A multi-location peak parking approach for calculation of second dimensional retention indices for improved volatile compound identification with cryogen-free comprehensive heart-cut two-dimensional gas chromatography†
Pannipa
Janta
a,
Duangkamol
Pinyo
b,
Yamonporn
Yodta
b,
Porames
Vasasiri
b,
Meinolf
Weidenbach
c,
Matthias
Pursch
d,
Xiuhan (Grace)
Yang
 *e and
Chadin
Kulsing
*e and
Chadin
Kulsing
 *af
*af
aDepartment of Chemistry, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand. E-mail: ckulsing@gmail.com; Tel: +66802971178
bThe Center for Advanced Analytical Technology, Dow Chemical Thailand Ltd, Map Ta Phut Industrial Estate, Rayong 21150, Thailand
cPolyurethanes Tech Center, Dow Deutschland Anlagen GmbH, 21677 Stade, Germany
dAnalytical Science, Dow Deutschland Anlagen GmbH, 21677 Stade, Germany
eDow Chemical China Investment Company, Shanghai 201203, China. E-mail: gxyang@dow.com; Tel: +862138513112
fChromatographic Separation and Flavor Chemistry Research Unit and Center of Molecular Sensory Science, Department of Chemistry, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand
First published on 15th December 2020
Abstract
Comprehensive heart-cut multidimensional gas chromatography (CH/C MDGC) without a cryogenic trapping device was developed with an established approach for calculation of first and second dimensional retention indices (1I and 2I) for improved compound identification. A first dimensional (1D) DB-1MS column (60 m) and a second dimensional (2D) DB-WAX column (60 m) were applied with a Deans switch (DS) using a constant H/C window of 0.2 min and a periodic multiple heartcut strategy comprising 225H/C throughout the CH/C. 1I was calculated based on comparison of the middle of the heartcut time with the alkane retention times on the 1D column. A multi-location peak parking approach using sixteen sets of automated injections of alkane references was also established with the least square curve fitting method for construction of the alkane isovolatility curves which were applied for 2I calculation. The untargeted compound analysis of a perfume sample was then performed according to comparison with the libraries of mass spectra, 1I and 2I. The CH/C MDGC system with a 25 h analysis time showed a peak capacity (nc) of 9198 and 128 separated peaks with 71 compounds successfully identified according to MS, 1I and 2I library match under the established error approximation criteria. Furthermore, relationship between the analysis time and number of separated peaks was proposed based on the set of 84 identifiable compounds. With the compensation of lower separation performance and greater I errors, the analysis time could be reduced by applying a 2.5 min H/C window with a total analysis time of 2 h and nc of 1134.
1. Introduction
Comprehensive two dimensional gas chromatography (GC × GC) offers separation with high resolution and enhanced peak capacity improving the analysis of complex samples such as petrochemicals, essential oils, and other food or biological samples.1–4 This technique solves the co-elution problem, enhances peak capacity and presents more reliable results than those obtained with conventional one dimensional GC.5,6 GC × GC hyphenated with mass spectrometry (GC × GC-MS) also provides untargeted compound analysis with higher confidence.7–9 This technique relies on an effective modulation process (using a cryogenic, valve based or flow modulator) which transfers all components obtained from a 1D to a 2D column with different selectivity.10,11 This can be generally performed by using a short 2D column (≤5 m) which enables fast 2D separation (<20 s) completing comprehensive analysis within a single injection.12,13Alternatively, multiple injection techniques can be applied to allow the use of a longer 2D column and longer analysis time improving separation and peak capacity. Such operation has been recognized as comprehensive heart-cut multidimensional GC (CH/C MDGC),14 and can be performed by using a heartcut device, Deans switch (DS) or switching valve, and a modulator.2,15,16 The modulator can also be replaced with a cryogenic trapping device with a lower cost.17,18 To this end, the DS heartcuts a selected pulse19,20 from a 1D column to be trapped at the cryogenic device located close to a 2D column inlet. This is followed by the pulse release within a narrow band of a sample undergoing 2D separation, which can be performed using an independent flow and temperature program for improved 2D separation. However, a cryogenic approach still involves additional devices (e.g. a trapping channel and a switching valve for automated on/off operation), cryogen consumption and system maintenance. With the compensation of lower separation efficiency, a system employing a DS without using a cryogenic trapping device (cryogen-free CH/C MDGC) has been reported.16 This approach applied a single DS with a H/C window of 0.2 min in order to transfer a narrow band of the sample from a 1D onto a 2D column without trapping, which improved the separation performance. A challenge is to develop a data analysis approach for improved compound identification with a cryogen-free CH/C MDGC-MS system.
Traditional methods used to identify untargeted compounds with GC × GC-MS are matched with MS spectra and 1I libraries.3 Calculation of 1I for a peak of interest is performed by additional injection of n-alkanes followed by comparison of the peak 1D retention time (1tR) with that of the alkanes according to van den Dool and Kratz relationship.21 Cryogen-free CH/C MDGC can also apply the same method for 1I calculation albeit the middle of the H/C is used instead of 1tR.16 Moreover, 2I can be taken into account to improve confidence in compound identification. For conventional GC × GC, 2I calculation can be performed by multiple injections of n-alkanes under the same temperature programs used for sample analysis or the temperature program with steps of different isothermal temperatures.3,9,22 This is performed to generate isovolatility curves (1tRvs.2tR plots) for alkanes on a 2D column, and the curves are used to calculate 2I for a peak. 2I calculation has also been reported with H/C MDGC using cryogenic trapping devices where all the trapped compounds simultaneously started eluting onto a 2D column under the same flow and temperature programs.3 This approach involves a single injection of alkanes onto the applied system. All the alkanes elute through a 1D column and are trapped at the inlet of the 2D column. The oven is then cooled down prior to the alkane release onto a 2D column under an identical 2D flow and temperature program to that applied for sample separation. This allows direct 2I calculation using the same approach as that applied for 1I calculation. However, this approach cannot be applicable for CH/C MDGC with different compounds eluting under different 2D conditions. In addition, an approach for 2I calculation with cryogen-free CH/C MDGC has not been reported.
