A double helical 4H assembly pattern with secondary hierarchical complexity in an Ag70 nanocluster crystal†
Tao
Chen
a,
Sha
Yang
b,
Qinzhen
Li
a,
Yongbo
Song
c,
Guang
Li
 *a,
Jinsong
Chai
*c and
Manzhou
Zhu
*a,
Jinsong
Chai
*c and
Manzhou
Zhu
 *bc
*bc
aSchool of Physics and Materials Science, Institute of Physical Science and Information Technology, Anhui Key Laboratory of Information Materials and Devices, Anhui University, Hefei, Anhui 230601, People's Republic of China. E-mail: liguang1971@ahu.edu.cn
bInstitutes of Physical Science and Information Technology, Key Laboratory of Structure and Functional Regulation of Hybrid Materials of Ministry of Education, Anhui University, Hefei, Anhui 230601, China. E-mail: zmz@ahu.edu.cn
cDepartment of Chemistry and Centre for Atomic Engineering of Advanced Materials, Anhui Province Key Laboratory of Chemistry for Inorganic/Organic Hybrid Functionalized Materials, Anhui University, Hefei, Anhui 230601, China. E-mail: chaijs@ahu.edu.cn
First published on 20th August 2021
Abstract
The hierarchical assemblies of well-defined structural nanoclusters can help to better understand those of biologically important molecules such as DNA and proteins. Herein, we disclose the synthesis and characterization of a new silver nanocluster, that is Ag70(SR)42(PPh3)5 (Ag70-TPP). Directed by the ligands, Ag70-TPP nanoclusters undergo self-hierarchical assembly into a highly space-efficient complex secondary structure of a double helical 4H (DH4H) close packing pattern. The chirality of Ag70-TPP, and the van der Waals forces interactions between the ligands are believed to drive its DH4H arrangement, and the observed interlocking of the phosphine ligands of adjacent Ag70-TPP nanoclusters also contributed. Overall, this work has yielded important and unprecedented insights into the internal structure and crystallographic arrangement of nanoclusters.
New conceptsNanoclusters with hierarchical assembly characteristics have potential applications in many scientific fields. Most known nanoclusters show simple packing patterns, which largely limits the research progress on the internal assembly of crystal structures. In this work, a new Ag70(TBBT)42(TPP)5 nanocluster was structurally determined by single crystal X-ray crystallography (SCXC), and a double helical 4H (DH4H) packing mode with complex secondary structure assembly is discovered in its crystal structure. By changing the phosphine ligands on the nanoclusters, the complex DH4H packing mode can be transformed into a simple 2H assembly. Multiple van der Waals forces (C–H⋯π, π⋯π, and H⋯H) between Ag70 molecules are considered to be the key factor in maintaining the DH4H packing mode. We believe that our study makes a significant contribution to the understanding of the origin of the arrangement of functional building blocks into ordered hierarchy architectures. |
Large and important biomolecules such as DNA, RNA, and proteins are characterized by primary, secondary, and/or more complex structures,1–3 achieved by self-assembly behavior, self-folding, and/or intermolecular interactions.4 Of these different assembly modes, hierarchical assembly offers the richest functionality, since secondary structures can result in functional groups that would otherwise be far apart being placed in close proximity, as is the case for enzymes. In this context, nanomaterials with a biomimetic hierarchical structure can be endowed with biomolecule-like properties. In addition, benefitted by the inherent properties of nanomaterials, this strategy will make it possible to construct biomimetic nanomaterials with more excellent performance than biomolecules.5,6 Such self-assembly can be driven by the entropic maximization of packing density, electrostatic attraction, and cDNA binding,7–9 and therefore surface modifications are usually made to discrete NPs to guide their assembly, based on these drivers.10 For example, gold-based NPs modified with amino acids or peptides can assemble into highly complex and well-defined nanoscale superstructures exhibiting exceptionally strong chiroptical activity.11,12 These achievements have advanced the field, but progress remains frustrated by the more fundamental problem that nanoparticles exist as polydispersions (no two are the same), and their surface layers and core structure are unclear.
