Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Capsid-like biodegradable poly-glycolic acid nanoparticles for a long-time release of nucleic acid molecules†
Clara
Guido‡
ab,
Mariangela
Testini‡
b,
Stefania
D’Amone
b,
Barbara
Cortese
 c,
Maria
Grano
d,
Giuseppe
Gigli
ab and
Ilaria E.
Palamà
c,
Maria
Grano
d,
Giuseppe
Gigli
ab and
Ilaria E.
Palamà
 *b
*b
aDep. of Mathematics and Physics, University of Salento, via Monteroni, Lecce, Italy. E-mail: Ilaria.palama@nanotec.cnr.it
bNanotechnology Institute, CNR-NANOTEC, via Monteroni, Lecce, Italy
cNanotechnology Institute, CNR-NANO TEC, c/o La Sapienza University, Piazzale A. Moro, Roma, Italy
dDep. of Emergency and Organ Transplantation (DETO), School of Medicine, University of Bari, Bari, Italy
First published on 13th November 2020
Abstract
Gene therapy can be described as the direct transfer of genetic material to cells or tissue for the treatment of inherited and acquired diseases. Viral vectors have long been proven to be the most efficient and stable vectors for transgene delivery into the cell, but they still have some safety issues. Non-viral nanosystems can overcome these safety problems and limitations of viral vectors. FDA-approved polymers are particularly attractive for gene delivery applications. Here, sustained release of plasmid DNA encoding the EGFP protein, used as a model, showed efficient delivery through new capsid-like biodegradable polyglycolic acid (PGA) nanoparticles (NPs). PGA NPs showed a mean size of 135 nm, with a polyhedron structure. Prior to loading into the PGA NPs, the pT7-EGFP plasmid was complexed with pH- and enzyme-responsive polycation polymers in order to guarantee high loading, stability, and controlled plasmid release over time. Our PGA NPs are hemocompatible, non-cytotoxic and have the ability to protect the gene cargo from DNase and serum action. Further, our PGA NPs exhibited a controlled and sustained transfection of cells that grow in suspension (human T lymphocytes, Jurkat) and adherent cell lines (human neuroblastoma cells, SH-SY5Y, and human cervix carcinoma cells, HeLa), with respect to transfection with commercial Lipofectamine 3000. In addition, our PGA NPs showed the ability to penetrate into 3D neurospheres, allowing transfection of inner cells. Our capsid-like NPs, thanks to their properties of biocompatibility, biodegradability, hemocompatibility, and sustained plasmid release, can be used as an efficient tool for transfection to overcome the problems of viral vectors.
1. Introduction
Gene therapy consists of genetic material delivery (as DNA, mRNA, miRNA, siRNA, etc.) in order to modulate critical cellular pathways that cause different diseases, such as cancer and inherited disorders.1 Genetic materials can be delivered using viral vectors, such as adenovirus, lentivirus, retrovirus, adeno-associated virus etc.,2 but despite some successes in clinical trials,3 viral vectors still have some safety issues, because they can evoke a strong cellular immune response. In addition, they have many disadvantages including limits in the size of the gene, vector antigenicity, inflammation and possible insertional mutagenesis.4,5In the last few years, researchers have focused on the development of non-viral vectors for gene delivery as an alternative to viral vectors.6–8
Non-viral vectors, such as inorganic nanoparticles (NPs), liposomes, and cationic polymeric NPs,9 may offer an alternative with low immunogenicity and the ability to accommodate large DNA plasmids, RNA or proteins, and may be produced at low cost on a large scale, but present low transfection efficacy,10,11 attributed to inefficient cellular penetration. So, the main obstacles are cellular uptake and penetration in three-dimensional (3D) complex structures of tissues and organs in vivo,12 preserving the integrity of the nucleic acid molecules. To overcome these issues, different nanoparticles with different physical–chemical characteristics12–15 such as size, morphology, and surface functionality have been explored, but efficient cellular uptake and penetration in a 3D system remain a key challenge.16
A non-viral vector that mimics the features of a virus can represent a key objective in gene delivery. Recently, some researchers have developed different nanoparticles that mimic some of the features of viruses, such as softness, roughness, needle-shape structure, etc. In particular, the ability to deform in a limited microenvironment, such as ECM, can represent a key factor for penetration in a 3D complex structure such as that of a tissue. It was observed that soft NPs were more efficient in uptake/penetration with respect to rigid NPs.17,18 Tao et al.19 demonstrated that viral-like inorganic mesoporous silica nanocapsules with soft domains and rough surfaces mimicking the envelopes of a virus can show increased uptake and penetration in cancers. Again, Wang et al.20 developed virus-like mesoporous silica NPs decorated with a needle-shape structure that showed improved cellular internalization. Moreover, Liu et al.21 synthesized dendrimer-like mesoporous silica NPs that mimic a virus, showing better cellular uptake.
Up till now, no polymeric nanoparticles with virus-like structural features, like a capsid shape and soft characteristics, have been developed. Usually, non-viral nanovectors synthesized starting with organic biomaterials (such as liposomes or polymeric NPs) are soft, with a spherical shape, but manipulation of their shape is an appealing approach22,23 due to the possibility to mimic the virus structure.
Recently, we have developed a versatile polymeric system for drug and gene delivery based on pH-responsive and enzyme-responsive core/shell NPs using the Food and Drug Administration (FDA) approved poly-caprolactone (PCL) polymer.24–27
Another attractive polymer for therapeutic molecule delivery applications is poly-glycolic acid (PGA), which has been approved by the FDA. PGA, after degradation, can be excreted in the metabolism of the tricarboxylic acid cycle. In addition, PGA presents a glass transition temperature (Tg) of 35–40 °C, and at the temperature of the human body, PGA is in a liquid-like state, becoming soft and flexible. Because of all the characteristics discussed above, PGA represents a promising candidate for the synthesis of new non-viral polymeric nanoparticles for gene delivery applications.
In this context, we have developed stimuli-responsive PGA NPs for delivery of DNA plasmids encoding EGFP used as a model. Our PGA NPs showed a virus capsid-like morphology and a mean size of 135 nm. Prior to DNA loading inside the PGA NPs, the plasmid molecules were complexed with poly-cation polymers, in order to ensure a high loading, good stability, and controlled plasmid release over time.25 For the DNA complexation, we have chosen stimuli-responsive biocompatible poly-cation polymers, in particular chitosan28 and poly-ethylenimine,29 as pH-sensitive polymers, and protamine30 as an enzyme-sensitive polymer. By merging the sensitivity of the poly-cations loaded into the core of the NPs with PGA surface degradation, it is possible to control, in an easy way, the release of payloads in order to achieve a sustained therapeutic effect over time.