In this study, a cryogen-free CH/C MDGC-MS system with a narrow H/C window of 0.2 min was applied, and relevant data analysis and experimental approaches for 1I and 2I calculation were established. Isovolatility curves were constructed according to a peak parking approach allowing the alkane peaks to park at different axial positions along the 1D column prior to elution onto the 2D column at different temperatures. This was adapted from the concept of multi-location peak parking (MLPP) which was reported in liquid chromatography and differently applied to investigate peak broadening after parking (stopped flow) for different periods prior to the peak elution and detection in order to determine the longitudinal diffusion coefficient of the analyte.23 An example application was demonstrated for perfume analysis where compounds were identified according to comparison with MS spectra, 1I and 2I from the libraries. The possible compounds observed in perfumes included aldehydes, alcohols, lactones, esters and terpene derivatives.24,25
2. Experimental
2.1 Materials and chemicals
Perfume was purchased from a local supermarket, Thailand, and kept in a refrigerator at 4 °C prior to use. n-alkane standards (C6–C22) and all the standard compounds were purchased from Sigma-Aldrich (St. Louis, MO).2.2 Instrumental
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A 1D nonpolar DB-1MS column (60 m × 0.25 mm I.D. × 0.25 μm; J&W Scientific, US) and a 2D polar DB-Wax column (60 m × 0.25 mm × 0.25 μm; J&W Scientific, US) were used for separation. The restrictor column (1.5 m × 0.1 mm; Agilent technologies Inc.) was applied to balance the flow with the 2D column. The outlets of the restrictor and the 2D columns were connected to a flame ionization detector (FID) and MS as reported,16 respectively. A Dean switch device (DS, Agilent technologies Inc.) is an interface connecting the 1D, 2D and restrictor columns. In this instrument, the DS was operated in off and on modes corresponding to analyte pulse transfer to the FID and MS, respectively. The flow rates of the carrier gas (99.999% He) in 1D and 2D separation were 2.0 and 4.0 mL min−1, respectively. The GC oven temperature program was set at 60 °C, increased to 250 °C with a rate of 4 °C min−1 and held at this temperature for 12.5 min. The FID temperature was set at 270 °C using flow rates of 40, 400 and 30 mL min−1 for hydrogen, air and N2 (makeup gas), respectively. For MS conditions, the ion source temperature was set at 250 °C, the electron ionization voltage was −70 eV and a mass range of 28–550 m/z was applied. The multiple H/C approach with a constant window of 0.2 min was applied. This required 25 injections in order to comprehensively sample all the peaks obtained from 1D separation. The H/C events are provided in Table S-1 (ESI).†
1. A 1D nonpolar DB-1MS column (60 m × 0.25 mm I.D. × 0.25 μm; J&W Scientific, US) and a 2D polar DB-Wax column (60 m × 0.25 mm × 0.25 μm; J&W Scientific, US) were used for separation. The restrictor column (1.5 m × 0.1 mm; Agilent technologies Inc.) was applied to balance the flow with the 2D column. The outlets of the restrictor and the 2D columns were connected to a flame ionization detector (FID) and MS as reported,16 respectively. A Dean switch device (DS, Agilent technologies Inc.) is an interface connecting the 1D, 2D and restrictor columns. In this instrument, the DS was operated in off and on modes corresponding to analyte pulse transfer to the FID and MS, respectively. The flow rates of the carrier gas (99.999% He) in 1D and 2D separation were 2.0 and 4.0 mL min−1, respectively. The GC oven temperature program was set at 60 °C, increased to 250 °C with a rate of 4 °C min−1 and held at this temperature for 12.5 min. The FID temperature was set at 270 °C using flow rates of 40, 400 and 30 mL min−1 for hydrogen, air and N2 (makeup gas), respectively. For MS conditions, the ion source temperature was set at 250 °C, the electron ionization voltage was −70 eV and a mass range of 28–550 m/z was applied. The multiple H/C approach with a constant window of 0.2 min was applied. This required 25 injections in order to comprehensively sample all the peaks obtained from 1D separation. The H/C events are provided in Table S-1 (ESI).†
2.3 Data analysis
