prevent the recovered fluorescence from being quenched and interfered by GO, further improving detection sensitivity and accuracy.As we know, adenosine plays an important role in regulating or mediating various neuronal phenomena, and different methods have been investigated to analyze adenosine. However, conventional strategies suffer from disadvantages such as low selectivity and accuracy. We explained the application of the aptamer/GO probe for ATP detection with improved sensitivity.95 In addition, the aptamer could be immobilized onto the inner surface of capillaries for separating and determining target analytes.96,97 The in-tube solid-phase microextraction (SPME) based on capillary has received considerable research interest due to their long service life, high target loadings, and low cost. To improve the specific recognition performance of in-tube SPME, it is necessary to develop desirable coating materials. We mentioned the tremendous advantages of aptamers in section 2.4. Preparing an aptamer-modified capillary can provide an efficient way for separating and enriching adenosine in bio-samples with enhanced affinity. Therefore, aptamer/gold nanoparticle (GNP) modified capillary was developed for in-tube SPME.37 The extraction amount of adenosine was able to be improved because of the increased loading of aptamers.
Monolithic columns are also considered desirable stationary phases for aptamer immobilization.98–103 For instance, an aptamer-modified hydrophilic polymer monolith was investigated for detecting human α-thrombin from the human plasma.104 A hybrid affinity monolithic column was developed by using GNPs as an intermediary.38 First, a (N-(2-Aminoethyl)-3-aminopropyl)tris-(2-ethoxy)silane (AEAPTES)-silica monolithic column was prepared (Fig. 10). The thiolated aptamer with a sequence of 5′-AGT CCG TGG TAG GGC AGG TTG GGG TGA CT-3′ was modified with GNPs. And then, the obtained aptamer-functionalized GNP solution was pumped through the hybrid monolith to synthesize an affinity monolith. Since nanoparticles possess a high surface-to-volume ratio and the monolithic matrix exhibited a specific surface area, the average coverage density of aptamers on the column could reach 342 pmol μL−1. Moreover, the affinity hybrid monolithic column was combined with the enzymatic chromogenic assay for enriching and determining thrombin proteins with improved sensitivity. As a result, a hybrid silica monolithic column with numerous active sites was able to enhance the binding capacity of the aptamer. Xie et al. prepared an innovative polyhedral oligomeric silsesquioxane (POSS)-based hybrid monolithic column for aptamer immobilization.105 Due to its abundant functional sites and highly stereoscopic nanocages, POSS was able to bind with branched polyethyleneimine (PEI) via a ring-opening reaction of POSS-epoxy with PEI linkers. Aptamer (5′-GATCGGGTGTGGGTGGCGTAAAGGG AGCAT-CGGACA-3′) modified GNPs were further coated onto the well-controlled 3D skeletal POSS-PEI monolith. The resultant POSS-based affinity monolith exhibited prominent response of ochratoxin A (OTA) with a detection limit of 0.06 ng mL−1. In addition to the post-modification method, the aptamer-based hybrid monolithic column could be prepared via a facile ‘one-pot’ process.106 For the fabrication of the aptamer-based hybrid monolith, a polymerization mixture containing the ternary porogenic system, POSS chemicals, acrylate-based monomers and SH-aptamer was introduced into the pretreated capillary. This strategy combined ‘free radical polymerization’ and ‘thiol-ene’ click reaction, providing a simpler preparation process for fabricating the aptamer-modified hybrid monolithic column. Attributing to the hydrophobic property of POSS, most POSS-containing hybrid monoliths were hydrophobic and nonspecific adsorption might occur. As a result, a hydrophilic monolithic column was prepared via the polymerization of hydrophilic monomers with aptamer and POSS-methacryl substituted (POSS-MA).107 The proposed column could achieve selective recognition of OTA, and the recovery yield of ochratoxin B (OTB) caused by nonspecific adsorption was only about 0.1%.