Ultrasmall nanoparticles (nanoclusters, < 2 nm in size) with well-defined crystal structures and unique quantum confinement effects have attracted increasing attention in the fundamental and applied sciences.13–15 Entire nanocluster crystal structures can be solved using single-crystal X-ray diffraction, which greatly informs about the forces and factors that determine assembly behavior at the atomic level.16,17 For example, most nanoclusters adopt either a hexagonal closest packed (HCP) or 2H arrangement with a packing sequence of “ABAB…”.18 However, the face-centered cubic (FCC) arrangement is also known, for example in the crystal of Au30(SR)18 reported by the Jin group, and 4H and 6H left-handed helical (6HLH) arrangements have been discovered.19–21 In addition, Au25(p-MBA)18− can be used as a building block to organize into well-defined nanoribbons through aurophilic interactions, which significantly enhance the luminescence intensity.22 However, current research has been mostly limited to first layered hierarchical assembly (helical single strand) with complex structures, and hierarchical assembly modes in second grade (assembly between helical strand, e.g. double helix structures in DNA or rhodopsin) are rarely reported in the context of nanoclusters. One exception to this is Zeng's study of atomically precise [Au246(p-MBT)80], which established that nanoclusters can have hierarchical structural complexities at the same level as biomolecules.23 X-ray diffraction analysis found the thiol ligands on the surface of this nanocluster to self-organize into rotational and parallel patterns via a C–H⋯π interaction. However, this phenomenon was confined to the internal structure of this nanocluster, and the secondary hierarchical assembly of gold or silver nanoclusters in their crystallographic arrangements has rarely been reported.
Herein, we disclose a chiral Ag nanocluster which spontaneously undergoes hierarchical assembly to give a complex secondary structure. This Ag cluster, the formula of which was determined to be Ag70(TBBT)42(TPP)5 (denoted as Ag70-TPP, TBBT = 4-tert-butylbenzenethiol, and TPP = triphenylphosphine), was synthesized using mixed ligands via the “NaSbF6-mediated two-phase ligand exchange method”.24 Based on the irregularity of the surface structure of Ag70-TPP, and its chirality, we speculated that the crystallographic arrangement is complex. Further analysis revealed a unique and unprecedented double helical 4H (DH4H) assembly pattern in the unit cells of Ag70-TPP, indicating that the cluster assembly forms a complex secondary structure. This unexpected DH4H arrangement of Ag70-TPP NCs in the crystal lattice was found to result from the chirality of the Ag70-TPP clusters and weak interactions between their ligands. Besides, the crystal structure of Ag70-TPTP (TPTP = tri(p-tolyl)phosphine) also pointed out that the ligand is the main influencing factor.
Ag70(TBBT)42(TPP)5 and Ag70(TBBT)42(TPTP)5 NCs were prepared in two steps: (i) the synthesis of water-soluble Agm(SG)n (H-SG = L-glutathione) clusters; (ii) two phase ligand exchange; in which a CH2Cl2 solvent containing excess TBBT was mixed with Agm(SG)n. Finally, TPP (or TPTP) was added to the above mixed solvent. After overnight reaction, the aqueous solution was removed, and the sample dissolved in CH2Cl2 was washed with methanol three times. Black crystals of the nanoclusters were obtained in CH2Cl2/CH3OH at room temperature after 5–7 days.