A contour plot for the CH/C MDGC-MS analysis covering 10–60 min of 1D separation was obtained using a combination of all the elution profiles (time vs. intensity profiles each of which belonged to each H/C event within every 5 min) into a single matrix (containing 1tRvs.2tRvs. intensity coordinates). The 1tR coordinate is the H/C center time (tH/Cmid) of the corresponding H/C event. 2tR coordinates of each profile were obtained from the time coordinates observed with MS (tobserved) subtracted with tH/Cmid of each H/C event. The matrix was then converted into a contour plot by using Fortner Transform 3.3 (Fortner, Inc., Savoy, IL). ChemStation software was used to process and identify separated peaks, and Excel 2016 was used for chromatographic parameter calculation and data visualisation. Peaks of interest were tentatively identified by comparison of their mass spectra with those from the NIST 14 library with MS match scores of >700, and further confirmed according to comparison of their experimental 1I and 2I values with those from the NIST 14 within the differences of ±37 and ±44 units, respectively; see the discussion of the errors associated with the I calculation in Section 3.4. Note that when there was no literature 1I available for a peak of interest (e.g. with a signal to noise ratio of >3), the peak was herein defined as a detected peak which was not identifiable.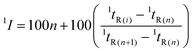 | (1) |
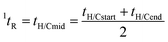 | (2) |
| 2tR = tobserved − 1tR | (3) |
| 2tR,cal = exp(exp(p1 × 2Te + p2) × N + exp(p3 × 2Te + p4) + p5) | (4) |
After calculation of 1tR and 2tR for a peak of interest, eqn (4) was applied to approximate 2tR of the n-alkane references eluting at 1tR of the peak under the same experimental conditions applied for the perfume sample. Kovatś relationship was then used to calculate 2I of this peak with the relationship:22
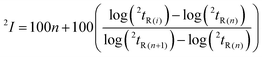 | (5) |
3. Results and discussion
3.1 Analysis with the cryogenic free comprehensive H/C MDGC approach
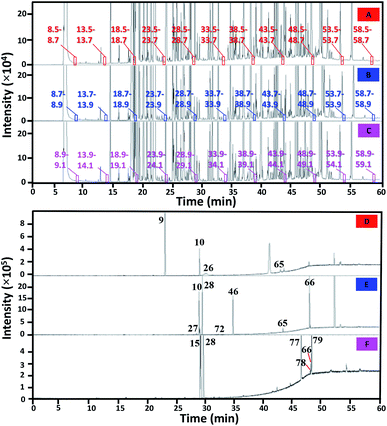
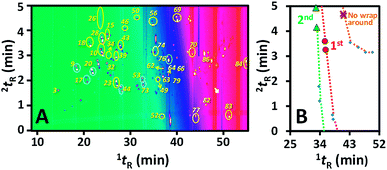
All peaks of the perfume sample in 1D separation were H/C with a constant window of 0.2 min prior to 2D separation. In order to shorten the total analysis time, a cyclic multiple H/C strategy was applied27 applying a 2D separation time of 5 min throughout the whole comprehensive analysis. This required 25 injections with a total analysis time of 25 h. Some of the H/C results are provided in Fig. 1.Fig. 2A shows the contour plot obtained from the overall comprehensive analysis with the compound profile shown in Table 1. From the contour plot, several peaks coeluting in the 1D separation were well separated onto the 2D polar column. For example, peaks 10, 26 and 28 co-eluting in 1D separation were clearly separated onto the 2D polar column. The enhanced separation was obtained according to the long 2D column (60 m) with a thick film of the stationary phase (0.25 μm) which increases separation performance and peak capacity.18 With this approach, several compounds underwent wraparound during the separation as indicated by the yellow circles in Fig. 2A, which allowed effective use of 2D separation space. In addition, 2I calculation could be more effective with a smaller error under wraparound conditions. Note that due to the non-linearly decreasing nature of the isovolatility curves (e.g. see Fig. 4), the isovolatility curves of different alkanes get closer at higher 1tR and lower 2tR leading to a high 2I calculation error.
| No. | 1 t R (min) | 2 t R (min) | Compound | %Area | Match | Rank in MS library match | Literature | 1 I difference | 2 I difference | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 I | 2 I | |||||||||
| a literature values obtained from the NIST 14 library (RI polar column). b literature values obtained from http://webbook.nist.gov/ (normal alkane RI, polar column, and temperature ramp). | ||||||||||
| 1 | 10.40 | 4.92 | 2-Methyl-2-buten-1-ol | 0.01 | 822 | 2 | 760 | 1320a | −5 | 39 |
| 2 | 12.95 | 4.53 | 1-Hexanol | 0.03 | 916 | 1 | 854 | 1348 | −5 | 21 |
| 3 | 14.60 | 3.54 | 3-Methyl-2-butenyl acetate | 0.69 | 948 | 1 | 902 | 1225 | −1 | 17 |
| 4 | 15.84 | 5.34 | Benzaldehyde | 0.02 | 919 | 2 | 933 | 1520 | 1 | 1 |
| 5 | 16.00 | 2.73 | α-Pinene | 0.09 | 920 | 1 | 933 | 1033 | 5 | −32 |
| 6 | 17.60 | 3.04 | β-Myrcene | 0.01 | 898 | 1 | 983 | 1169 | −2 | −9 |
| 7 | 17.62 | 2.90 | β-Pinene | 0.08 | 922 | 1 | 973 | 1128 | 8 | −23 |
| 8 | 17.84 | 3.51 | cis-3-Hexenyl acetate | 0.04 | 934 | 1 | 986 | 1317 | 1 | −16 |
| 9 | 18.45 | 4.46 | 1-Methyl-4-methoxybenzene | 5.45 | 939 | 1 | 1003 | 1461 | 0 | 21 |
| 10 | 18.62 | 10.28 | Benzenemethanol | 0.42 | 921 | 1 | 1012 | 1864 | −5 | 18 |
| 11 | 19.41 | 3.14 | Limonene | 0.33 | 924 | 2 | 1023 | 1216 | 3 | 10 |
| 12 | 19.49 | 3.15 | Eucalyptol | 0.01 | 805 | 1 | 1022 | 1227 | 6 | 5 |
| 13 | 19.80 | 8.41 | α-Methylbenzyl alcohol | 0.01 | 795 | 1 | 1037 | 1801 | −1 | 32 |
| 14 | 19.94 | 3.09 | trans-β-Ocimene | 0.01 | 848 | 1 | 1038 | 1251 | 1 | −31 |
| 15 | 19.54 | 10.29 | Dipropylene glycol | 5.36 | 907 | 1 | 1056 | 1892b | −26 | 18 |
| 16 | 20.60 | 3.09 | 1,4-Cyclohexadiene, 1-methyl-4-(1-methylethyl)- | 0.02 | 894 | 1 | 1050 | 1259 | 6 | −27 |
| 17 | 20.74 | 3.92 | 2,6-Dimethyl-7-octen-2-ol | 0.50 | 929 | 1 | 1062 | 1473 | −3 | −27 |
| 18 | 21.40 | 5.44 | Methyl benzoate | 4.32 | 956 | 1 | 1072 | 1654 | 3 | 13 |
| 19 | 21.31 | 4.00 | trans-Linalool oxide | 0.06 | 885 | 1 | 1074 | 1461 | −1 | 14 |
| 20 | 21.90 | 4.28 | Linalool | 9.84 | 972 | 1 | 1086 | 1541 | 2 | −5 |
| 21 | 22.20 | 4.14 | 3,5-Dimethylanisole | 0.02 | 874 | 4 | 1105 | 1533b | −10 | −14 |
| 22 | 22.40 | 3.45 | trans-Rose oxide | 0.03 | 903 | 1 | 1121 | 1383 | −21 | −4 |
| 23 | 22.10 | 8.82 | Phenylethyl alcohol | 6.56 | 957 | 1 | 1088 | 1895 | 5 | 26 |
| 24 | 22.80 | 4.57 | Fenchol | 0.02 | 855 | 1 | 1105 | 1580 | 5 | 22 |
| 25 | 23.20 | 4.00 | 1,2-Dihydrolinalool | 0.04 | 864 | 1 | 1121 | 1520 | −1 | −2 |
| 26 | 23.60 | 6.42 | β-Phenethyl formate | 0.41 | 908 | 2 | 1149 | 1771a | −19 | 43 |
| 27 | 23.80 | 4.75 | β-Terpineol | 0.05 | 895 | 1 | 1137 | 1627a | −3 | 22 |
| 28 | 23.97 | 5.65 | Benzyl ethanoate | 12.41 | 975 | 1 | 1139 | 1762 | 0 | −6 |
| 29 | 24.35 | 3.75 | cis-p-Menthan-3-one | 0.02 | 911 | 1 | 1148 | 1528 | 0 | −32 |
| 30 | 24.51 | 5.01 | Ethyl benzoate | 0.05 | 901 | 1 | 1150 | 1698 | 2 | 4 |
| 31 | 25.00 | 7.45 | 2,6-Dimethyl-3,7-octadiene-2,6-diol | 0.02 | 885 | 1 | 1185 | 1945a | −21 | −16 |
| 32 | 25.01 | 4.60 | 1-Methyl-4-(1-methylethyl) cyclohexanol | 0.92 | 933 | 2 | 1156 | 1650 | 8 | 10 |
| 33 | 25.20 | 4.56 | Isomenthol | 0.02 | 755 | 4 | 1186 | 1667 | −17 | −8 |
| 34 | 25.20 | 5.00 | α-Methylbenzyl acetate | 0.23 | 938 | 1 | 1166 | 1687a | 3 | 31 |
| 35 | 25.60 | 5.83 | Methyl salicylate | 0.25 | 952 | 1 | 1174 | 1795 | 5 | 24 |
| 36 | 25.70 | 4.40 | Dihydrocitronellol | 0.03 | 783 | 1 | 1182 | 1680 | −1 | −33 |
| 37 | 25.64 | 4.90 | α-Terpineol | 0.38 | 924 | 2 | 1175 | 1705 | 5 | 13 |
| 38 | 26.00 | 4.78 | γ-Terpineol | 0.14 | 897 | 1 | 1178 | 1684a | 11 | 27 |
| 39 | 26.93 | 4.89 | Citronellol | 5.48 | 953 | 1 | 1211 | 1761 | 1 | −11 |
| 40 | 27.01 | 5.25 | cis-Geraniol | 0.36 | 928 | 1 | 1213 | 1815 | 1 | −17 |
| 41 | 27.29 | 5.20 | Isogeraniol | 0.10 | 826 | 1 | 1237 | 1812 | −16 | −12 |
| 42 | 27.60 | 5.50 | β-Phenethyl acetate | 0.09 | 928 | 2 | 1228 | 1822 | 1 | 19 |
| 43 | 27.96 | 5.32 | trans-Geraniol | 4.93 | 953 | 1 | 1237 | 1854 | 1 | −22 |
| 44 | 28.07 | 3.62 | Linalyl acetate | 2.08 | 955 | 1 | 1241 | 1563 | −1 | −12 |
| 45 | 28.30 | 4.63 | α-Citral | 0.03 | 899 | 1 | 1249 | 1732a | −3 | 18 |
| 46 | 28.80 | 6.02 | Hydroxycitronellal | 1.03 | 935 | 1 | 1266 | 1929a | −7 | 1 |
| 47 | 29.20 | 4.93 | Anethole | 0.20 | 936 | 2 | 1264 | 1817a | 5 | 2 |
| 48 | 29.40 | 4.76 | Benzyl 2-methylpropanoate | 0.01 | 881 | 1 | 1273 | 1784a | 1 | 17 |
| 49 | 31.00 | 8.95 | Methyl anthranilate | 0.22 | 962 | 1 | 1326 | 2194 | −11 | 17 |
| 50 | 31.40 | 6.44 | Dihydro-5-pentyl-2(3H)-furanone | 0.22 | 942 | 1 | 1324 | 2024b | 2 | 23 |
| 51 | 31.56 | 3.66 | Citronellol acetate | 0.70 | 894 | 1 | 1335 | 1668 | −5 | −20 |
| 52 | 31.80 | 7.47 | Eugenol | 0.84 | 947 | 1 | 1335 | 2168 | 2 | −25 |
| 53 | 32.71 | 3.92 | Geranyl acetate | 1.70 | 956 | 1 | 1361 | 1764 | −1 | −25 |
| 54 | 33.00 | 4.15 | 1-(2,6,6-Trimethyl-1-cyclohexen-1-yl)-2-buten-1-one | 0.03 | 680 | 3 | 1395 | 1830a | −27 | −32 |
| 55 | 33.20 | 5.29 | 3-Methyl-2-(cis-2-penten-1-yl)-2-cyclopenten-1-one | 0.02 | 833 | 1 | 1368 | 1964 | 5 | 19 |
| 56 | 34.79 | 6.23 | 3-Phenyl-2-propenyl acetate | 2.90 | 951 | 1 | 1418 | 2176 | −2 | −39 |
| 57 | 34.80 | 4.00 | Nopyl acetate | 0.05 | 809 | 1 | 1413 | 1777a | 3 | 40 |
| 58 | 35.40 | 6.19 | 5-Hexyldihydro-2(3H)-furanone | 0.04 | 847 | 1 | 1428 | 2119 | 5 | 35 |
| 59 | 36.00 | 3.57 | β-Chamigrene | 0.04 | 725 | 26 | 1475 | 1737 | −26 | 1 |
| 60 | 36.28 | 4.67 | Pentyl benzoate | 0.07 | 828 | 1 | 1456 | 1987 | 1 | 1 |
| 61 | 36.80 | 3.53 | α-Selinene | 0.02 | 677 | 33 | 1491 | 1751 | −19 | −3 |
| 62 | 36.90 | 4.47 | β-Ionone | 0.77 | 922 | 1 | 1491 | 1971a | −17 | 0 |