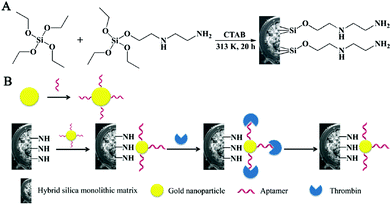 | ||
| Fig. 10 Scheme for the preparation of AEAPTES- (A) and aptamer@GNP@AEAPTES- (B) silica hybrid monolithic capillary columns (Reproduced from ref. 38 with permission from [Royal Society of Chemistry], copyright [2016].). | ||
Various DNA aptamer/NP probes can be used to improve the performance of CE-based affinity analysis. CdTe/CdS core–shell QDs and GO were chosen to enhance the detection sensitivity of the CE assay. Moreover, the state-of-the-art applications of NPs for preparing aptamer-modified OT and monolithic capillaries with increased binding capacity are described.
4. Conclusions
The aim of this review is to summarize various approaches by which DNA meets capillary-based microsystems, including capillary-based methods for DNA assay and DNA-based methods for fabricating capillary-based analysis systems. This ‘meeting’ facilitates the combination of high affinity of DNA and the advantages of capillary-based microsystems such as reduced reagent consumption, improved analysis speed, and high automation. Capillary-based microsystems provide desirable platforms for DNA–ligand interaction assay, DNA translocation study, DNA separation, DNA aptamer selection, DNA amplification assay, and DNA digestion. Meanwhile, DNA can be utilized to construct capillary-based biosensors and enzymatic reactors and enhance CE performance.Investigating effective techniques for DNA–ligand interaction assay is important in the biological analysis field. We reported multiple modes of CE applied for exploring binding properties and DNA–biomolecule interactions. Moreover, the CE-MS strategy also presents tremendous merits in the quantification and characterization of the binding of DNA to biomolecules/metallodrugs. As an alternative to CE, HDC possesses specific sensitivity and quantification ability for DNA–biomolecule interaction assay. Efforts have been made on developing CE-based methods to separate DNA depending on molecular size or sequence differences. Studies indicate that the CE-LIF system offers a simple and robust platform for sequencing the same-length strands and exploring base pair mutations. Currently, the rapidly expanding development of glass capillary nanopores provides novel opportunities for the DNA translocation study. Innovative orifice glass nanopores applied for multiarm DNA concatemer sensing are described in detail. Since DNA presents specific affinity to various molecules, the DNA hydrogel can also be used for the fabrication of capillary-based biosensors. We believe that much more attention should be paid on the development of novel capillary-based microsystems for biomolecule sensing applications. Compared with traditional SELEX, the CE-SELEX technique displays specific advantages for DNA aptamer selection. Interestingly, DNA aptamers are suitable ligands for capillary-based affinity analysis as well. We foresee state-of-the-art applications of NPs for enhancing the performance of DNA aptamer-based affinity analysis. The influences of different capillary-based methods on the DNA amplification assay have been discussed in detail. New capillary-based bioreactors employed for DNA digestion are described, and DNA is also an efficient linker for the fabrication of capillary-based enzymatic reactors. Furthermore, DNA has been used as mobile phase additives in the CE separation of enantiomers or biomolecules with improved resolution. We believe that noteworthy efforts will continue to be made to combine DNA and capillary-based microsystems for use in chemical and biological analyses.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful for the funds from the National Natural Science Foundation of China (81803495); Natural Science Foundation of Hunan Province (2019JJ50749); Jiangsu Key Research and Development Plan (Society Development, No. BE2018639); the International Scientific Cooperation Project of Changzhou Scientific Bureau (grant numbers CZ20190004, CZ20190009); the Science & Technology Support Program of Changzhou (Application Basic Research CJ20190033); the QingLan Project of Jiangsu Province.References
- S. Bernardo-Bermejo, E. Sánchez-López, M. Castro-Puyana and M. L. Marina, Trends Anal. Chem., 2020, 124, 115807 CrossRef CAS
.