As shown in Fig. S1 (ESI†), the UV-vis spectrum of Ag70-TPP exhibits two distinct absorption peaks at ∼390 and ∼510 nm. According to previous reports, the peak at ∼390 nm (high-energy absorption peak) is mainly attributed to the mixing of metal-to-metal and metal-to-ligand charge-transfer processes (MMCT and MLCT), and the ∼510 nm (low-energy absorption peak) may come from charge transfer within the metal core.25,26 The Ag70-TPP crystals were dissolved in a CH2Cl2/CH3OH mixed solvent for electrospray ionization mass spectrometry (ESI-MS) tests. Two sets of +2 peaks were found in Fig. S2 (ESI†); the main peak at m/z = 7922.2 Da corresponds to the full molecular formula of [Ag70(TBBT)42(TPP)5 + K+ + H+]2+ (Cal. 7922.0 Da), and a fragment peak labelled by an asterisk corresponding to the loss of one –PPh3 ligand.27,28 We undertook an X-ray photoelectron spectroscopy (XPS) study of Ag70-TPP, and the results are shown in Fig. S3a (ESI†). In addition, the binding energies of the Ag 3d peaks in Ag70-TPP and Ag(0), Ag(I) species were also compared. The binding energy of Ag 3d5/2 in Ag70-TPP is 368.6 eV (Fig. S3b and c), which is higher than that of Ag(0) (367.9 eV), and closer to the binding energy of a Ag(I)-complex (368.7 eV), indicating that the valence of Ag70-TPP is between 0 and +1. 1H, 31P, and 2H-NMR spectroscopy were also used to probe the chemical composition and the ligand environments of Ag70-TPP (Fig. S4–S6, ESI†), and a detailed description is shown in the ESI.†
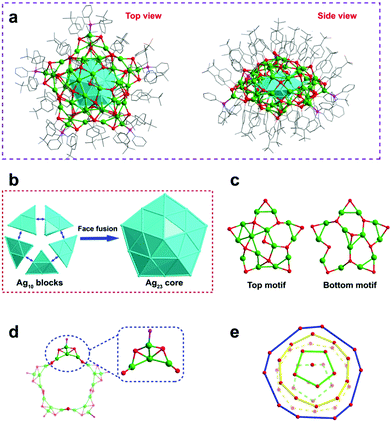
Single crystal X-ray crystallography (SCXC) analysis showed that Ag70-TPP crystallized in an orthorhombic Pbca space group and is a pseudo-five-fold symmetric pentagram-like nanocluster (Fig. 1a). The anatomical diagram of the total structure (Fig. 1b) shows that Ag70-TPP has a five-fold symmetric Ag23 kernel with 10 (111) faces, which can be described as five conjoined tetrahedral domains of FCC Ag10 (i.e., 4 (111) faces), indicating that the Ag23 kernel in Ag70-TPP is a complete decahedral. This kind of decahedral silver core can also be found in large-sized clusters with more than 100 atoms.29,30 Atomically precise nanoclusters with a decahedral core are summarized in Table S1 (ESI†). Previous reports indicate that nanoclusters with a general size of 20 to 100 metal atoms tend to have icosahedral, FCC, body-centered cubic (BCC) or HCP cores; few nanoclusters of this size have decahedral cores, although some contain incomplete decahedral structures, and it is expected that as the size (number of metal atoms) increases, the partial decahedral structure may evolve to a complete decahedral structure, as is found in Ag70-TPP. Furthermore, the Ag–Ag bond lengths of the Ag23 kernel range from 2.740 to 3.194 Å (2.904 Å on average).
The top and bottom of the Ag23 kernel are capped by two large pentagram-like shells (Ag14S11, Ag13S11) (Fig. 1c). As shown in Fig. S7 (ESI†), one Ag atom disordered in two positions, occupies 0.85 and 0.15, respectively. Additionally, a Ag20S20P5 circular motif (consists of five Ag4S4P units) terminated by phosphine was found to encircle the decahedron Ag23 core in the outermost shell (Fig. 1d). All the phosphine ligands form AgS3P tetrahedrons (Fig. S8, ESI†) with Ag–P bonds ranging from 2.416 to 2.447 Å (2.429 Å on average, Fig. S9, ESI†). The surface of the flat spherical Ag70-TPP cluster is covered by 42 peripheral thiolate ligands, which can be divided into three categories (Fig. S10, ESI†). The Ag–S bond lengths in Ag70-TPP range from 2.403 Å to 2.812 Å (average: 2.588 Å). On the other hand, these ligands exhibit a layer-by-layer assembly pattern arranged from pole to equator to opposite pole of the Ag70-TPP, containing 1, 5, 10, 10, 5, and 1 thiolate ligands for each layer (Fig. 1e).