| 63 | 37.00 | 4.07 | α-Cetone | 1.33 | 933 | 1 | 1484 | 1877 | −7 | 19 |
| 64 | 38.20 | 4.14 | 1-(2,6,6-Trimethylcyclohex-2-en-1-yl)-1-pentene-3-one | 0.45 | 902 | 3 | 1503 | 1933 | 8 | 11 |
| 65 | 38.70 | 4.70 | Isoamyl salicylate | 0.12 | 884 | 1 | 1515 | 2033a | 10 | 35 |
| 66 | 38.90 | 4.17 | β-Methyl ionone | 1.86 | 736 | 2 | 1557 | 1988b | −26 | −15 |
| 67 | 39.30 | 5.23 | α-(Trichloromethyl)benzyl acetate | 0.35 | 856 | 1 | 1538 | 2197a | 4 | −28 |
| 68 | 39.60 | 4.24 | 3,7,11-Trimethyl-1,6,10-dodecatrien-3-ol | 0.03 | 730 | 1 | 1551 | 2020 | 0 | −9 |
| 69 | 39.90 | 6.46 | Diethyl phthalate | 9.14 | 946 | 1 | 1564 | 2378b | −5 | −41 |
| 70 | 42.20 | 5.19 | Methyl (2-pentyl-3-oxocyclopentyl) acetate | 0.61 | 896 | 1 | 1649 | 2264a | −23 | −1 |
| 71 | 43.23 | 5.08 | cis-3-Hexenyl salicylate | 0.28 | 898 | 1 | 1644 | 2280a | 13 | 3 |
| 72 | 23.80 | 8.50 | 3,7-Dimethyl-1-octen-3-ol | 0.01 | 761 | 1 | 1122 | Not available | 12 | — |
| 73 | 32.76 | 3.76 | 4-tert-Butylcyclohexyl acetate | 0.83 | 932 | 1 | 1346 | Not available | 16 | — |
| 74 | 35.20 | 5.17 | 3,4-Dihydro-β-ionone | 0.25 | 729 | 7 | 1460 | Not available | −33 | — |
| 75 | 35.65 | 4.91 | 3-(4-Isopropylphenyl)-2-methylpropionaldehyde | 0.25 | 921 | 1 | 1424 | Not available | 16 | — |
| 76 | 38.19 | 4.73 | Lilial | 6.94 | 909 | 1 | 1500 | Not available | 10 | — |
| 77 | 39.00 | 7.50 | α-Methyl-3,4-methylenedioxy-hydrocinnamic aldehyde | 0.61 | 940 | 1 | 1542 | Not available | −8 | — |
| 78 | 39.00 | 9.06 | Methyl 2-formamidobenzoate | 0.03 | 878 | 1 | 1564 | Not available | −30 | — |
| 79 | 39.05 | 4.23 | 6-Methyl-β-ionone | 0.43 | 763 | 2 | 1566 | Not available | −31 | — |
| 80 | 45.00 | 3.99 | Caryophyllene acetate | 0.08 | 792 | 1 | 1720 | Not available | −10 | — |
| 81 | 47.45 | 4.72 | 9-Acetyl-2,6,6,8-tetramethyltricyclo (5.3.1.01,5) undec-8-ene | 2.38 | 952 | 1 | 1780 | Not available | 5 | — |
| 82 | 41.20 | 7.95 | 1-Acetonaphthone | 0.39 | 926 | 3 | 1592 | 2471b | 5 | — |
| 83 | 46.17 | 7.48 | Benzyl benzoate | 0.97 | 935 | 1 | 1732 | 2272 | 14 | — |
| 84 | 49.84 | 9.65 | Benzyl salicylate | 1.87 | 956 | 1 | 1836 | 2784a | 20 | — |
It should be noted for the CH/C approach that separate injections (runs) were performed resulting in several independent data sets prior to the combination into the CH/C analysis. Thus, the wraparound only occurred within the individual set having a time interval of 5n min (n is times of wraparounds = 1, 2, 3, …) between each H/C event. This is isolated from the adjacent H/C events performed in a different run.
In other words, wraparound occurs within the next adjacent modulation event(s) in conventional GC × GC using single injection whilst the wraparound in the CH/C analysis here solely affects the H/C event(s) within the next 5n min (not the adjacent event). Thus, the apparent 1tR value of a peak with wraparound in Fig. 2A should be subtracted with 5n min with the 2tR added with 5n before reporting the actual 1tR and 2tR data in Table 1. As a result, the possible 1I values for a peak with vs. without wraparound are different with the 5n min intervals. This facilitates identification of n leading to actual 1tR and 2tR for the wraparound peaks with the CH/C analysis according to the 1I information for each peak observed with MS.
Take the example for a peak with a tobserved (= 1tR + 2tR) of 28.90 min (see compound 10 in Table 1, 1tR = 18.62 min, 2tR = tobserved − 1tR = 10.28 min on the contour plot). This compound was identified by comparison with the MS library as benzenemethanol (the match score of 921) and the literature I on the 1D column of 1012. According to the 5n min concept, the possible H/C interval before this compound could be either 18.5–18.7 or 23.5–23.7. The former was assigned as the correct interval since this resulted in an experimental 1I of 1009 being closer to the database value of 1012. The actual 1tR and 2tR of this peak identified according to the weighted average approach were 18.70 and 10.20 min, respectively. In the case that the MS library provided incorrect compound identity (e.g. not matched with the 2I data), a lower rank identity with the next highest MS library match score and available I information was proposed. The calculation process to obtain the actual 1tR and 2tR of this peak was then repeated until the compound identity showed both 1I and 2I matched with the library data. It should be noted that compound wraparound can also be confirmed by performing a single H/C experiment solely focusing on a peak of interest.
In addition, several split peaks of the same compounds were observed; see most of the circled peaks in the contour plot (Fig. 2A). These split peaks (sub-peaks) resulted from a peak of a compound in the 1D separation sampled into different H/C windows. With the high 2D separation efficiency and the great difference between 2D separation temperature of any two adjacent H/C events, these sub-peaks showed significantly different 2tR (e.g. baseline separation) leading to the split peaks of the same compounds in the contour plot. 1tRvs.2tR data along the isovolatility curve of alkane C19 experimentally obtained using the peak parking approach were provided as an example to confirm such a peak splitting mechanism (Fig. 2B). With the focus on three sets of two adjacent sub-peaks undergoing no wraparound, 1st wraparound and 2nd wraparound, the sub-peaks of the alkane were separated better at the higher order of wraparound (more clearly split peaks at the lower temperature). Note that the presence of these split peaks is not problematic in data analysis since the individual sub peaks could be identified as the same compounds, and the overall peak areas were obtained by the summation of all the sub-peak areas. Also note that 1tR and 2tR profiles were obtained based on the weighted averages of all the sub-peak times and their corresponding peak areas. After the determination of actual 1tR and 2tR based on comparison with the MS and 1I library, the compound identity was further confirmed from 2I data. Calculation of 2I was performed by using isovolatility curves.
3.2 Multi-location peak parking and construction of isovolatility curves
Generation of n-alkane isovolatility curves in this study is based on multiple injections and variation of an alkane elution temperature (or time) on the 2nd column. This was adapted from the multi-location peak parking (MLPP) technique applying a stopped flow and multiple injections within a single analysis. This technique was previously reported for investigation of longitudinal diffusion at different axial positions along the columns in liquid chromatography.23 Application of this technique here provided the advantage of isovolatility curve construction without the use of a cryogenic trap. However, the approach has been adapted where a compound was injected under a continuous flow with the increasing oven temperature program. The program was stopped at different times with a subsequent temperature drop to 35 °C. This parked a peak of an alkane at different axial positions along the 1D column. To this end, n-alkane standards were injected under the temperature program starting from 35 °C which was increased with a rate of 20 °C min−1 and stopped at 100, 120, …, 220, 240 °C and 240 °C held for different times as the parking process for the alkanes (see Fig. 3 and Section 2.2.2 with details for the 16 sets of the parking approach provided in Table S-2†). There are 4 runs applied within each parking-eluting set. For example, the first set employed 1st and 3rd runs (for the peak parking) with the temperature program from 35 °C to 100 °C whilst the temperature program applied in 2nd and 4th runs (for the peak eluting) was the same as that applied in the perfume sample separation (see also Set 1 in Table S-2†). This corresponds to generation of one data point along each isovolatility curve for all the alkanes. The second parking-eluting set employed 1st and 3rd (parking) runs with the temperature program from 35 °C to 120 °C using the same temperature program applied in 2nd and 4th (eluting) runs as that applied in the perfume sample separation. Note that different alkanes were parked at different positions along the capillary (as a result of an alkane undergoing different temperature programs), and the smaller alkanes were not parked (eluting to the detector during the parking) when the elution was stopped at higher oven temperature (e.g. alkane C8 not parked inside the 1D column with the parking process reaching 240 °C). With the additional blank injection, the parked peaks were then eluted through either the restrictor to the FID or the 2D column to the MS under the same temperature program used for the sample separation; see the example for alkane C17 provided in Fig. 4A. Different data sets of 1tR and 2tR along the isovolatility curve of the alkane can be calculated from the alkane peaks at the FID and MS (Fig. 3). A regression model used for calculating the isovolatility curves for all the alkanes is shown in eqn (4) with the values of p1, p2, p3, p4 and p5 being −0.0039, −0.6181, −26.4775, 5.5559 and −3.9524, respectively. The curve fitting results are provided in Table S-3 (ESI†). All the alkane isovolatility curves are plotted in Fig. 4B, with each alkane showing abrupt decrease in 2tR at higher 1tR. This corresponded to the parking steps with the increasing final temperature from 100 °C to 240 °C. To validate the parking approach, the constructed isovolatility curves were applied to calculate 2I (2ICal) for 10 standard compounds with different functionalities. The 2ICal values were compared with I from the NIST 14 library on the polar columns (2ILit) showing differences (2ILit − 2ICal) ranging from −35 to 41. This is within the expected value of ±44 units indicating the reliability of the method. The related data are provided in Table S-4 (ESI†). These curves were further used to calculate 2I for the volatile compounds in the perfume sample.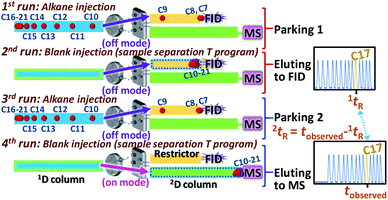 | ||
| Fig. 3 Diagram illustrating the peak parking and eluting approach for different alkanes (one parking-eluting event consisting of 4 runs). | ||
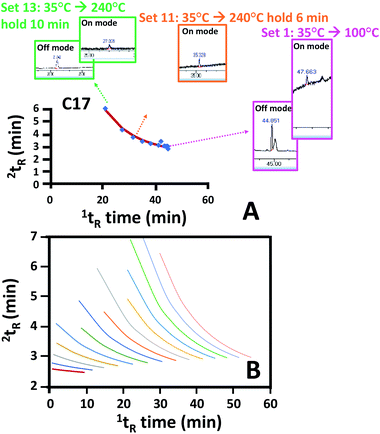 | ||
| Fig. 4 (A) The example of raw FID and MS chromatograms (with the DS off and on, respectively) to obtain the data points along the isovolatility curve of alkane C17, and (B) the generated isovolatility curves of all the n-alkanes obtained from 16 sets of 64 automated runs. Sets 1, 11 and 13 represent the 1st, 11th and 13th parking-eluting events provided in Table S-2.† | ||
3.3 Tentative identification of compounds in perfume samples