Our PGA NPs are hemocompatible, non-cytotoxic and have the ability to protect the gene cargo from DNase and protease action. In addition, our PGA NPs displayed a good and sustained transfection of cells that grow in suspension (human T lymphocyte cells, Jurkat) and adherent cell lines (human neuroblastoma cells, SH-SY5Y, and human cervix carcinoma cells, HeLa) with respect to transfection with the commercial Lipofectamine 3000. Again, our PGA NPs revealed the ability to penetrate into 3D neurospheres, allowing transfection of the inner core.
The combination of the polyhedron structure of our PGA NPs similar to the capsid of viral vectors and the glass transition temperature of the PGA polymer that at human body temperature gives a liquid-like state producing soft and flexible nanoparticles allows us to increase the efficiency of uptake, thanks to the improved cell adhesion, penetration and distribution in complex 3D structures. To the best of our knowledge, no other studies have reported polymeric NPs with capsid-like structures for which it is possible to combine in one system the sensitivity of poly-cations used to complex the DNA with surface degradation of the outer layers of the NPs.
2. Materials and methods
2.1 Materials
All tissue culture media and chemical reagents were purchased from Sigma-Aldrich. All chemical reagents were used without additional purification. Cell lines were purchased from American Tissue Type Collection (ATTC). Lyso-tracker and SYBR-safe were purchased from Thermo Fisher. Coomassie brilliant blue (R-250) was obtained from BioRad. Plasmid (pEGFP-N3, 4.7 kb) encoding the enhanced green fluorescence protein (EGFP) was obtained from Addgene, USA. Whole blood for protein corona and hemolytic analysis was collected in tubes with heparin from adult C57BL/6j male mice. The animal procedures were approved by La Sapienza University (Rome) ethics committee and the Italian Ministry of Health in accordance with the European Community Council (2010/63/UE), and D. L. 26/2014 of Italian Ministry of Health rules.2.2 Plasmid DNA preparation
The pT7-EGFP plasmid (4726 bp) contains the human cytomegalovirus (CMV) immediate-early promoter, a ColE1 type origin of replication and the kanamycin resistance gene for bacterial selection. Plasmid DNA (pDNA) was obtained by growing E. coli DH5α strain cultures, transformed with pT7-eGFP overnight, in 2 L shake-flasks containing 250 mL of LB medium and antibiotics (30 μg mL−1 of kanamycin). Then, the plasmids were purified using a GenElute™ Plasmid Maxiprep EndoFree plasmid purification Kit (Sigma-Aldrich). The purified plasmid was diluted using tris-EDTA (TE) buffer solution and stored at −20 °C. The integrity of the plasmid was confirmed by agarose gel electrophoresis. The purity and concentration of the plasmid were determined by UV absorbance at 260/280 nm.2.3 Polymer–DNA complex formation
Before loading into PGA NPs, pDNA was complexed with poly-cationic polymers such as chitosan (CH, medium MW, 70% deacetylated), branched poly-ethylenimine (PEI, MW 25 kDa) and protamine sulphate salt from herring-grade III (PRM). CH was dissolved in 2% acetic acid, while PEI and PRM were dissolved in 0.1 M NaCl. Plasmid DNA-pT7-EGFP (100 ng μL−1) was mixed with the cationic polymers in acetate buffer (pH 7.2) at concentrations corresponding to different ratios of DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) complexing polymer (1
complexing polymer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, 1
5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 1
10, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 1
20, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and 1
50, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) and immediately vortexed. The complexes of pT7-EGFP–polymer were incubated for 1 hour at RT to allow formation. DNA–polymer complexes were analyzed on a 1.2% agarose gel (90 V, 1 h) in TAE 1× buffer. Bands were visualized with SYBR-safe staining under a UV transilluminator. Changes in zeta potential were determined using a Malvern Zetasizer Nano. DNA–polymer complexes were used for PGA NP synthesis.
100) and immediately vortexed. The complexes of pT7-EGFP–polymer were incubated for 1 hour at RT to allow formation. DNA–polymer complexes were analyzed on a 1.2% agarose gel (90 V, 1 h) in TAE 1× buffer. Bands were visualized with SYBR-safe staining under a UV transilluminator. Changes in zeta potential were determined using a Malvern Zetasizer Nano. DNA–polymer complexes were used for PGA NP synthesis.
2.4 Empty and DNA-loaded PGA NP synthesis
Empty poly-glycolic acid (PGA) NPs were prepared using nanoprecipitation/desolvation methods. Briefly, the PGA polymer (MW 20 kDa, final concentration 5 mg mL−1) was dissolved in THF by vigorous stirring for 3 days at RT and filtered. Then, 50 μL of this solution was added slowly into 500 μL of polysorbate 80 solution (0.5% w/v in milliQ water) at room temperature (RT) under vigorous stirring and incubated for 10 minutes. Subsequently 300 μL of 96% ethanol was added and incubated overnight (ON) at RT under slow stirring in order to allow the slow evaporation of THF and the solidification of NP structures. Next, the NP suspension was dialyzed against distilled water for 6 hours using cellulose membrane tubing (6–8 kDa MWCO). For the fluorescent PGA NPs, a TRITC solution in methanol was added to the starting organic PGA solution, while FITC dissolved in water was added to the aqueous polysorbate 80 solution and dialyzed against distilled water for 6 hours. In order to determine the NP concentration (w/v), the NP suspension was lyophilized before weighing. DNA-loaded PGA NPs were prepared using the same procedure employed for the preparation of empty PGA NPs, but adding CH–DNA complexes or PEI–DNA complexes or PRM–DNA complexes to the aqueous polysorbate 80 solution to a final concentration of DNA of 2 ng μL−1.2.5 PGA NP characterization
2.6 Agarose gel retardation assay
DNA–polymer complexes and DNA-loaded PGA NPs were analyzed on a 1.2% agarose gel (90 V, 1 h) in TAE 1× buffer. Bands were visualized with SYBR-safe staining under a UV transilluminator.2.7 SYBR-safe displacement assay
The DNA condensation degree was evaluated as a function of the DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratio by a displacement assay using SYBR-safe. In the displacement assay, the binding of an agent to DNA displaces the intercalated SYBR-safe and quenches the fluorescence emitted by the SYBR-safe/DNA complex. After measuring the background fluorescence intensity of a SYBR-Safe 1× solution in HEPES/NaCl buffer, DNA/complexes/NPs at 20 ng μL−1 were mixed with SYBR-safe 1× and stirred for 30 minutes. Each aliquot of DNA–polymer complex or DNA-loaded NP solution was excited at 509 nm, and the fluorescence intensity at 530 nm was read using a Cary Eclipse Fluorimeter. Sample fluorescence was determined after subtracting the baseline fluorescence of SYBR-safe in the absence of DNA.
polymer ratio by a displacement assay using SYBR-safe. In the displacement assay, the binding of an agent to DNA displaces the intercalated SYBR-safe and quenches the fluorescence emitted by the SYBR-safe/DNA complex. After measuring the background fluorescence intensity of a SYBR-Safe 1× solution in HEPES/NaCl buffer, DNA/complexes/NPs at 20 ng μL−1 were mixed with SYBR-safe 1× and stirred for 30 minutes. Each aliquot of DNA–polymer complex or DNA-loaded NP solution was excited at 509 nm, and the fluorescence intensity at 530 nm was read using a Cary Eclipse Fluorimeter. Sample fluorescence was determined after subtracting the baseline fluorescence of SYBR-safe in the absence of DNA.