The overall structure of Ag70-TPP exhibits C1 symmetry, indicating its chirality. The Ag23 core of Ag70 is achiral, which shows a D5h symmetry. Two chiral Ag14S11 and Ag13S11 networks were found to cap on the two poles of the decahedral Ag23 kernel. Finally, a twisted pentagonal Ag20S20P5 motif coats on the equator of the Ag23 kernel, forming the total chiral framework of the Ag70 clusters (Fig. S11, ESI†).31
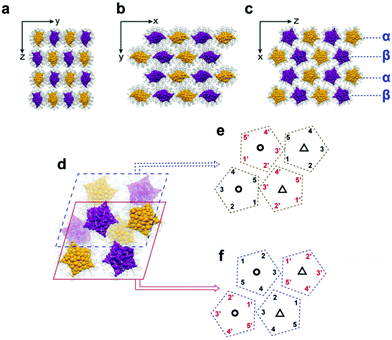
The Ag70-TPP unit cell contains four pairs of enantiomers (Fig. S12, ESI†). The arrangements of the Ag70-TPP superlattices are shown in Fig. 2(a–c). As shown in Fig. 2a, enantiomers are arranged along the y axis and z axis, organized into a rectangle superlattice in the (100) plane. In the (001) and (010) planes (Fig. 2b and c), Ag70-TPP molecules with the same chiral configuration are neatly assembled according the polyline mode (R-configuration Ag70-TPP NCs are shown in purple, and the S-configuration NCs are shown in golden). The molecules in the β layer can be regarded as the directional arrangement of the α layer through a flip operation, making the crystal a layer-by-layer manner (Fig. 2c). As shown in Fig. 2d, eight Ag70-TPP molecules in one unit cell can be divided into upper and lower layers. Interestingly, conventional NCs are typically packed into superlattices with simple translational or rotational symmetry,32 while the eight Ag70-TPP molecules in one unit cell are arranged differently (Fig. 2e and f) and occupy a more complex orthorhombic lattice via monomer rotational, flip, mirror symmetry, and translational operation (Fig. S13, ESI†), such that the eight constituent nanoclusters within an individual unit are symmetrically different and closely packed. This complex arrangement increases the packing efficiency, and the molecules are arranged as closely as possible in the crystal lattice.
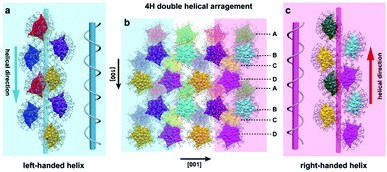
Fig. 3 shows the combined observations on the upper and lower layers of the (010) plane. The eight Ag70-TPP structures within the same unit cell are depicted in eight colours. In the crystal lattice, a complete helix contains four Ag70-TPP molecules, which arrange around the helical axis along the [100] and [![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 00] directions, respectively (Fig. 3a and c). Along the [100] direction, the stacking sequence is “A B C D”, which is characteristic of 4H (four layered hexagonal) close packing (Fig. 3b).33 However, in contrast to traditional 4H the four pairs of Ag70-TPP are arranged into two helical arrangements (Fig. S14, ESI†), wherein the blue shadow is left-handed and the other in pink shadow is a right-handed helix alternately arranged in adjacent locations (Fig. S14, ESI†), termed as a double helical 4H (DH4H) arrangement, and the average pitch is 26.1 Å. This type of DH4H assembly mode is reminiscent of the helix structure characteristics of biological DNA, but different. The DNA helix biopolymer of nucleic acids is held together by nucleotides which base pair together, with a unique axis, while Ag70-TPP possesses two parallel helix axes with opposite directions, and connected by weak interactions. (Fig. S15, ESI†). The comparison of the related parameters of the helix of Ag70-TPP with other helical biological molecules (Alpha-keratin, TMV) is shown in Table S2 (ESI†). Its discovery proves that the self-assembly of nanoclusters such as Ag70-TPP can be as intricate as is observed for biological molecules.
00] directions, respectively (Fig. 3a and c). Along the [100] direction, the stacking sequence is “A B C D”, which is characteristic of 4H (four layered hexagonal) close packing (Fig. 3b).33 However, in contrast to traditional 4H the four pairs of Ag70-TPP are arranged into two helical arrangements (Fig. S14, ESI†), wherein the blue shadow is left-handed and the other in pink shadow is a right-handed helix alternately arranged in adjacent locations (Fig. S14, ESI†), termed as a double helical 4H (DH4H) arrangement, and the average pitch is 26.1 Å. This type of DH4H assembly mode is reminiscent of the helix structure characteristics of biological DNA, but different. The DNA helix biopolymer of nucleic acids is held together by nucleotides which base pair together, with a unique axis, while Ag70-TPP possesses two parallel helix axes with opposite directions, and connected by weak interactions. (Fig. S15, ESI†). The comparison of the related parameters of the helix of Ag70-TPP with other helical biological molecules (Alpha-keratin, TMV) is shown in Table S2 (ESI†). Its discovery proves that the self-assembly of nanoclusters such as Ag70-TPP can be as intricate as is observed for biological molecules.