The CH/C analysis of the perfume sample revealed 128 separated compounds which were successfully identified solely using the MS library with a match score of >700. 1I and 2I were then applied to confirm the compounds obtained from the MS library. The results are summarized in Table 1 showing 84 identifiable compounds which can be divided into 3 groups according to the identification criteria. The first group is confirmed from the MS match scores of >700, and 1I and 2I differences of within ±37 and ±44 units, respectively, from literature values. The second group is confirmed by only the MS and 1I matches with “2I not available in the literature”. The third group is confirmed by only the MS and 1I matches with “their 2tR values above the investigated range of the isovolatility curves (where 2I could not be calculated)”. There were 71 compounds in the first group with five major compounds of benzyl ethanoate (12.41% peak area), linalool (9.84%), diethyl phthalate (9.14%), phenylethyl alcohol (6.56%) and citronellol (5.48%). In the second group, there were 10 compounds with three major compounds of Lilial (6.94%), 9-acetyl-2,6,6,8-tetramethyltricyclo(5.3.1.01,5)undec-8-ene (2.38%) and α-methyl-3,4-methylenedioxy-hydrocinnamic aldehyde (0.83%) whilst 1-acetonaphthone (0.39%), benzyl benzoate (0.97%) and benzyl salicylate (1.87%) were identified for the last group. For high confidence identification of compounds in the first group, most of the compounds with the highest MS match scores were correctly identified based on the confirmation from 1I and 2I. However, 18 compounds were incorrectly identified solely by the MS match such as α-selinene (compound 61 in Table 1) with the 33rd rank of the MS match. This compound was correctly identified according to both 1I and 2I match.3.4 Effects of the H/C window and analysis time on the number of identified peaks and retention index calculation errors in comprehensive H/C analysis
For a given set of 84 identified compounds (Table 1), the number of successfully separated peaks (which could be identified according to comparison with the 1I, 2I and/or MS database) using different H/C windows was approximated by using a previously established method.2 In this study, simulation of 1tR and 2tR is not required since they could be readily obtained from the comprehensive H/C analysis experiment above with a 0.2 min H/C window and a total analysis time of 25 h. The same compounds sampled with different H/C windows are expected to show almost the same 1tR + 2tR (= tobserved detected by MS in this study) since compound elution has not been modulated or stopped by a cryogenic trap. Note that the small variation of tobserved could be caused by different H/C positions along the peak as well as the shift in peak time caused by intensity change according to the peak splitting using different H/C windows. Thus, 1tR and 2tR can be recalculated into 1tR,recal and 2tR,recal from tobserved of each compound for a given H/C window by using eqn (2) and (3). Also note in this section that 1tR,recal and 2tR,recal of each compound were approximated from the sub-peak with the highest area (not the weighted average approach as that performed in the above section). The 1tR,recal and 2tR,recal of the 84 identified compounds were then converted into grid-scale coordinates2 (containing the data points of 1tR,recal,grid-scale and 2tR,recal,grid-scale of all the peaks obtained from their 1tR,recal and 2tR,recal divided by the 1D and 2D widths, respectively). These coordinates represent the separated peaks with a resolution of ≥1 (with ‘1’ defined as baseline separation) in 1D or 2D separation using average 1D and 2D widths (1wb,ave and 2wb,ave) of 0.2–2.5 min and 0.07–0.10 min, respectively, depending on the applied H/C window. The number of separated compounds could then be approximated by counting the number of grid-scale coordinates of the separated peaks. The related data of 1tR,recal,grid-scale and 2tR,recal,grid-scale and number of separated peaks in the CH/C analysis with different H/C windows and analysis times are provided in ESI Microsoft Excel.†Fig. 5A shows the number of identified peaks slightly decreased (by 4%) with the increasing H/C window from 0.2 to 2.5 min. However, the 1D peak capacity (1nc, approximated according to the approach proposed in (ref. 16) dropped more significantly (by 83%, see ( ) in Fig. 5B) with the increasing H/C window from 0.2 to 2.5 min. The corresponding 2D peak capacity (2nc) only decreased by 26% (see (
) in Fig. 5B) with the increasing H/C window from 0.2 to 2.5 min. The corresponding 2D peak capacity (2nc) only decreased by 26% (see ( ) in Fig. 5B). The better correlation between 2nc and the number of identified peaks suggests that the separation performance was mainly governed by 2D separation using the long 2D column (60 m) with a high phase ratio (0.25 μm stationary phase film thickness). In addition, the number of identified peaks slightly increased with the total comprehensive analysis time until reaching the upper limit (around 84 peaks, see Fig. 5C). This indicates that there were no more compounds in this perfume sample or the selectivity of the selected column set reached the maximum limit.2 As a result, application of a 2.5 min H/C window with a total analysis time of 2 h could result in a total peak capacity of 1134 with 81 identified peaks.
) in Fig. 5B). The better correlation between 2nc and the number of identified peaks suggests that the separation performance was mainly governed by 2D separation using the long 2D column (60 m) with a high phase ratio (0.25 μm stationary phase film thickness). In addition, the number of identified peaks slightly increased with the total comprehensive analysis time until reaching the upper limit (around 84 peaks, see Fig. 5C). This indicates that there were no more compounds in this perfume sample or the selectivity of the selected column set reached the maximum limit.2 As a result, application of a 2.5 min H/C window with a total analysis time of 2 h could result in a total peak capacity of 1134 with 81 identified peaks.
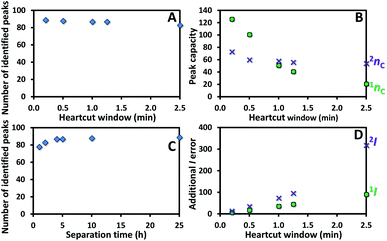 | ||
Fig. 5 Effect of the H/C window with the CH/C MDGC analysis on the (A) number of identified peaks and (B) 1nc ( ) and 2nc ( ) and 2nc ( ), with the corresponding plots of (C) total separation time vs. number of identified peaks and (D) additional errors in 1I ( ), with the corresponding plots of (C) total separation time vs. number of identified peaks and (D) additional errors in 1I ( ) and 2I ( ) and 2I ( ) approximated from the average 1tR and 2tR values (in Table 1). ) approximated from the average 1tR and 2tR values (in Table 1). | ||
However, additional errors (to the conventional limit of ±30) in 1I and 2I calculation were significantly increased with a wider H/C window (Fig. 5D), due to the 1tR and 2tR errors calculated using eqn (1) and (2) discussed above. To this end, application of the 2.5 min H/C window led to errors of ±91 and ±318 in 1I and 2I calculation, respectively, compared with the ±7 and ±14 obtained with a 0.2 min H/C window. This can be compared with the cryogenic H/C MDGC system which is expected to show the same 1I additional errors whilst there is a much smaller additional 2I error (e.g. caused by the compound that could not be effectively trapped/released). This is due to the fact that all the H/C compounds were trapped to start eluting on the 2D column at the same time.
4. Conclusion
A simple peak parking approach was established and successfully applied for construction of alkane isovolatility curves and further 2I calculation. This was demonstrated with one of the most difficult MDGC cases applying comprehensive H/C MDGC without a cryogenic device. This approach applied a narrow H/C window of 0.2 min which required 25 injections to complete the overall comprehensive analysis. According to the developed peak parking technique, 2I values were calculated for 71 compounds in the perfume sample where identities of 15 compounds can be corrected based on the 2I information. The compromise between analysis confidence (error) and analysis time was also proposed depending on the application goal. To this end, the developed approach is expected to be useful for high confidence untargeted analysis of complex samples and applicable with any CH/C MDGC operation in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support from the Chulalongkorn University's 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarship and the 90th Anniversary of Chulalongkorn University, Ratchadaphiseksomphot Fund, and the research facility from the center for advanced analytical technology, Dow chemical Thailand Ltd, Map Ta Phut Industrial Estate, Rayong. CK acknowledges the Second Century Fund (C2F), Chulalongkorn University.Notes and references
- P. J. Marriott, S.-T. Chin, B. Maikhunthod, H.-G. Schmarr and S. Bieri, TrAC, Trends Anal. Chem., 2012, 34, 1–21 CrossRef CAS.
- C. Kulsing, Y. Nolvachai, P. Rawson, D. J. Evans and P. J. Marriott, Anal. Chem., 2016, 88, 3529–3538 CrossRef CAS PubMed.
- Y. Nolvachai, C. Kulsing and P. J. Marriott, TrAC, Trends Anal. Chem., 2017, 96, 124–137 CrossRef CAS.
- C. Mallet, LC·GC Eur., 2017, 13, 28–33 Search PubMed.
- P. Q. Tranchida, D. Sciarrone, P. Dugo and L. Mondello, Anal. Chim. Acta, 2012, 716, 66–75 CrossRef CAS PubMed.
- D. Barden and L. M. Gregor, LC·GC Eur., 2017, 13, 14–20 Search PubMed.
- B. J. Peter, J. Anal. Bioanal. Tech., 2016, 7, 2 Search PubMed.
- V. Ponsin, T. E. Buscheck and D. Hunkeler, J. Chromatogr. A, 2017, 1492, 117–128 CrossRef CAS PubMed.
- M. Jiang, C. Kulsing and P. J. Marriott, Anal. Bioanal. Chem., 2018, 410, 3185–3196 CrossRef CAS PubMed.
- M. Adahchour, J. Beens, R. J. J. Vreuls and U. A. Th. Brinkman, TrAC, Trends Anal. Chem., 2006, 25, 540–553 CrossRef CAS.
- M. Camenzuli, LC·GC Eur., 2017, 30, 346–351 CAS.
- S. E. Prebihalo, K. L. Berrier, C. E. Freye, H. D. Bahaghighat, N. R. Moore, D. K. Pinkerton and R. E. Synovec, Anal. Chem., 2018, 90, 505–532 CrossRef CAS PubMed.
- Z. Liu and J. B. Phillips, J. Chromatogr. Sci., 1991, 29, 227–231 CAS.
- C. Kulsing, Y. Nolvachai and P. J. Marriott, TrAC, Trends Anal. Chem., 2020, 130, 115995 CrossRef CAS.
- L. F. de Alencastro, D. Grandjean and J. Tarradellas, Chimia, 2003, 57, 499–504 CrossRef CAS.
- N. Thongdorn, T. Nhujak, P. Janta, A. Rueangthaweep, N. Hinchiranan and C. Kulsing, Anal. Methods, 2020, 12, 5160–5167 RSC.
- C. Kulsing, Y. Nolvachai, Y. F. Wong, M. I. Glouzman and P. J. Marriott, J. Chromatogr. A, 2018, 1546, 97–105 CrossRef CAS PubMed.
- H. D. Waktola, C. Kulsing, Y. Nolvachai and P. J. Marriott, J. Chromatogr. A, 2018, 1549, 77–84 CrossRef CAS PubMed.
- D. R. Deans, J. Chromatogr. A, 1965, 18, 477–481 CrossRef CAS PubMed.
- P. Boeker, J. Leppert, B. Mysliwietz and P. S. Lammers, Anal. Chem., 2013, 85, 9021–9030 CrossRef CAS PubMed.
- B. Girard, J. Chromatogr. A, 1996, 721, 279–288 CrossRef CAS.
- M. Jiang, C. Kulsing, Y. Nolvachai and P. J. Marriott, Anal. Chem., 2015, 87, 5753–5761 CrossRef CAS PubMed.
- F. Gritti and G. Guiochon, J. Chromatogr. A, 2011, 1218, 896–906 CrossRef CAS PubMed.
- A. Amenduni, F. Massari, J. Palmisani, G. de Gennaro, M. Brattoli and M. Tutino, Environ. Eng. Manag. J., 2016, 15, 1963–1969 CrossRef CAS.
- A. Chisvert, L.-N. Marina, P. Miralles and A. Salvador, in Analysis of cosmetic products, 2nd edn, 2018, Elsevier, Boston. pp. 225–248 Search PubMed.
- B. Wang, H. Shen, A. Fang, D.-S. Huang, C. Jiang, J. Zhang and P. Chen, J. Chromatogr. A, 2016, 1451, 127–134 CrossRef CAS PubMed.
- B. Mitrevski, R. L. Webster, P. Rawson, D. J. Evans, H.-K. Choi and P. J. Marriott, J. Chromatogr. A, 2012, 1224, 89–96 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables S-1–S-4 and supplementary information Microsoft Excel. See DOI: 10.1039/d0ay01976c |
| This journal is © The Royal Society of Chemistry 2021 |