2.8 DNase and serum protection assay
DNA–polymer complexes or DNA-loaded PGA NPs were incubated with DNase I 1U or 1% fetal bovine serum (FBS) overnight at 37 °C with constant stirring (600 rpm). The reaction was stopped with SDS 1% and samples were analyzed using 1.2% agarose gel (90 V, 1 h) in TAE 1× buffer. Bands were visualized with SYBR-safe staining under a UV transilluminator.2.9 DNA loading efficacy and in vitro release
DNA loading efficacy into the PGA NPs and DNA in vitro release were assessed as described in our previous work.25 In particular, for DNA in vitro release, the different PGA NP formulations were incubated in solutions of PBS 1× at pH 7.4 and 5.0 for different times at 37 °C with constant stirring (600 rpm) and the supernatants were analyzed after removing the NPs by centrifugation (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes). In order to assess DNA-release by degradation of enzyme sensitive PRM, DNA-loaded PGA NPs were incubated in the presence of protease (protease type XIV from Streptomyces griseus, 5 mg mL−1) at pH 7.4 and 37 °C with stirring.
000 rpm for 30 minutes). In order to assess DNA-release by degradation of enzyme sensitive PRM, DNA-loaded PGA NPs were incubated in the presence of protease (protease type XIV from Streptomyces griseus, 5 mg mL−1) at pH 7.4 and 37 °C with stirring.
2.10 Hemocompatibility
Hemolysis assays were performed as described in our previous work.26 Briefly, different DNA-loaded PGA NP formulations and DNA-Lipofectamine 3000 were incubated with 2% mouse blood for 24 hours at 37 °C with shaking (600 rpm). After centrifugation to remove the NPs, the supernatant absorption at a wavelength of 540 nm was evaluated using a UV-visible spectrophotometer (Varian Cary® 300 Scan; Varian Instruments, CA, USA) and the hemolytic rate was calculated.2.11 Protein corona analysis
The protein corona was formed by exposure of PGA NPs to 10% FBS and 2% mouse blood at 37 °C for 1 h and ON with shaking (600 rpm). Proteins from the soft corona were separated from the hard corona and NPs via subsequent centrifugation steps (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes). Proteins were collected and quantified using the Bradford method. The profiles of the hard corona were visualized by SDS-PAGE electrophoresis. A 5–12% polyacrylamide gel was employed to separate the proteins and Coomassie R-250 was used to stain the gels. In addition, zeta potential was measured by DLS analysis.
000 rpm for 30 minutes). Proteins were collected and quantified using the Bradford method. The profiles of the hard corona were visualized by SDS-PAGE electrophoresis. A 5–12% polyacrylamide gel was employed to separate the proteins and Coomassie R-250 was used to stain the gels. In addition, zeta potential was measured by DLS analysis.
2.12 In vitro cellular analysis
A human T cell line (Jurkat), human neuroblastoma cell line (SH-SY5Y) and human cervical carcinoma cell line (HeLa) were maintained in culture in RPMI 1640 (Jurkat) and DMEM (SH-SY5Y, HeLa) supplemented with 10% FBS, penicillin (50 U mL−1), streptomycin (0.05 mg mL−1), and glutamine (4 mM) in an incubator at 37 °C with 5% CO2.2.13 Cytotoxicity assay
Jurkat, SH-SY5Y and HeLa cells (105 cells per mL) were treated with DNA (100 ng) loaded in Lipofectamine 3000 and in CH–, PEI–, PRM–PGA NPs for 48 hours. Untreated (CTR) samples were used as the control groups. Cell viability was evaluated by the MTT assay (Sigma-Aldrich) according to the manufacturer's instructions.2.14 Analysis of uptake, intracellular localization and cellular entry modality of PGA NPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ungated cells using a Flow Cytometer (Accuri C6, BD, USA).
000 ungated cells using a Flow Cytometer (Accuri C6, BD, USA).
2.15 Transfection assessment in 2D and 3D
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ungated cells using a Flow Cytometer (Accuri C6, BD, USA).
000 ungated cells using a Flow Cytometer (Accuri C6, BD, USA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ungated cells.
000 ungated cells.
2.16 Statistical analysis
Three independent experiments were performed, and the results were expressed as mean ± standard deviation. Statistical analysis was performed using the t-student test or one-way ANOVA.3. Results and discussion
3.1 Synthesis and characterization of DNA-loaded PGA nanoparticles
The empty and DNA loaded-PGA NPs were prepared using a nanoprecipitation–desolvation method, as illustrated in Scheme 1.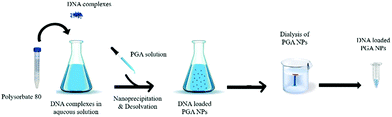 | ||
| Scheme 1 Schematic illustration of DNA loaded PGA NPs obtained using a nanoprecipitation–desolvation method. DNA refers to the pT7-EGFP plasmid. | ||
The nanoprecipitation–desolvation method allowed us to obtain in an easy way polymeric nanoparticles with a small size and specific structure. In addition, in this way, it is possible to avoid degradation of the payload, which in this case is the DNA.
A PGA solution in THF (10% in water) was added drop by drop to an aqueous solution containing DNA complexed with poly-cations under constant stirring followed by addition of ethanol and the slow evaporation of THF. Recently, Wong et al.31 reported the synthesis of polyhedron polymersomes using a PEG43-b-P(NIPAM23-co-PDMI19) polymer dissolved in 65–75% THF/water. In contrast, by changing the starting polymer, we obtained small NPs with a shape that resembles the structure of a viral capsid using a small quantity of THF and preserving the integrity of their payload.
SEM and AFM images (see the ESI,† Fig. S1) revealed a polyhedron structure of our PGA NPs mimicking the structure of viral vectors, similar to those used for gene delivery. From the 3D AFM images, it is possible to observe a polyhedron structure displayed by the PGA NPs, with an arithmetic average roughness (Ra) of 15.36 ± 0.05 nm and a root mean square roughness (Rq) of 18.34 ± 0.47 nm. Line profiles in Fig. S1E and F (ESI†) confirmed a rather smooth surface, and that such particles were fully covered by the polymer coating. In addition, the nanotopographical roughness of our NP surface decreases the repulsive interactions with the cell membrane, improving cell adhesion and uptake. This confirmed that our PGA NPs mimic the structure of viral vectors, similar to those used for gene delivery.32
In addition, thanks to the presence of Polysorbate 80, used as a surfactant in the synthesis, we obtained unaggregated NPs, as shown in the SEM images (ESI,† Fig. S1).