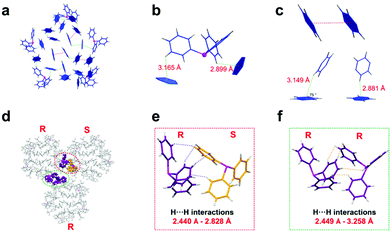
The secondary structures of proteins are mainly maintained by hydrogen bonds. To explore the factors governing the DH4H-type assembly in Ag70-TPP nanoclusters, we sought to understand the extent and nature of the interactions between clusters.34,35 On the surface of the nanocluster, C–H⋯π and π⋯π interactions between ligands (Fig. 4a) were found. These interactions can be roughly divided into two types: (i) the interaction between the H atoms of the phosphine and the phenyl ring of the nearby thiolate ligands (Fig. 4b); and (ii) the interaction between multiple thiolate ligands (C–H⋯π and π⋯π interactions (Fig. 4c)). The C–H⋯π distances range from 2.6 Å to 3.2 Å; these are weak interactions, widely distributed in the entire crystal structure, which presumably contribute to its stability.36,37
As shown in Fig. 4d and Fig. S16 (ESI†), adjacent NCs are narrowly spaced and C–H⋯π and H⋯H interactions are evident, resulting in an intercluster permutation pattern (Fig. 4d). The shortest distance between neighbouring clusters is only 2.440 Å, suggesting their intermolecular interaction to be strong. The phenyl rings of adjacent phosphines interlock (Fig. 4e and f).38 The interaction between phosphines is likely to influence the distortion of the cluster. From Fig. S17 (ESI†), the phosphine ligand at the border of the cluster has a large angle of distortion, which is similar to the same poles of a magnet repelling. These intra- and inter-molecular interactions greatly influence the arrangement of clusters in this crystal lattice.
In order to verify whether the phosphine ligand plays an important role in maintaining the DH4H assembly mode.39,40 Tri(p-tolyl)phosphine (TPTP) was selected to replace the TPP ligand during the synthesis of the clusters. The UV-vis spectrum (Fig. S18, ESI†) and TGA (Fig. S19, ESI†) confirmed that the cluster is still Ag70 (denoted as Ag70-TPTP). In addition, the crystal structure of Ag70-TPTP was determined by SCXC. As shown in Fig. S20 (ESI†), not only the crystal system (an orthorhombic Pbca to a triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group), but also the assembly pattern (complex DH4H to simple 2H) of Ag70-TPTP had changed when compared with the Ag70-TPP. The stacking sequence of Ag70-TPTP is “A·B·A·B”, and Ag70-TPTP clusters in A-layer and B-layer show opposite chirality (Fig. S20(d–f)). In one unit cell, we found that two groups of phosphine ligands in R-, and S-Ag70-TPTP interlocked each other (Fig. S21a, ESI†) through the C–H⋯π and H⋯H interactions. While in the adjacent unit cells, only one pair of phosphine ligand interations was found (Fig. S21b, ESI†), which is different from that of the Ag70-TPP.
space group), but also the assembly pattern (complex DH4H to simple 2H) of Ag70-TPTP had changed when compared with the Ag70-TPP. The stacking sequence of Ag70-TPTP is “A·B·A·B”, and Ag70-TPTP clusters in A-layer and B-layer show opposite chirality (Fig. S20(d–f)). In one unit cell, we found that two groups of phosphine ligands in R-, and S-Ag70-TPTP interlocked each other (Fig. S21a, ESI†) through the C–H⋯π and H⋯H interactions. While in the adjacent unit cells, only one pair of phosphine ligand interations was found (Fig. S21b, ESI†), which is different from that of the Ag70-TPP.
Conclusions
In summary, the pseudo-five-fold symmetric pentagram-like silver nanocluster Ag70(TBBT)42(TPP)5 was synthesized, and its composition was determined by SCXC, ESI-MS, and NMR analysis. Analysis of the crystal structure of Ag70-TPP determined its structure to incorporate a complete decahedral Ag23 kernel. Due to the coordination preferences and electronic or steric hindrance effects of the mixed ligands (i.e. thiolate and phosphine ligands), the intramolecular and intermolecular C–H⋯π, π⋯π, and H⋯H interactions can account for the complex arrangement of Ag70-TPP in the unit cells. Furthermore, the steric hindrance of the phosphine ligand can be changed to control the way of assembly between molecules to achieve the transformation from complex to simple packing. Collectively, these results show that the secondary structures of sub-nanometer-sized noble metal nanoclusters can be as complex as those found in biological systems, and that structures assembled from nanoclusters can reach not only atomic accuracy but also the same hierarchical assembly as biomolecules levels.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
We acknowledge financial support from the National Natural Science Foundation of China (21631001, 21871001, and 22001002), the Ministry of Education, the Education Department of Anhui Province, Anhui Provincial Natural Science Foundation (2008085QB82), and the University Synergy Innovation Program of Anhui Province (GXXT-2020-053).References
- M. J. Adams, E. N. Baker, T. L. Blundell, E. J. Dodson, G. G. Dodson, M. Vijayan, M. M. Harding, D. C. Hodgkin, B. Rimmer and S. Sheat, Nature, 1969, 224, 491–495 CrossRef CAS.