- X. Shen, Z. Yang, E. N. McCool, R. A. Lubeckyj, D. Chen and L. Sun, Trends Anal. Chem., 2019, 120, 115644 CrossRef CAS
.
- K. Petr, D. Miloš and K. Pavel, Anal. Chim. Acta, 2019, 1075, 1–26 CrossRef
.
- S. C. Lam, E. S. Rodriguez, P. R. Haddad and B. Paull, Analyst, 2019, 144, 3464–3482 RSC
.
- R. L. C. Voeten, I. K. Ventouri, R. Haselberg and G. W. Somsen, Anal. Chem., 2018, 90, 1464–1481 CrossRef CAS
.
- C. Chen, W. Liu and T. Hong, Analyst, 2019, 144, 3912–3924 RSC
.
- S. Ranallo, A. Porchetta and F. Ricci, Anal. Chem., 2019, 91, 44–59 CrossRef CAS
.
- E. Duffy, J. Florek, S. Colon and A. E. Gerdon, Anal. Chim. Acta, 2020, 11108, 115–121 CrossRef
.
- C. Y. Lee, H. Kim, K. S. Park and H. G. Park, Anal. Chim. Acta, 2019, 10604, 30–44 CrossRef
.
- Y. Liu, Z. Zhu, C. Wang, R. Gao, X. Yang and S. Liu, Analyst, 2019, 144, 2130–2137 RSC
.
- C. J. Kristoff, L. Bwanali, L. M. Veltri, G. P. Gautam, P. K. Rutto, E. O. Newton and L. A. Holland, Anal. Chem., 2020, 92, 49–66 CrossRef CAS
.
- R. K. Harstad, A. C. Johnson, M. M. Weisenberger and M. T. Bowser, Anal. Chem., 2016, 88, 299–319 CrossRef CAS
.
- R. L. C. Voeten, I. K. Ventouri, R. Haselberg and G. W. Somsen, Anal. Chem., 2018, 90, 1464–1481 CrossRef CAS
.
- I. O. Neaga, E. Bodoki, S. Hambye, B. Blankert and R. Oprean, Talanta, 2016, 148, 247–256 CrossRef CAS
.
- J. W. Guthrie, R. T. Limmer, E. A. Brooks, C. C. Wisnewski, N. D. Loggins-Davis and A. Bouzid, Anal. Chim. Acta, 2015, 8531, 676–681 CrossRef
.
- C. Artner, H. U. Holtkamp, W. Kandioller, C. G. Hartinger, S. M. Meier and B. K. Keppler, Chem. Commun., 2017, 53, 8002–8005 RSC
.
- L. H. Chung and V. Murray, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1008, 87–97 CrossRef CAS
.
- J. Wang, Y. Qin, S. W. Jiang, L. Liu, Y. Lu, J. Li, L. Qiu and P. Jiang, Sens. Actuators, B, 2016, 237, 106–112 CrossRef CAS
.
- S. M. Friedrich, R. Bang, A. Li and T. H. Wang, Anal. Chem., 2019, 91, 2822–2830 CrossRef CAS
.
- X. Xu, C. Li, Y. Zhou and Y. Jin, ACS Sens., 2017, 2, 1452–1457 CrossRef CAS
.
- T. Kubo, N. Nishimura, H. Furuta, K. Kubota, T. Naito and K. Otsuka, J. Chromatogr., A, 2017, 1523, 107–113 CrossRef CAS
.
- L. Duan, Z. Cao and L. Yobas, Anal. Chem., 2017, 89, 10022–10028 CrossRef CAS
.
- X. Zhang and L. B. McGown, Electrophoresis, 2016, 37, 2017–2024 CrossRef CAS
.
- Z. E. Hughes and T. R. Walsh, ACS Sens., 2017, 2, 1602–1611 CrossRef CAS
.