In agreement with different simulations33,34 and experimental data,19,31 we assumed that our PGA NPs are composed of soft and hard structures and this can be beneficial for the enhanced uptake and diffusion of the NPs in a 3D complex structure.
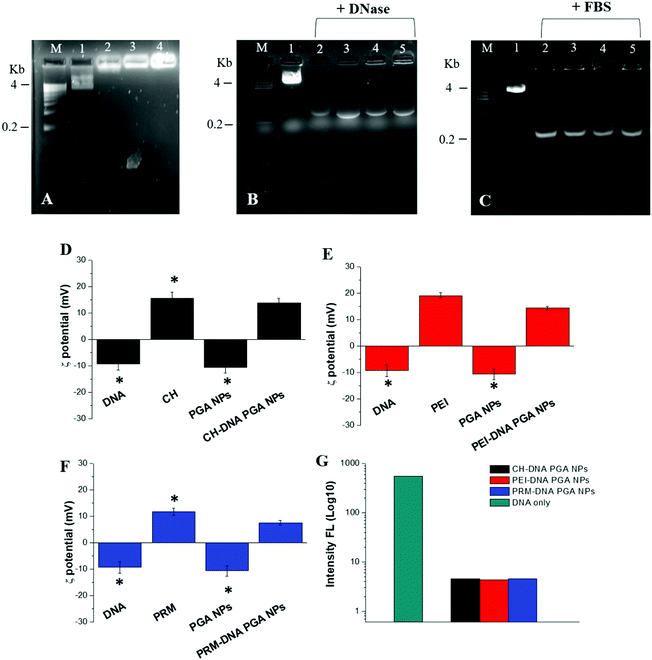
In Table 1, the mean size, polydispersion index (PDI) and zeta potential (ζ) of the empty and DNA-loaded PGA NP formulations are reported. We obtained empty PGA NPs with a homogeneous size of 135 nm, confirmed by the low PDI value, and a negative ζ-potential of −10 mV. For the DNA-loaded PGA NPs, the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 of plasmid DNA (pT7-EGFP) to the complexing polymer (CH, PEI or PRM) proved the best combination. This optimum mixture showed a size ranging from 132 nm to 138 nm with a PDI < 0.2 and a ζ-potential ranging from −10.69 mV for the empty PGA NPs to +14.4 mV for the DNA-loaded PGA NPs. The change in ζ-potential from negative (empty NPs) to positive (DNA-loaded NPs) is likely due to the cationic polymer charge used for DNA complexation.
100 of plasmid DNA (pT7-EGFP) to the complexing polymer (CH, PEI or PRM) proved the best combination. This optimum mixture showed a size ranging from 132 nm to 138 nm with a PDI < 0.2 and a ζ-potential ranging from −10.69 mV for the empty PGA NPs to +14.4 mV for the DNA-loaded PGA NPs. The change in ζ-potential from negative (empty NPs) to positive (DNA-loaded NPs) is likely due to the cationic polymer charge used for DNA complexation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio of plasmid DNA to complexing polymer. DNA refers to the pT7-EGFP plasmid
100 ratio of plasmid DNA to complexing polymer. DNA refers to the pT7-EGFP plasmid
| Sample | Size (nm) | PDI | Z (mV) |
|---|---|---|---|
| PGA NPs empty | 135 | 0.14 | −10.69 |
| PRM–DNA PGA NPs | 138 | 0.13 | +7.59 |
| CH–DNA PGA NPs | 137 | 0.16 | +13.98 |
| PEI–DNA PGA NPs | 132 | 0.18 | +14.4 |
In order to establish the best formulation for cellular transfection analysis, DNA–polycation complexes were prepared by varying the mass ratio of DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer. The complexation efficacy was assessed using an agarose retardation assay (ESI,† Fig. S2). At all ratios, no naked DNA was observed. This was confirmed by the change in ζ-potential and in SYBR-safe displacement (see the ESI,† Fig. S3). The zeta potential was near neutral for the 1
polymer. The complexation efficacy was assessed using an agarose retardation assay (ESI,† Fig. S2). At all ratios, no naked DNA was observed. This was confirmed by the change in ζ-potential and in SYBR-safe displacement (see the ESI,† Fig. S3). The zeta potential was near neutral for the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes and increased with the mass ratio (see Fig. S3A–C, ESI†). We chose to encapsulate complexes formed with a mass ratio of 1
1 complexes and increased with the mass ratio (see Fig. S3A–C, ESI†). We chose to encapsulate complexes formed with a mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 because they revealed the largest increase in zeta potential (about +15 mV), suitable for the formation of stable electrostatic interaction with the PGA polymer and a controlled long release of plasmid DNA.
100 because they revealed the largest increase in zeta potential (about +15 mV), suitable for the formation of stable electrostatic interaction with the PGA polymer and a controlled long release of plasmid DNA.
The effective encapsulation into PGA NPs was assessed by agarose retardation assay (Fig. 1A). No band corresponding to free pT7-EGFP DNA could be detected for any of the formulations, and the stable DNA-loaded PGA NPs remained in the loading well. This was confirmed by the analysis of the ζ-potential (Fig. 1D, E and F) and of SYBR-safe displacement (Fig. 1G). In addition, FT-IR analysis confirmed the effective loading of DNA–polymer complexes inside the PGA NPs (see Fig. S4, ESI†).
3.2 DNA protection analysis of PGA NPs
Another important factor regards the capability of our NPs to offer protection from DNase I and serum, which are able to degrade DNA. In order to assess this protection, qualitative DNase I and serum (FBS) protection assays were performed. As shown in Fig. 1B, PGA NPs prepared with the different polycations as complexing agents at a high DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer ratio (1
polymer ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) were capable of maintaining the integrity of the plasmid DNA in the presence of DNase I for 90 min (Fig. S5, ESI†); only after overnight (ON) incubation was it possible to observe a reduction in band intensity in the starting zone of the gel. The same experiments were performed in the presence of FBS (Fig. 1C), and only after incubation ON was it possible to observe a decrease of the bands in the starting zone of the gel. The presence of fluorescent bands in the DNase (Fig. 1B) and FBS (Fig. 1C) protection assays at 0.2 kb is due to the action of DNAse I and DNAse present in the FBS, which cause partial DNA fragmentation.