- J. D. Watson and F. H. C. Crick, Nature, 1953, 171, 737–738 CrossRef CAS PubMed.
- J. C. Kendrew, Science, 1963, 139, 1259–1266 CrossRef CAS PubMed.
- W. W. Grabow and L. Jaeger, Acc. Chem. Res., 2014, 47, 1871–1880 CrossRef CAS PubMed.
- Y. Zhao, S. Zhuang, L. Liao, C. Wang, N. Xia, Z. Gan, W. Gu, J. Li, H. Deng and Z. Wu, J. Am. Chem. Soc., 2020, 142, 973–977 CrossRef CAS PubMed.
- J. Teyssier, S. V. Saenko, D. V. D. Marel and M. C. Milinkovitch, Nat. Commun., 2015, 6, 6368 CrossRef CAS PubMed.
- P. F. Damasceno, M. Engel and S. C. Glotzer, Science, 2012, 337, 453–457 CrossRef CAS PubMed.
- R. Huang, Y. Wei, X. Dong, X. Wu, C. Du, S. Zang and T. C. W. Mak, Nat. Chem., 2017, 9, 689–697 CrossRef CAS PubMed.
- S. Y. Park, A. K. R. Lytton-Jean, B. Lee, S. Weigand, G. C. Schatz and C. A. Mirkin, Nature, 2008, 451, 553–556 CrossRef CAS PubMed.
- R. Nidetz and J. Kim, Nanotechnology, 2012, 23, 045602 CrossRef PubMed.
- A. D. Merg, J. C. Boatz, A. Mandal, G. Zhao, S. Mokashi-Punekar, C. Liu, X. Wang, P. Zhang, P. C. A. van der Wel and N. L. Rosi, J. Am. Chem. Soc., 2016, 138, 13655–13663 CrossRef CAS PubMed.
- H. Fakhouri, M. Perić, F. Bertorelle, P. Dugourd, X. Dagany, I. Russier-Antoine, P. Brevet, V. Bonačić-Koutecký and R. Antoine, Phys. Chem. Chem. Phys., 2019, 21, 12091–12099 RSC.
- R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS PubMed.
- I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- R. S. Dhayal, J. Liao, Y. Liu, M. Chiang, S. Kahlal, J. Saillard and C. W. Liu, Angew. Chem., Int. Ed., 2015, 54, 3702–3706 CrossRef CAS PubMed.
- P. D. Jadzinsky, G. Calero, C. J. Ackerson and D. A. Bushnell, Science, 2007, 318, 430–433 CrossRef CAS PubMed.
- C. P. Joshi, M. S. Bootharaju, M. J. Alhilaly and O. M. Bakr, J. Am. Chem. Soc., 2015, 137, 11578–11581 CrossRef CAS PubMed.
- Y. Yang, T. Jia, Y. Han, Z. Nan, S. Yuan, F. Yang and D. Sun, Angew. Chem., Int. Ed., 2019, 58, 12280–12285 CrossRef CAS PubMed.
- T. Higaki, C. Liu, C. Zeng, R. Jin, Y. Chen, N. L. Rosi and R. Jin, Angew. Chem., Int. Ed., 2016, 55, 6694–6697 CrossRef CAS PubMed.
- L. Liao, J. Chen, C. Wang, S. Zhuang, N. Yan, C. Yao, N. Xia, L. Li, X. Bao and Z. Wu, Chem. Commun., 2016, 52, 12036–12039 RSC.
- Z. Gan, J. Chen, J. Wang, C. Wang, M. Li, C. Yao, S. Zhuang, A. Xu, L. Li and Z. Wu, Nat. Commun., 2017, 8, 14739 CrossRef CAS PubMed.
- Z. Wu, Y. Du, J. Liu, Q. Yao, T. Chen, Y. Cao, H. Zhang and J. Xie, Angew. Chem., Int. Ed., 2019, 58, 8139–8144 CrossRef CAS PubMed.
- C. Zeng, Y. Chen, K. Kirschbaum, K. J. Lambright and R. Jin, Science, 2016, 354, 1580–1584 CrossRef CAS PubMed.
- T. Chen, S. Yang, Y. Song, J. Chai, Q. Li, X. Ma, G. Li, H. Yu and M. Zhu, Chem. Commun., 2020, 56, 7605–7608 RSC.
- F. Hu, J. Li, Z. Guan, S. Yuan and Q. Wang, Angew. Chem., Int. Ed., 2020, 59, 5312–5315 CrossRef CAS PubMed.
- J. Li, Z. Guan, Z. Lei, F. Hu and Q. Wang, Angew. Chem., Int. Ed., 2019, 58, 1083–1087 CrossRef CAS PubMed.
- L. G. AbdulHalim, M. S. Bootharaju, Q. Tang, S. D. Gobbo, R. G. AbdulHalim, M. Eddaoudi, D. Jiang and O. M. Bakr, J. Am. Chem. Soc., 2015, 137, 11970–11975 CrossRef CAS PubMed.
- I. Russier-Antoine, F. Bertorelle, R. Hamouda, D. Rayane, P. Dugourd, Ž. Sanader, V. Bonačić-Koutecký, P. Brevet and R. Antoine, Nanoscale, 2016, 8, 2892–2898 RSC.
- T. K. Sau and A. L. Rogach, Adv. Mater., 2010, 22, 1781–1804 CrossRef CAS PubMed.
- H. Yang, Y. Wang, X. Chen, X. Zhao, L. Gu, H. Huang, J. Yan, C. Xu, G. Li, J. Wu, A. J. Edwards, B. Dittrich, Z. Tang, D. Wang, L. Lehtovaara, H. Häkkinen and N. Zheng, Nat. Commun., 2016, 7, 12809 CrossRef CAS PubMed.
- H. Han, Y. Yao, A. Bhargava, Z. Wei, Z. Tang, J. Suntivitch, O. Voznyy and R. D. Robinson, J. Am. Chem. Soc., 2020, 142, 14495–14503 CrossRef CAS PubMed.
- A. Desireddy, B. E. Conn, J. Guo, B. Yoon, R. N. Barnett, B. M. Monahan, K. Kirschbaum, W. P. Griffith, R. L. Whetten, U. Landman and T. P. Bigioni, Nature, 2013, 501, 399–402 CrossRef CAS PubMed.
- M. I. Novgorodova, A. I. Gorshkov and A. V. Mokhov, Zap. Vses. Mineral. O-va., 1979, 108, 552–563 CAS.
- Y. Li and R. Jin, J. Am. Chem. Soc., 2020, 142, 13627–13644 CrossRef CAS PubMed.
- T. Chen, S. Yang, J. Chai, Y. Song, J. Fan, B. Rao, H. Sheng, H. Yu and M. Zhu, Sci. Adv., 2017, 3, e1700956 CrossRef PubMed.
- X. Liu, G. Saranya, X. Huang, X. Cheng, R. Wang, M. Chen, C. Zhang, T. Li and Y. Zhu, Angew. Chem., Int. Ed., 2020, 59, 13941–13946 CrossRef CAS PubMed.
- A. Nag, P. Chakraborty, M. Bodiuzzaman, T. Ahuja, S. Antharjanam and T. Pradeep, Nanoscale, 2018, 10, 9851–9855 RSC.
- J. Yan, S. Malola, C. Hu, J. Peng, B. Dittrich, B. K. Teo, H. Häkkinen, L. Zheng and N. Zheng, Nat. Commun., 2018, 9, 3357 CrossRef PubMed.
- Q. Li, J. C. Russell, T. Luo, X. Roy, N. L. Rosi, Y. Zhu and R. Jin, Nat. Commun., 2018, 9, 3871 CrossRef PubMed.
- L. He, Z. Gan, N. Xia, L. Liao and Z. Wu, Angew. Chem., Int. Ed., 2019, 58, 9897–9901 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the syntheses, crystallization, X-ray analysis, supporting figures, and tables. Crystallographic information for Ag70 (CIF). CCDC 2053542 and 2053704. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1nh00332a |
| This journal is © The Royal Society of Chemistry 2021 |