- C. Zhu, L. Li, G. Yang, S. Fang, M. Liu, M. Ghulam, C. Hao, Y. Chen and F. Qu, Anal. Chim. Acta, 2019, 1070, 112–122 CrossRef CAS
.
- X. Li, Y. He, Y. Ma, Z. Bie, B. Liu and Z. Liu, Anal. Chem., 2016, 88, 9805–9812 CrossRef CAS
.
- C. Liu, Y. Yamaguchi, S. Sekine, Y. Ni and X. Dou, Sens. Actuators, B, 2018, 258, 263–269 CrossRef CAS
.
- X. Qiu, J. I. Shu, O. Baysal, J. Wu, S. Qian, S. Ge, K. Li, X. Ye, N. Xia and D. Yu, Microfluid. Nanofluid., 2019, 23, 1–8 CrossRef CAS
.
- X. Li and A. Manz, Sens. Actuators, B, 2019, 288, 678–682 CrossRef CAS
.
- T. Hong, W. Liu, M. Li and C. Chen, Anal. Chim. Acta, 2019, 1067, 31–47 CrossRef CAS
.
- B. Derkus, Biosens. Bioelectron., 2016, 79, 901–913 CrossRef CAS
.
- W. Xue, X. Tan, M. K. K. Oo, G. Kulkarni, M. A. Ilgen and X. Fan, Analyst, 2020, 145, 1346–1354 RSC
.
- Y. X. Luo, Q. J. Li, L. Zhou, Y. S. Li and X. F. Gao, Analyst, 2018, 143, 700–708 RSC
.
- C. Jiang, Y. Li, H. Wang, D. Chen and Y. Wen, Sens. Actuators, B, 2020, 307, 127625 CrossRef
.
- N. Wu, S. Wang, Y. Yang, J. Song, P. Su and Y. Yang, Int. J. Biol. Macromol., 2018, 113, 38–44 CrossRef CAS
.
- Y. Bai, Y. Li, D. Zhang, H. Wang and Q. Zhao, Anal. Chem., 2017, 89, 9467–9473 CrossRef CAS
.
- Y. Ma, L. Hao, X. Lin, X. Liu, X. Qiu, X. Zhang and X. Hu, J. Chromatogr. A, 2020, 1611, 460617 CrossRef CAS
.
- J. Zhao, Q. Zhu, L. Zhao, H. Lian and H. Chen, Analyst, 2016, 141, 4961–4967 RSC
.
- M. Kanoatov and S. N. Krylov, Anal. Chem., 2016, 88, 7421–7428 CrossRef CAS
.
- X. Wang, Y. Hu, F. Qu and R. U. Khan, J. Chromatogr. A, 2017, 1501, 161–166 CrossRef CAS
.
- H. J. Kim, K. Y. Lee, Y. C. Kim, S. C. Kim, S. Y. Lee, T. W. Jang, M. K. Lee, K. C. Shin, G. H. Lee, J. C. Lee, J. E. Lee and S. Y. Kim, Lung Cancer, 2012, 75, 321–325 CrossRef
.
- B. Macadangdang, N. Zhang, P. E. Lund, A. H. Marple, M. Okabe, M. M. Gottesman, D. H. Appella and C. Kimchi-Sarfaty, PLoS One, 2011, 6, e17981 CrossRef CAS
.
- C. Przybylski, J. M. Benito, V. Bonnet, C. O. Mellet and J. M. García Fernández, Anal. Chim. Acta, 2018, 1002, 70–81 CrossRef CAS
.
- L. S. Foteeva, M. Matczuk, K. Pawlak, S. S. Aleksenko, S. V. Nosenko, V. K. Karandashev, M. Jarosz and A. R. Timerbaev, Anal. Bioanal. Chem., 2017, 409, 2421–2427 CrossRef CAS
.
- Y. L. Ying, Y. X. Hu, R. Gao, R. J. Yu, Z. Gu, L. P. Lee and Y. T. Long, J. Am. Chem. Soc., 2018, 140, 5385–5392 CrossRef CAS
.
- C. Gao, S. Ding, Q. Tan and L. Q. Gu, Anal. Chem., 2009, 81, 80–86 CrossRef CAS
.
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos and E. Birney, Nature, 2013, 494, 77–80 CrossRef CAS
.
- G. Zhu, R. Hu, Z. Zhao, Z. Chen, X. Zhang and W. Tan, J. Am. Chem. Soc., 2013, 135, 16438–16445 CrossRef CAS
.
- S. Modi, C. Nizak, S. Surana, S. Halder and Y. Krishnan, Nat. Nanotechnol., 2013, 8, 459–467 CrossRef CAS
.
- Y. Zhou, R. Wu, D. Wang, P. Hu and Y. Jin, ACS Sens., 2019, 4, 3119–3123 CrossRef CAS
.
- C. Giovannoli, L. Anfossi, C. Tozzi, G. Giraudi and A. Vanni, J. Sep. Sci., 2004, 27, 1551–1556 CrossRef CAS
.
- V. Villareal, Y. Zhang, C. Zurita, J. Moran, I. Silva and F. A. Gomez, Anal. Lett., 2003, 36, 451–463 CrossRef CAS
.
- Z. Cao and L. Yobas, Anal. Chem., 2014, 86, 737–743 CrossRef CAS
.
- Z. Cao and L. Yobas, ACS Nano, 2015, 9, 427–435 CrossRef CAS
.
- M. Fraňo, K. Džganová, P. Koiš and M. Masár, Electrophoresis, 2016, 37, 3084–3088 CrossRef
.
- T. Sakurai, H. Hoshino and T. Takahashi, J. Sep. Sci., 2017, 40, 3153–3160 CrossRef CAS
.
- J. Zhou and J. Rossi, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS
.
- K. Wakui, T. Yoshitomi, A. Yamaguchi, M. Tsuchida, S. Saito, M. Shibukawa, H. Furusho and K. Yoshimoto, Mol. Ther. – Nucleic Acids, 2019, 16, 348–359 CrossRef CAS
.
- K. Hirose, M. Tsuchida, H. Asakura, K. Wakui, K. Yoshimoto, K. Iida, M. Sato, M. Shibukawa, M. Suganuma and S. Saito, Analyst, 2017, 142, 3997–4152 RSC
.
- Z. Li, X. Wang, M. Chen, Q. Li, H. Qu, T. Shao, R. Sun, Y. Zhang and Z. Xia, Anal. Biochem., 2019, 564–565, 47–53 CrossRef CAS
.
- N. S. Hamedani and J. Müller, Methods Mol. Biol., 2016, 1380, 61–75 CrossRef CAS
.
- A. T. H. Le, S. M. Krylova, M. Kanoatov, S. Desai and S. N. Krylov, Angew. Chem., Int. Ed., 2019, 58, 2739–2743 CrossRef CAS
.
- B. Lou, E. Chen, X. Zhao, F. Qu and J. Yan, J. Chromatogr. A, 2016, 1437, 203–209 CrossRef CAS
.
- T. Houssin, J. Cramer, R. Grojsman, L. Bellahsene, G. Colas, H. Moulet, W. Minnella, J. Pannetier, M. Leberre, A. Plecis and Y. Chen, Lab Chip, 2016, 16, 1401–1411 RSC
.
- M. J. Jian, C. L. Perng, J. R. Sun, Y. H. Cheng, H. Y. Chung, Y. H. Cheng, S. Y. Lee, S. C. Kuo and H. S. Shang, Sci. Rep., 2019, 9, 1–10 CrossRef CAS
.
- C. Zhou, M. Li, C. Sun, H. Zou, X. Wu, L. Zhang, S. Tao, B. Wang and Y. Li, Anal. Biochem., 2016, 494, 68–75 CrossRef CAS
.
- G. Xu, H. Zhao, J. M. Cooper and J. Reboud, Chem. Commun., 2016, 52, 12187–12190 RSC
.
- Y. Zhang, L. Zhang, J. Sun, Y. Liu, X. Ma, S. Cui, L. Ma, J. J. Xi and X. Jiang, Anal. Chem., 2014, 86, 7057–7062 CrossRef CAS
.
- L. Zhang, Y. Zhang, C. Wang, Q. Feng, F. Fan, G. Zhang, X. Kang, X. Qin, J. Sun, Y. Li and X. Jiang, Anal. Chem., 2014, 86, 10461–10466 CrossRef CAS
.
- R. Li, J. Chen, X. Zhang, J. Cui, S. Tao and L. Yang, J. Agric. Food Chem., 2020, 68, 899–906 CrossRef CAS
.
- M. Cheng, R. Wang, B. Zhang, Z. Mao and Z. Chen, J. Pharm. Biomed., 2019, 165, 129–134 CrossRef CAS
.
- S. Štěpánová and V. Kašička, J. Sep. Sci., 2016, 39, 198–211 CrossRef
.
- J. Yin, T. Xu, N. Zhang and H. Wang, Anal. Chem., 2016, 88, 7730–7737 CrossRef CAS
.
- J. Li, L. Mo, C. H. Lu, T. Fu, H. H. Yang and W. Tan, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC
.
- B. Xiang, K. He, R. Zhu, Z. Liu, S. Zeng, Y. Huang, Z. Nie and S. Yao, ACS Appl. Mater. Interfaces, 2016, 8, 22801–22807 CrossRef CAS
.
- Y. Li, Y. Ma, X. Jiao, T. Li, Z. Lv, C. J. Yang, X. Zhang and Y. Wen, Nat. Commun., 2019, 10, 1036 CrossRef
.
- J. Song, P. Su, R. Ma, Y. Yang and Y. Yang, Ind. Eng. Chem. Res., 2017, 56, 5127–5137 CrossRef CAS
.
- J. Song, T. Lei, Y. Yang, N. Wu, P. Su and Y. Yang, New J. Chem., 2018, 42, 8458–8468 RSC
.
- Y. Lin, P. Yu and L. Mao, Analyst, 2015, 140, 3781–3787 RSC
.
- W. G. Koh and M. Pishko, Sens. Actuators, B, 2005, 106, 335–342 CrossRef CAS
.
- M. Li, H. Shen, Z. Zhou, W. He, P. Su, J. Song and Y. Yang, Electrophoresis, 2020, 41, 335–344 CrossRef CAS
.
- C. Pan, W. Lv, X. Niu, G. Wang, H. Chen and X. Chen, J. Chromatogr. A, 2018, 1541, 31–38 CrossRef CAS
.
- T. Hong, C. Chi and Y. Ji, J. Sep. Sci., 2014, 37, 3377–3383 CrossRef CAS
.
- S. Xu, R. Mo, C. Jin, X. Cui, R. Bai and Y. Ji, J. Pharm. Biomed., 2017, 140, 190–198 CrossRef CAS
.
- L. Tohala, F. Oukacine, C. Ravelet and E. Peyrin, Anal. Chem., 2015, 87, 5491–5495 CrossRef CAS
.
- J. Ruta, S. Perrier, C. Ravelet, B. Roy, C. Perigaud and E. Peyrin, Anal. Chem., 2009, 81, 1169–1176 CrossRef CAS
.
- R. Huang, W. M. Xiong, D. F. Wang, L. H. Guo, Z. Y. Lin, L. S. Yu, K. D. Chu, B. Qiu and G. N. Chen, Electrophoresis, 2013, 34, 254–259 CrossRef CAS
.
- L. Tohala, F. Oukacine, C. Ravelet and E. Peyrin, Electrophoresis, 2017, 38, 1383–1390 CrossRef CAS
.
- D. W. Wegman, F. Ghasemi, A. S. Stasheuski, A. Khorshidi, B. B. Yang, S. K. Liu, G. M. Yousef and S. N. Krylov, Anal. Chem., 2016, 88, 2472–2477 CrossRef CAS
.
- L. Hu, M. Anand, S. M. Krylova, B. B. Yang, S. K. Liu, G. M. Yousef and S. N. Krylov, Anal. Chem., 2018, 90, 14610–14615 CrossRef CAS
.
- N. Padhan, J. Yan, A. Boge, E. Scrivener, H. Birgisson, A. Zieba, M. Gullberg, M. Kamali-Moghaddam, L. Claesson-Welsh and U. Landegren, Sci. Rep., 2017, 7, 1–9 CrossRef CAS
.
- S. Alsudir and E. P. C. Lai, Anal. Bioanal. Chem., 2017, 409, 1857–1868 CrossRef CAS
.
- L. Sun, Y. Li, H. Wang and Q. Zhao, Talanta, 2019, 204, 182–188 CrossRef CAS
.
- T. Tang, J. Deng, M. Zhang, G. Shi and T. Zhou, Talanta, 2016, 146, 55–61 CrossRef CAS
.
- B. Y. Fang, M. H. Yao, C. Y. Wang, C. Y. Wang, Y. D. Zhao and F. Chen, Colloids Surf., B, 2016, 140, 233–238 CrossRef CAS
.
- M. Mahmoud, S. Laufer and H. P. Deigner, Microchim. Acta, 2019, 186, 1–8 CrossRef
.
- L. Liu, K. Yang, X. Zhu, Y. Liang, Y. Chen, F. Fang, Q. Zhao, L. Zhang and Y. Zhang, Talanta, 2017, 175, 189–193 CrossRef CAS
.
- Y. Chen, X. Ding, D. Zhu, X. Lin and Z. Xie, Talanta, 2019, 200, 193–202 CrossRef CAS
.
- C. Acquah, Y. W. Chan, S. Pan, L. S. Yon, C. M. Ongkudon, H. Guo and M. K. Danquah, Sci. Rep., 2019, 9, 14501 CrossRef
.
- H. Lyu, H. Sun, Y. Zhu, J. Wang, Z. Xie and J. Li, Anal. Chim. Acta, 2020, 1103, 97–105 CrossRef CAS
.
- Q. Zhao, M. Wu, X. Chris Le and X. F. Li, Trends Anal. Chem., 2012, 41, 46–57 CrossRef CAS
.
- H. P. Jiang, J. X. Zhu, C. Peng, J. Gao, F. Zheng, Y. X. Xiao, Y. Q. Feng and B. F. Yuan, Analyst, 2014, 139, 4940–4946 RSC
.
- A. Marechal, F. Jarrosson, J. Randon, V. Dugas and C. Demesmay, J. Chromatogr. A, 2015, 1406, 109–117 CrossRef CAS
.
- Y. Chen, N. Deng, C. Wu, Y. Liang, B. Jiang, K. Yang, Z. Liang, L. Zhang and Y. Zhang, Talanta, 2016, 154, 555–559 CrossRef CAS
.
- X. Yu, H. Song, J. Huang, Y. Chen, M. Dai, X. Lin and Z. Xie, J. Mater. Chem. B, 2018, 6, 1965–1972 RSC
.
- Y. Chen, M. Chen, J. Chi, X. Yu, Y. Chen, X. Lin and Z. Xie, J. Chromatogr. A, 2018, 1563, 37–46 CrossRef CAS
.
- Y. Chen, D. Zhu, X. Ding, G. Qi, X. Lin and Z. Xie, Analyst, 2019, 144, 1555–1564 RSC
.
Footnote |
| † Co-first author. |
| This journal is © The Royal Society of Chemistry 2021 |