100) were capable of maintaining the integrity of the plasmid DNA in the presence of DNase I for 90 min (Fig. S5, ESI†); only after overnight (ON) incubation was it possible to observe a reduction in band intensity in the starting zone of the gel. The same experiments were performed in the presence of FBS (Fig. 1C), and only after incubation ON was it possible to observe a decrease of the bands in the starting zone of the gel. The presence of fluorescent bands in the DNase (Fig. 1B) and FBS (Fig. 1C) protection assays at 0.2 kb is due to the action of DNAse I and DNAse present in the FBS, which cause partial DNA fragmentation.
3.3. Encapsulation efficacy, release, stability and hemocompatibility
The efficiency of encapsulation of plasmid DNA inside the PGA NPs was about 70%. Plasmid DNA release from our NPs was analysed for a time window of 96 hours under two different conditions, pH 7.4 and 5.0, that mimic physiological circumstances, such as blood and cellular cytoplasm at pH 7.4 and intracellular endosomes and lysosomes at pH 5.0. We observed a low plasmid DNA release (up to 9% for all DNA-loaded PGA NP formulations) over time at pH 7.4 (Fig. S6A, ESI†). At this neutral pH value, the release of plasmid DNA happens by passive diffusion through the polymeric wall of the NPs. As expected, the incubation of PRM–DNA PGA NPs with a solution supplemented with protease indicated that protamine was efficiently degraded following the release of DNA. On the other hand, at pH 5.0, we observed a low release in the first hours of incubation, which increased with time until 36% release (Fig. S6B, ESI†) was observed at 96 hours. Under acid conditions, the polymeric wall of the NPs is degraded, increasing the NP porosity and the release of plasmid DNA.In addition, our NPs preserve their hydrodynamic size, zeta potential and release profiles when lyophilized, supporting their stability. This represents another important result, because usually the NP formulations administered in pre-clinical models (i.e. rats, mice, etc.) are freshly prepared for any set of experiments;35 this is not appropriate in a clinical setting and storage for a long-time can be necessary under sterile conditions, therefore lyophilization of the NP formulations represents a good form of preservation.36
The use of in vivo NP formulations depends on an additional crucial parameter that is their hemocompatibility. The hemocompatibility of our formulations (Fig. S7, ESI†) was significantly higher (3–6% hemolysis) when compared to Lipofectamine 3000 (about 18% hemolysis). Our NPs are thus hemocompatible with mouse blood and can be administrated in vivo.
3.4 Protein corona study
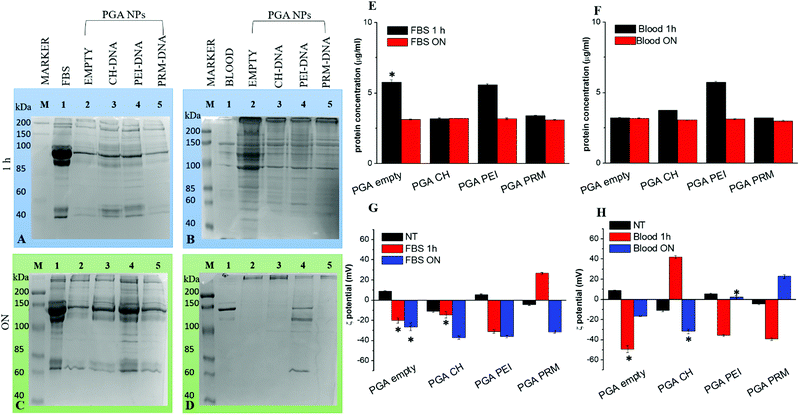
NPs for drug/gene delivery often show therapeutic efficacy in vitro but not in vivo.37 This is due to the binding of different proteins on the NP surface, which forms a unique biological environment. It is important to define the protein corona composition around NPs for the development of useful therapeutic nano-agents for drug and gene delivery applications.38In our study, we have focused our attention on studies of protein corona formation on PGA NPs, after incubation in FBS and mouse blood at 37 °C for 1 hour or ON with shaking. Proteins from the hard corona were separated from the soft corona and the NPs by subsequent centrifugation steps. The profiles of the hard corona were visualized by electrophoresis on a 12% polyacrylamide gel followed by staining with Coomassie R-250. The intensities of the bands were evaluated by densitometric analysis and protein concentration was determined using the Bradford assay. The protein patterns of PGA NPs after incubation with FBS or mouse blood are shown in Fig. 2A–D. In FBS and mouse blood, the most dominant protein bands are visible, while the protein patterns recovered from the different PGA NPs change with time. There are different proteins observed after 1 hour of incubation with respect to the ON case; these results suggest that the protein corona is a structure that is susceptible to changes in composition when exposed to biological media for different time intervals. In addition, as shown in Fig. 2E and F, we observed a high quantity of protein on the empty and PEI PGA NPs after 1 hour of incubation with FBS with respect to the ON incubation, and for PEI PGA NPs this was also evident upon incubation with blood. On the other hand, the quantity of protein adsorbed by CH or PRM PGA NPs after incubation with FBS was similar after 1 hour and overnight (and for empty PGA NPs with blood), as evident in the SDS-PAGE results (Fig. 2A and D), which showed dissimilar bands of proteins.
Probably, proteins with different concentrations and properties are first bound to the NP surface, and then they are gradually replaced by higher affinity proteins (Vroman effect).39 This result may have deep biological implications for the application of gene delivery both in vitro and in vivo because the protein corona can potentially alter the NP's surface properties, changing the fate of the NPs as well as their application outcome. It is important to note that the general adsorption pattern is independent of the polymer used for the DNA complexation and loading inside PGA NPs.
Zeta-potential analysis, after incubation in FBS and mouse plasma, of different formulations of NPs indicated that the adsorption of serum proteins induced a change in their zeta potential to a favorable negative zeta potential value (Fig. 2G and H). It is well known that a higher absolute value of zeta potential means a more stable state of a system.
3.5 In vitro cell viability, uptake and intracellular localization analysis
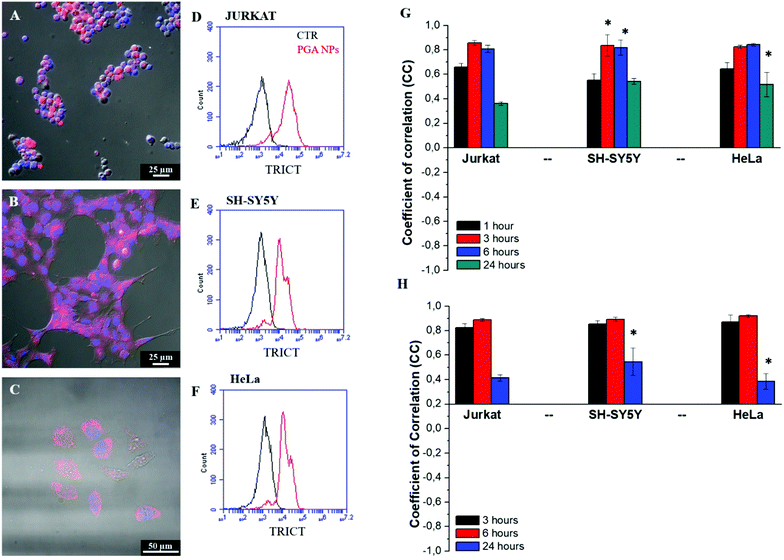
In vitro cell toxicity of the DNA (100 ng) loaded in Lipofectamine 3000 or different PGA NP formulations was tested by performing an MTT assay on Jurkat, SH-SYS5Y and HeLa cells after 48 hours of incubation. As shown in Fig. S8 (ESI†), the cell viability of all cell lines was significantly higher with different DNA-loaded PGA NPs (range 90–98%) than with DNA loaded Lipofectamine 3000 (range 70–86%). This different viability may be due to the fact that cationic liposomes, such as Lipofectamine 3000, have a stronger interaction with cell membranes, which may damage them and cause cell death.40Qualitative and quantitative analyses were performed in order to evaluate the PGA NP uptake, endocytosis and intracellular localization. As shown in the confocal images of Fig. 3A–C, after 6 hours of incubation with fluorescent TRITC–PGA NPs, it was possible to note a strong red fluorescence inside the cytoplasm associated with the uptake of our NPs. Quantitative uptake was evaluated by FACS analysis (Fig. 3D–F) after 6 hours of incubation with TRITC–PGA NPs and the efficacy of uptake was found to be about 87% for a human T cell line (Jurkat) and 78% for a human neuroblastoma cell line (SH-SY5Y).
An important objective is to understand how NPs are taken up and transported within the cells. There are different types of endocytosis used for the internalization of materials, such as micropinocytosis/phagocytosis,41 or clathrin-42 or caveolin-43 mediated endocytosis. Due to their size (ranging from 132 nm to 138 nm), our NPs were internalized inside the cells by a clathrin-mediated endocytosis mechanism. In particular, we performed immunofluorescence analysis from 1 hour to 24 hours of the co-localization of fluorescent PGA-NPs with clathrin or caveolin pits. As shown in Fig. 3G, the coefficient of correlation (CC) for PGA NPs and clathrin in all cell lines tested showed an increase until 6 hours from 0.65 (1 hour) to 0.84 (6 hours) and decreased after 24 hours to around 0.36 for Jurkat and 0.51 and 0.54 for HeLa and SH-SY5Y, respectively. These data indicate a high co-localization with clathrin pits in the first hours of uptake followed by redistribution at 24 hours (see the qualitative CLSM images in Fig. S9, S10 and S11, ESI†). In contrast, we observed a low CC between PGA NPs and caveolin (Fig. S12 (ESI†) and qualitative CLSM images in Fig. S13–S15, ESI†).
To identify the intracellular localization of fluorescent PGA NPs, a co-localization assay with Lysotracker was performed on Jurkat, SH-SY5Y and HeLa cells in order to label the lysosomes. As shown in Fig. 3H, for both cell lines, a high CC was observed after 3 and 6 hours of incubation and a low CC was observed after 24 hours between the PGA NPs and Lysotracker. These data show that PGA NPs were primarily carried to lysosomes and after 24 hours, the PGA NPs start disappearing from the lysosomal compartment (see qualitative CLSM images in Fig. S16–S18, ESI†). In the acidic environment of lysosomes, the polymeric shell of the PGA NPs is degraded, increasing their porosity, which allowed the release in the cytoplasm of DNA–polycation complexes, thanks to the proton-sponge effect due to the presence of the polycationic polymers.44
3.6 Transfection analysis in 2D and 3D cultures
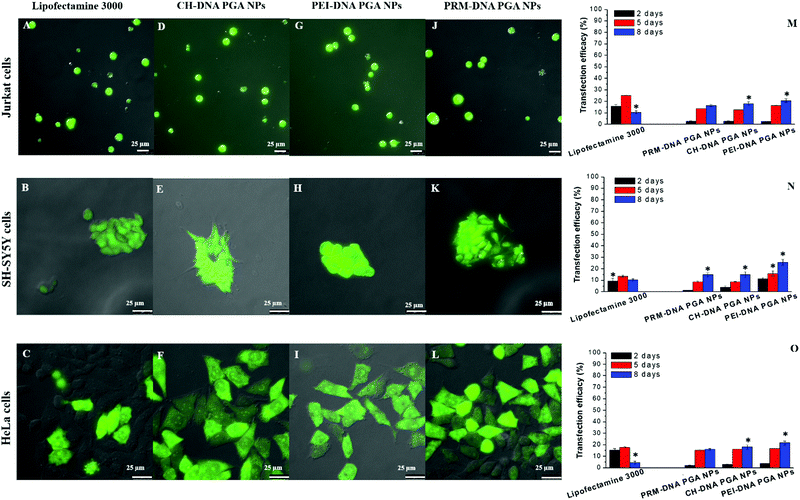
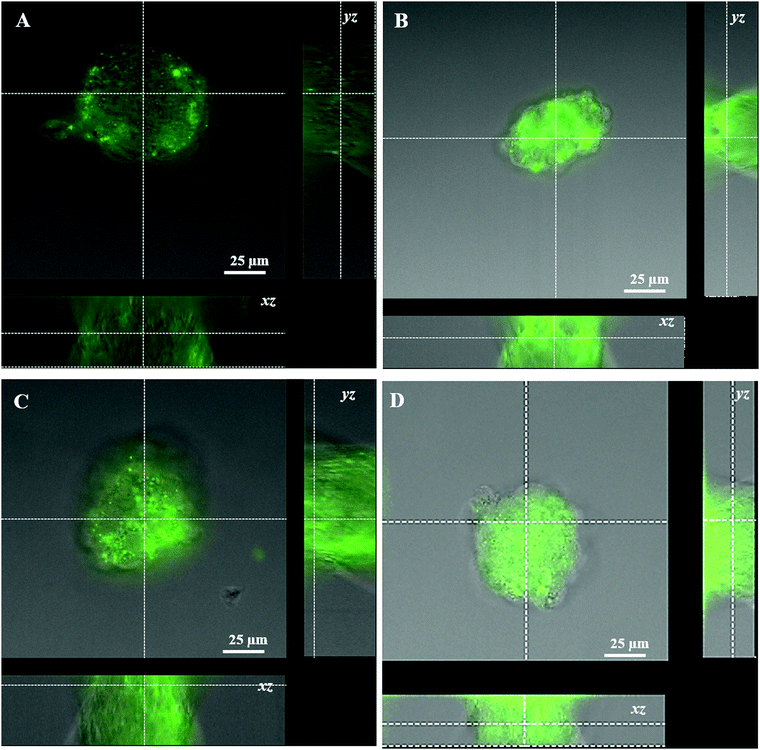
Transfection analysis was performed in vitro in 2D and 3D cultures. Qualitative analysis of the transfection efficiency was achieved in the former case via confocal laser scanning microscopy. The images showed strong green fluorescence associated with the expression of EGFP protein in all the cell lines transfected with DNA-loaded PGA NP formulations in comparison to commercial Lipofectamine 3000 (Fig. 4A–L). Quantitative transfection efficiency analysis of different DNA-PGA NP formulations was conducted using a flow cytometer. As shown in Fig. 4M–O, Lipofectamine 3000 showed a transfection yield of 15% in the first hours of treatment, which decreased with time in all cell lines tested. In contrast, PGA NPs loaded with the DNA plasmid showed a transfection efficiency that increased over time, reaching 25% after 8 days of treatment. These data confirm the prolonged release of the plasmid over time by the nanoparticles compared to the commercial agent. As reported in the release analysis of the DNA plasmid by PGA NPs, it is possible to observe a dissimilar profile of release at different pH values that suggests a structural change in the wall of the NPs allowing sustained DNA release in the cells. In contrast, the lipidic molecules that constitute Lipofectamine, when in contact with the cell membrane, tend to fuse with it, freeing all their contents in the cell and consequently leading to the total expression of the plasmid in the first hours of transfection, which is not observed with the NPs.We next examined the capability of our NPs to penetrate into a 3D structure, such as spheroids. Spheroids derived from human neuroblastoma (SH-SY5Y) cells were used as a model for the study of penetration, spreading and transfection of our DNA loaded PGA NPs in comparison with Lipofectamine 3000. Qualitative observations using confocal microscopy evidenced that our NPs showed a better capacity to penetrate the spheroid structure, and to induce transfection in comparison with lipofectamine 3000 after 5 days (Fig. S19, ESI†) and 8 days of treatment (Fig. 5). Flow cytometric analysis on disaggregated spheroids incubated with different formulations of NPs (Fig. S20, ESI†) confirmed the confocal results (Fig. 5). Different factors,13 such as size,14 superficial charge,14etc. of the NPs and Lipofectamine, influence the cellular uptake and penetration in a complex cellular structure. Of note, the size of NPs influences their uptake in 2D cellular systems and penetration in 3D cellular systems,12,15 therefore the small size of the DNA-loaded PGA NPs (about 135 nm) permits a better penetration in comparison with the lipo-complexes obtained with Lipofectamine 3000 with a mean size of 300 nm. In addition, the surface charges of different formulations strongly influenced their penetration. In fact, the positive charge permits an easy infiltration in the 3D structures, thanks to electrostatic interactions with the spheroid's negatively charged surfaces. In addition, thanks to the glass transition temperature (Tg) of 35–40 °C, PGA can assume a liquid-like form at physiological temperatures, therefore our NPs become soft and flexible, allowing an efficient uptake and penetration in complex structures.
Despite the similar positive charges of DNA-Lipofectamine 3000 and different formulations of DNA-loaded PGA NPs, we observed an efficiency of transfection of about 4.5% for Lipofectamine 3000, and of 6%, 10% and 12%, respectively, for the PRM–DNA, CH–DNA and PEI–DNA-loaded PGA NPs. This dissimilar efficiency of transfection in the 3D spheroids may be due to the fact that DNA lipo-complexes obtained with Lipofectamine 3000, when internalized into cells, release quickly their plasmid cargos (see 2D transfection), while PGA NPs allow a sustained release of plasmid DNA for a long time.
4. Conclusions
We synthetized capsid-like, biodegradable PGA nanoparticles using a nanoprecipitation and desolvation method, encapsulating plasmid DNA encoding EGFP proteins as a model nucleic acid. Prior to NP assembly, the pT7-EGFP plasmid was complexed with pH-(chitosan and poly-ethyleneimine) or enzymatic-(protamine) responsive polycationic polymers. By combining the sensitivity of the core polymer with the slow degradation of surface PGA, we were able to obtain a simple and easy way to control the release of the active agent and to improve its transfection efficiency. The mean size of our PGA NPs was 135 nm, with a polyhedron shape that mimics the capsid structure of viral vectors, typically used for gene transfection. In addition, our PGA NPs were hemocompatible, non-cytotoxic and have the ability to protect their gene cargo from DNase and serum action.Compared to commercial Lipofectamine 3000, our PGA NPs showed a sustained transfection efficacy that increased with time, in suspension and adherent cell lines, such as human T lymphoma (Jurkat), human neuroblastoma (SH-SY5Y) and human cervical carcinoma (HeLa). In addition, our PGA NPs showed the ability to penetrate into and allow the transfection of a 3D spheroid.
Our capsid-like PGA NPs, thanks to their properties, can be used as an efficient tool for transfection and to overcome problems associated with the use of viral vectors.
Conflicts of interest
All authors collected and discussed the material, providing experimental data. IEP conceived the research, supervised the experimental work, and undertook the writing, review and editing. CG, MT, SDA, BC, and IEP performed the synthesis and physico-chemical characterization of the PGA NPs used in the work. MT and IEP performed the in vitro experiments. MG and GG reviewed the manuscript. The authors declare no known competing financial interests.Acknowledgements
We thank Dr Patrizia Ratano and Prof. Silvia Di Angelantonio for mouse blood supply and Dr Francesco Calabi for helpful discussion. This study was supported by the FISR project “Tecnopolo di Nanotecnologia e Fotonica per la Medicina di Precisione” and “Progetto Tecnopolo per la Medicina di precisione, Delibera della Giunta Regionale no. 2117 del 21/11/2018”.References
- Y. K. Sung and S. W. Kim, Biomater. Res., 2019, 23, 8 CrossRef CAS.
- M. A. Kotterman, T. W. Chalberg and D. V. Schaffer, Annu. Rev. Biomed. Eng., 2015, 17, 63–89 CrossRef CAS.
- S. L. Ginn, A. K. Amaya, I. E. Alexander, M. Edelstein and M. R. Abedi, J. Gene Med., 2018, 20, e3015 CrossRef.
- L. S. Young, P. F. Searle, D. Onion and V. Mautner, J. Pathol., 2006, 208, 299–318 CrossRef CAS.
- J. Jeevanandam, K. Pal and M. K. Danquah, Biochimie, 2019, 157, 38–47 CrossRef CAS.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS.
- M. E. Davis, Curr. Opin. Biotechnol., 2002, 13, 128–131 CrossRef CAS.
- P. Wu, H. Chen, R. Jin, T. Weng, J. K. Ho, C. You, L. Zhang, X. Wang and C. Han, J. Transl. Med., 2018, 16, 29 CrossRef CAS.
- M. Ramamoorth and A. Narvekar, J. Clin. Diagn. Res., 2015, 9, 1–6 CrossRef.
- C. L. Hardee, L. M. Arevalo-Soliz, B. D. Hornstein and L. Zechiedrich, Genes, 2017, 8, 65, DOI:10.3390/genes8020065.
- R. Kircheis, L. Wightman, A. Schreiber, B. Robitza, V. Rössler, M. Kursa and E. Wagner, Gene Ther., 2001, 8, 28 CrossRef CAS.
- A. Tchoryk, V. Taresco, R. H. Argent, M. Ashford, P. R. Gellert, S. Stolnik, A. Grabowska and M. C. Garnett, Bioconjugate Chem., 2019, 30, 1371–1384 CrossRef CAS.
- N. Ma, C. Ma, C. Li, T. Wang, Y. Tang, H. Wang, X. Mou, Z. Chen and N. He, J. Nanosci. Nanotechnol., 2013, 13, 6485–6498 CrossRef CAS.
- R. Gupta and B. Rai, Sci. Rep., 2017, 7, 45292 CrossRef CAS.
- J. Zhao, H. Lu, S. Wong, M. Lu, P. Xiao and M. H. Stenzel, Polym. Chem., 2017, 8, 3317–3326 RSC.
- C. Kinnear, T. L. Moore, L. Rodriguez-Lorenzo, B. Rothen-Rutishauser and A. Petri-Fink, Chem. Rev., 2017, 117, 11476–11521 CrossRef CAS.
- S. Wang, H. Guo, Y. Li and X. Li, Nanoscale, 2019, 11, 4025–4034 RSC.
- R. K. Jain and T. Stylianopoulos, Nat. Rev. Clin. Oncol., 2010, 7, 653–664 CrossRef CAS.
- J. Tao, K. Chen, X. Su, L. Ren, J. Zhang, L. Bao, H. Dong, G. Lu, Z. Teng and L. Wang, Biomater. Sci., 2020, 8, 2227–2233 RSC.
- W. Wang, P. Wang, X. Tang, A. A. Elzatahry, S. Wang, D. Al-Dahyan, M. Zhao, C. Yao, C.-T. Hung, X. Zhu, T. Zhao, X. Li, F. Zhang and D. Zhao, ACS Cent. Sci., 2017, 3, 839–846 CrossRef CAS.
- Z. Liu, J. Ru, S. Sun, Z. Teng, H. Dong, P. Song, Y. Yang and H. Guo, J. Mater. Chem. B, 2019, 7, 3446–3454 RSC.
- J. A. Champion, Y. K. Katare and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11901–11904 CrossRef CAS.
- D. Klinger, C. X. Wang, L. A. Connal, D. J. Audus, S. G. Jang, S. Kraemer, K. L. Killops, G. H. Fredrickson, E. J. Kramer and C. J. Hawker, Angew. Chem., Int. Ed., 2014, 53, 7018–7022 CrossRef CAS.
- B. Cortese, S. D'Amone, G. Gigli and I. E. Palama, MedChemComm, 2015, 6, 212–221 RSC.
- I. E. Palamà, B. Cortese, S. D'Amone and G. Gigli, Biomater. Sci., 2015, 3, 144–151 RSC.
- B. Cortese, S. D'Amone, M. Testini, P. Ratano and I. E. Palama, Cancers, 2019, 11, 18 Search PubMed.
- B. Cortese, S. D'Amone and I. E. Palama, Pharmaceutics, 2018, 10, 18 CrossRef.
- Y. Cao, Y. F. Tan, Y. S. Wong, M. W. J. Liew and S. Venkatraman, Mar. Drugs, 2019, 17, 381 CrossRef CAS.
- A. Zakeri, M. A. J. Kouhbanani, N. Beheshtkhoo, V. Beigi, S. M. Mousavi, S. A. R. Hashemi, A. Karimi Zade, A. M. Amani, A. Savardashtaki, E. Mirzaei, S. Jahandideh and A. Movahedpour, Nano Rev. Exp., 2018, 9, 1488497 CrossRef.
- Y. Tsuchiya, T. Ishii, Y. Okahata and T. Sato, J. Bioact. Compat. Polym., 2006, 21, 519–537 CrossRef CAS.
- C. K. Wong, A. D. Martin, M. Floetenmeyer, R. G. Parton, M. H. Stenzel and P. Thordarson, Chem. Sci., 2019, 10, 2725–2731 RSC.
- C. S. Lee, E. S. Bishop, R. Zhang, X. Yu, E. M. Farina, S. Yan, C. Zhao, Z. Zeng, Y. Shu, X. Wu, J. Lei, Y. Li, W. Zhang, C. Yang, K. Wu, Y. Wu, S. Ho, A. Athiviraham, M. J. Lee, J. M. Wolf, R. R. Reid and T.-C. He, Genes Dis., 2017, 4, 43–63 CrossRef.
- G. Vernizzi, R. Sknepnek and M. Olvera de la Cruz, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4292–4296 CrossRef CAS.
- R. Sknepnek, G. Vernizzi and M. Olvera de la Cruz, Soft Matter, 2012, 8, 636–644 RSC.
- N. Dan and D. Danino, Adv. Colloid Interface Sci., 2014, 205, 230–239 CrossRef CAS.
- R. L. Ball, P. Bajaj and K. A. Whitehead, Int. J. Nanomed., 2017, 12, 305–315 CrossRef CAS.
- V. Kumar, N. Sharma and S. S. Maitra, Int. Nano Lett., 2017, 7, 243–256 CrossRef CAS.
- V. Gorshkov, J. A. Bubis, E. M. Solovyeva, M. V. Gorshkov and F. Kjeldsen, Environ. Sci.: Nano, 2019, 6, 1089–1098 RSC.
- S. Angioletti-Uberti, M. Ballauff and J. Dzubiella, Mol. Phys., 2018, 116, 3154–3163 CrossRef CAS.
- K. Romøren, B. J. Thu, N. C. Bols and Ø. Evensen, Biochim. Biophys. Acta, Biomembr., 2004, 1663, 127–134 CrossRef.
- R. Misra and S. K. Sahoo, Eur. J. Pharm. Sci., 2010, 39, 152–163 CrossRef CAS.
- O. Harush-Frenkel, N. Debotton, S. Benita and Y. Altschuler, Biochem. Biophys. Res. Commun., 2007, 353, 26–32 CrossRef CAS.
- J. Lee, M. Twomey, C. Machado, G. Gomez, M. Doshi, A. J. Gesquiere and J. H. Moon, Macromol. Biosci., 2013, 13, 913–920 CrossRef CAS.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ma00353k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |