Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Post-translational modifications soften vimentin intermediate filaments†
Julia
Kraxner
a,
Charlotta
Lorenz
a,
Julia
Menzel
b,
Iwan
Parfentev
c,
Ivan
Silbern
 cd,
Manuela
Denz
a,
Henning
Urlaub
cd,
Blanche
Schwappach
be and
Sarah
Köster
cd,
Manuela
Denz
a,
Henning
Urlaub
cd,
Blanche
Schwappach
be and
Sarah
Köster
 *aef
*aef
aInstitute for X-Ray Physics, University of Göttingen, 37077 Göttingen, Germany. E-mail: sarah.koester@phys.uni-goettingen.de
bDepartment of Molecular Biology, University Medical Center Göttingen, 37073 Göttingen, Germany
cBioanalytical Mass Spectrometry, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany
dBioanalytical Group, Institute of Clinical Chemistry, University Medical Center Göttingen, 37075 Göttingen, Germany
eCluster of Excellence “Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells” (MBExC), University of Göttingen, Germany
fMax Planck School “Matter to Life”, Germany
First published on 10th December 2020
Abstract
The mechanical properties of biological cells are determined by the cytoskeleton, a composite biopolymer network consisting of microtubules, actin filaments and intermediate filaments (IFs). By differential expression of cytoskeletal proteins, modulation of the network architecture and interactions between the filaments, cell mechanics may be adapted to varying requirements on the cell. Here, we focus on the intermediate filament protein vimentin and introduce post-translational modifications as an additional, much faster mechanism for mechanical modulation. We study the impact of phosphorylation on filament mechanics by recording force-strain curves using optical traps. Partial phosphorylation softens the filaments. We show that binding of the protein 14-3-3 to phosphorylated vimentin IFs further enhances this effect and speculate that in the cell 14-3-3 may serve to preserve the softening and thereby the altered cell mechanics. We explain our observation by the additional charges introduced during phosphorylation.
1 Introduction
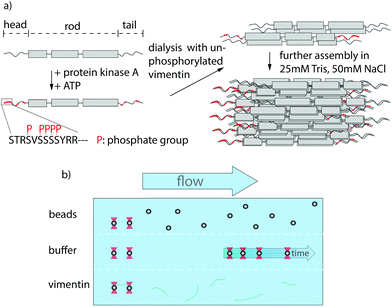
The cytoskeleton of eukaryotes consists of three filament systems – microtubules, actin filaments and intermediate filaments (IFs) – which form a complex biopolymer network. The exact composition of the cytoskeleton and the interplay between the three filament types largely influence the mechanical properties of different cell types.1 Microtubules and actin filaments are highly conserved throughout cell types, whereas more than 70 human genes encode for IFs,2 yet they all share the same hierarchical assembly pathway from monomers to filaments. The secondary structure of the monomers consists of an α-helical rod domain, flanked by intrinsically disordered head and tail domains.3,4 During assembly, two monomers align and form parallel coiled-coil dimers, two dimers form antiparallel, half-staggered tetramers, and multiple tetramers constitute a unit-length filament (ULF) (see Fig. 1a). Subsequent longitudinal annealing yields elongated filaments with a diameter of about 10 nm.5 This hierarchical structure of IFs, in contrast to polar microtubules or actin filaments, grants IFs their unique mechanical properties.6–8The most abundant IF protein, vimentin, is found in cells of mesenchymal origin.5,9 Single vimentin filaments are highly extensible and can be elongated up to at least 4.5 fold.7,10,11 During elongation, three regimes are observed in the force-strain curves: an initial linear (elastic) increase, a plateau region and a subsequent stiffening.6,7,12,13 These different stretching regimes have been linked to structural changes in the rod domain of IFs12 such as the opening of α helices during the plateau regime.6,14,15 Additionally, vimentin filaments are able to dissipate large amounts of the input energy while being stretched.14 Consequently, vimentin IFs act as shock-absorbers for mechanical protection of the cell as well as scaffolds that help to maintain cell shape and organization of the cytoskeleton.9,16
An effective way to tune the mechanics of IFs is the variation of the charges of the amino acids constituting the protein.17 One cellular mechanism for such charge variation are post-translational modifications (PTMs). Within the IF cytoskeleton, several types of PTMs have been described18,19 and the most abundantly studied PTM in IFs is phosphorylation, which is involved in regulation of IF dynamics by leading to disassembly and in providing binding sites to signaling proteins.20,21 It has been shown that the phosphorylation of vimentin by protein-kinase A (PKA) leads to various phosphorylated sites, most of which are positioned in the head region (see Fig. 1a).21 The importance of the head domain in the assembly process was shown in ref. 3 and 22, stressing the obvious link between phosphorylation and assembly dynamics. Although such changes in the molecular interactions during assembly are likely to influence the behavior of the fully assembled filaments, the influence of phosphorylations on filament mechanics is not yet resolved.
A further interesting aspect of phosphorylation is the ability of certain proteins to bind to the modified sites. One such protein is 14-3-3,23 which is involved in several cellular processes like signal transduction, adhesion and inhibition of tumorigenesis.24 The role of this protein depends on the interaction partner, e.g. it binds to keratin during the cell cycle25 and affects the assembly dynamics of neurofilaments.26 However, the role of 14-3-3 for vimentin is unknown.
Here we investigate the effect of phosphorylation and 14-3-3 on vimentin mechanics by studying precise force-strain curves from optical trap experiments. We find that the filaments soften with increasing amount of phosphorylated protein within the filament and that interaction with the protein 14-3-3 further enhances this effect. By combining our mechanical measurements with mass spectrometry, cross-linking, phosphomimicry and numeric modeling, we are able to attribute the softening to reduced lateral coupling of monomers within the filaments.
2 Experimental
An extended version of the Methods section can be found in the ESI.†2.1 Experimental design
Carboxylated polystyrene beads (Kisker Biotech GmbH & Co. KG, Steinfurt, Germany) with a diameter of 4.4 μm were functionalized with maleimide according to ref. 30.
2.2 Experimental setup and data analysis
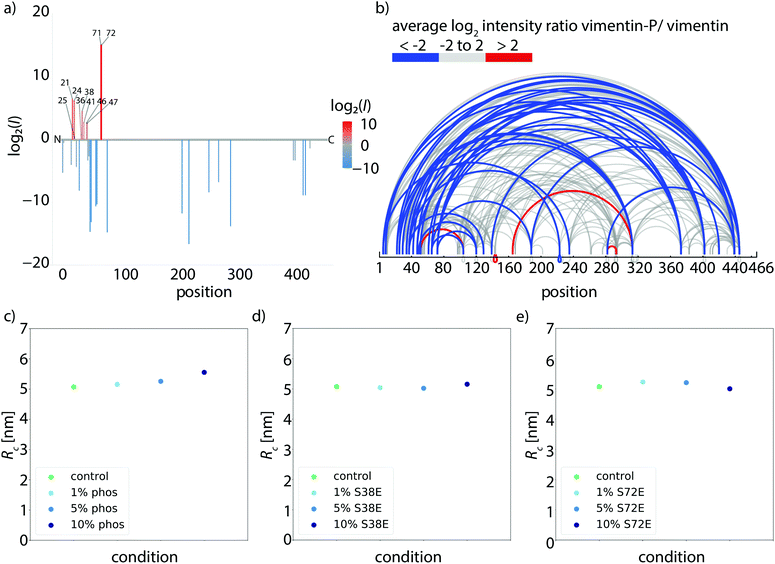
Data were analyzed with self-written Python codes. The calculation of the mean curves for each conditions was adapted from ref. 8. For the calculation of the Young's modulus, a linear fit up to a force of 130 pN was performed for the initial slope in the linear regime of the mean force-strain curves. The Young's modulus was calculated via the ratio of stress and strain, Y = σ/ε = F/(πR2ε). For the radius R, the corresponding values for the radius of gyration of the cross-section, Rc, obtained from the small-angle X-ray scattering experiments, were used.
3 Results and discussion
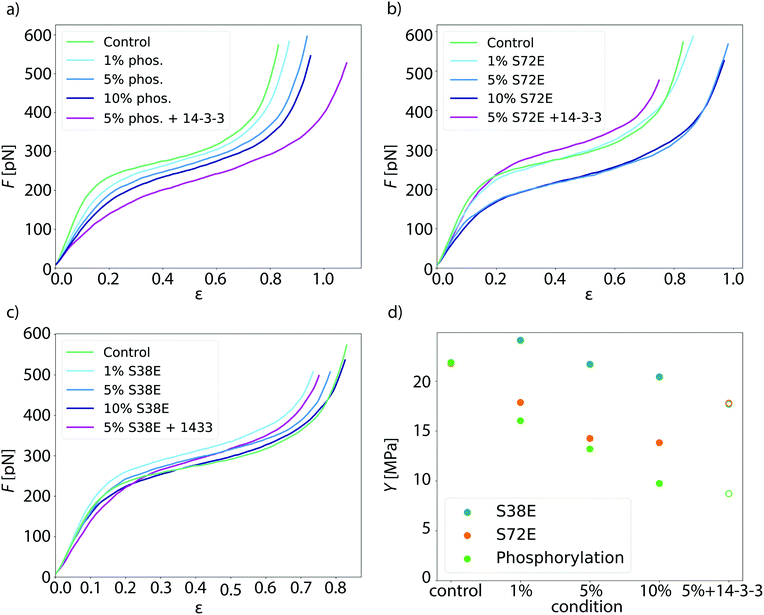
To investigate whether phosphorylation of vimentin influences filament mechanics, we record force-strain curves using optical traps. As complete phosphorylation of vimentin filaments leads to disassembly,21 we perform the stretching experiments on partially phosphorylated vimentin filaments with varying percentages of 1, 5 or 10%, as sketched in Fig. 1a. The incorporation of the phosphorylated vimentin is confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-page) and by fluorescence microscopy, shown in Fig. S1 and S2.† The single filaments are covalently bound to two polystyrene beads and each bead is captured in an optical trap for precise force measurements. Filaments with 10% phosphorylated protein incorporated assemble to the same total length as unphophorylated filaments, see Fig. S3.† Additionally, we determine the distance between both beads and calculate the strain as ε = ΔL/L0, where L0 is the original filament length and ΔL the additional extension upon application of a force F. We perform all measurements in the “buffer” channel, i.e. without any disturbance from surrounding beads or protein, see Fig. 1b. During stretching, we observe the characteristic response of vimentin to the force, including the linear increase for small strains, the plateau region, where the α helices open up6,7,14 and the subsequent stiffening of the filaments. We perform all measurements in 25 mM Tris-HCl, 50 mM NaCl, pH 7.5 and the curves are comparable to the ones previously recorded in phosphate buffer,7,14 see Fig. S4.† The pH of the measuring buffer is chosen such that it is comparable to most in vitro studies on vimentin filaments. For a lower pH, we expect the same shape of the curve, but slightly shifted to higher forces, as shown recently in ref. 17. Such a typical curve for our standard condition is shown in Fig. 2a, green. We show an average curve here for clarity, however, the individual data sets are shown in the Fig. S5.† The measurements end when the force becomes too large for the optical traps, i.e. at roughly 550–600 pN. Even though strong phosphorylation leads to disassembly of vimentin filaments,21 we do not observe an increase in the number of breakage events upon increasing phosphorylation. Fig. 2a also shows corresponding data for filaments containing a certain percentage of phosphorylated protein (shades of blue, for color code see legend). To quantify the force-strain data, we focus on the Young's modulus, which we calculate from the initial slope in the linear regime up to a force of 130 pN of each curve, as a measure of the filament stiffness. The Young's modulus for the unphosphorylated vimentin filaments is in good agreement with the results found in ref. 7. The analysis procedure is visualized in Fig. S6.† The Young's moduli plotted in Fig. 2d (green) show a strong decrease with increasing percentage of phosphorylated protein. Further analysis of the force-strain curves shows a decrease of the force at the onset of the plateau and an increase of the maximum strain reached with an increasing amount of phosphorylation, shown in Fig. S7.†Upon phosphorylation of vimentin by PKA, several amino acids are modified. To determine these sites, we perform an LC-MS (liquid chromatography mass spectrometry) analysis. Fig. 3a shows that phosphorylated sites are dispersed throughout the whole protein, but the most abundant ones (log2(I) > 0, red lines) are all found in the head region of vimentin. This is in agreement with the head domain of vimentin being a serine rich region and containing multiple motives required for kinases.31 Strikingly, the positions S71 and S72 are always phosphorylated simultaneously and show the highest degree of phosphorylation. Hence, we speculate that these two positions are the main phosphorylation sites, whereas all other positions occur only sporadically and therefore differ from monomer to monomer. We employ phosphomimicry to investigate the effect of defined phosphorylated sites and choose two of the most abundantly phosphorylated sites that also occur in vivo, S38 and S72.21 We perform the same force-strain measurements as described above. For the mutant S72E, we observe a similar trend as in the phosphorylation data including enhanced softening, see Fig. 2b, green to dark blue. On the contrary, Fig. 2c, green to dark blue, shows filaments containing the mutation S38E and no systematic trend is observed. The individual data sets are provided in Fig. S8 and S9.† When comparing these three different conditions, i.e. phosphorylation, mutation S72E and mutation S38E, we observe that the Young's modulus, and therefore the filament stiffness, decreases with an increasing amount of phosphorylation (green) and mutation S72E (orange) but stays fairly constant for the mutation S38E (blue), see Fig. 2d. These results suggest that the phosphorylation at position S72 influences the filament mechanics whereas at position S38 it does not have any effect. As the filament radius is a critical parameter for calculation of the Young's modulus, we perform small angle X-ray scattering (SAXS) experiments to determine the radius of gyration of the cross-section Rc, see Fig. 3c–e. No changes in radius are observed. The individual SAXS curves are shown in Fig. S10–S12.†
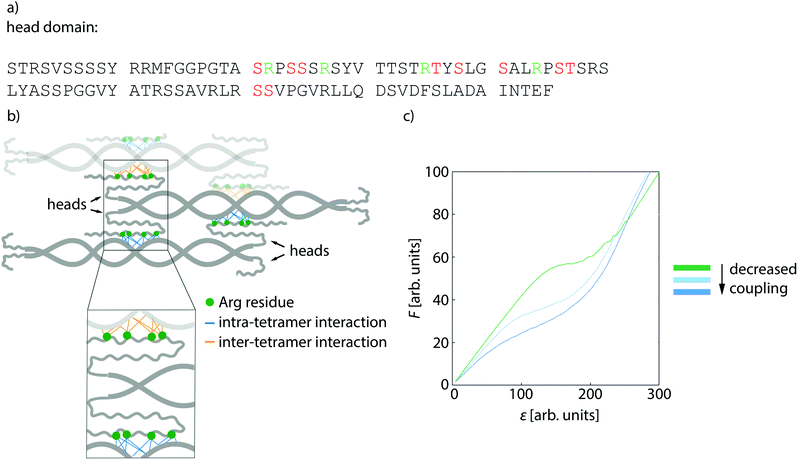
To explain the softening of vimentin filaments with increasing amount of phosphorylation, we consider previous studies that have shown neighboring dimers to be coupled by electrostatic interactions between specific positively charged amino acids in the head, namely R23, R28, R36 and R45, and the negatively charged coiled-coils3,27,32 as sketched in Fig. 4b. When vimentin becomes phosphorylated, the positive charges of the head domain are flanked by negative charges of the phosphorylated amino acids as shown in Fig. 4a, which diminishes the electrostatic attraction between the head and the coiled-coils. This observation raises the question of whether the shift in filament stiffness can be explained by weaker coupling. Therefore, we run Monte-Carlo simulations to understand how the coupling of dimers affects the force-strain curves of vimentin by extending the model from ref. 8 and 14 as described in detail in the ESI.† We model a vimentin monomer by including the α helices in the rod domain as the spring constant κα and as elements, which extend in length at a certain force, thereby transitioning to an unfolded state u.8,14,15 The model in ref. 8 assumes a strong coupling within dimers, tetramers or any other subunit of the ULF and weaker coupling between these subunits. In that work, the force-strain curves of filaments with smaller coupled subunits exhibit a lower plateau than the force-strain curves of filaments with larger coupled subunits. As demonstrated in ref. 17, the model can also explain different maximum strains of filaments in different conditions either by a different number of α helices which unfold or a different extension of single α helices. Yet, this previous model does not explain a pronounced decrease in Young's modulus as observed here.
We thus supplement the model from ref. 8 by spring constants κbt, which represent bonds between tetramers, and κbd for bonds between dimers within the tetramer. In case of more phosphorylated monomers in the filament, the filament subunits do not interact as strongly as without phosphorylated monomers because the additional negative charges repel each other. Thus, not all bonds between tetramers and dimers can form, so that the spring constants κbd and κbt do not contribute to the overall spring constant and the filaments get softer as shown for the simulated force-strain curves in Fig. 4d. Consequently, the initial slope of the force-strain curves decreases with more phosphorylated monomers as shown in Fig. 2c. Our results show that the negative charge at position S72, in particular, has an effect on filament mechanics: the phosphomimetic mutant shows a very similar trend compared to the phosphorylation data (Fig. 2d). Indeed, this position is also found to be the mayor phosphorylation site (see Fig. 3a). By contrast, an additional negative charge at position S38 has no effect on the filament mechanics. Therefore, we conclude that a decreased coupling around position S72 is crucial for the shift in mechanics whereas a decreased coupling at position S38 has no effect.
We confirm these numerical findings with mass spectrometry cross-linking experiments. When comparing cross-linked phosphorylated vimentin tetramers and unphosphorylated vimentin tetramers in Fig. 3b, fewer cross-links are found in the phosphorylated state, blue lines. All individual cross-linking positions can be found in Fig. S13.† This supports our proposal that there are decreased interactions between neighboring dimers in phosphorylated vimentin which indicates that the lateral coupling of dimers is reduced. Despite these decreased interactions, we do not observe an increase in filament radius for increasing amount of phosphorylation, see Fig. 3c–e. We speculate that the softening effect due to phosphorylation is reversible when the phosphate group is removed again by phosphatases and filaments would recover their initial stiffness.
Besides the fact that phosphorylation modifies the protein itself, it also creates binding sites for other proteins, such as 14-3-3 in the case of phosphorylated vimentin.23 This raises the question of whether the binding of the protein 14-3-3 to phosphorylated vimentin also has an effect on the mechanics and we perform optical trap measurements of the vimentin/14-3-3 complex. We confirm the interaction between vimentin and 14-3-3 by performing a streptavidin pulldown assay as shown in Fig. S14.†Fig. 2d, open green circle, shows that for phosphorylated filaments the interaction with 14-3-3 softens the filaments even more. By contrast, the original stiffness of the unphosphorylated filaments is recovered for the mutant S72E, Fig. 2d, open orange circle, and we observe only a slight influence of 14-3-3 for the mutant S38E, Fig. 2d, open blue circle. The individual curves for these experiments are provided in Fig. S15.† We show that 14-3-3 binds to the phosphorylated sites in the head domain of vimentin, as confirmed by cross-linking the complex and analyzing it with mass spectrometry, see Fig. S16.† Assumedly, because of this interaction and the similar size of 14-3-3 compared to vimentin, it forces vimentin subunits further apart and thus enhances the decoupling and softening of vimentin filaments. Mostly, vimentin is cross-linked to position 78 of the amino acid sequence of 14-3-3 as shown in Fig. S16.† By contrast, we cannot unambiguously determine the cross-linking position in the amino acid sequence of vimentin. However, our data show that the amino acids S38 and S72 in vimentin are not the binding sites for 14-3-3 as the complex of the two proteins is retrieved after phosphorylation of the phosphomimetic mutants as confirmed by cross-linking the complex, see Fig. S17.† Additionally, we find that no complex is formed between 14-3-3 and the phosphomimetic mutants S38E and S72E. The corresponding SDS gel is shown in Fig. S17.† Despite this lack of complex formation between 14-3-3 and the phosphomimetic mutant S72E, we observe a stiffening of the filaments upon addition of 14-3-3, indicating some kind of interaction between the proteins, such as electrostatic interactions. Such electrostatic interaction could possibly diminish the effect of the negative charges and thus lead to a recovery of the initial stiffness. Taking these results together, we propose that 14-3-3 protects the protein from phosphatases by binding to strategic phosphorylated sites as suggested in ref. 33. Due to the large size of the protein 14-3-3, it sterically hinders the phosphatases, which can reverse the effect of the protein kinases, from binding to neighboring amino acid positions within the vimentin monomer. Thereby the protein 14-3-3 could be able to keep the vimentin filaments in the soft state for extended times, which might be important for in vivo situations.
Previous studies have shown that increased vimentin phosphorylation is required for efficient cellular migration34 and that it is relevant in metastasis35,36 which may be linked to a softer vimentin network and therefore render the cells more deformable. In general, phosphorylation controls the assembly and disassembly dynamics of vimentin21 and in particular the phosphorylation of vimentin leads to disassembly. Phosphatase activity enables recovery to assembled filaments. If 14-3-3 binds to phosphorylated vimentin and inhibits the phosphatase activity, vimentin remains in the phosphorylated state. Such an effect would slow down the assembly dynamics, which is, however, crucial for cell adhesion, migration and signaling.37 In addition, the vimentin/14-3-3 complex builds a larger ensemble with the phosphorylated protein beclin1, which then promotes tumorigenesis.38 It was already suggested that vimentin might be a key regulator of tumorigenic pathways as this complex, namely 14-3-3, vimentin and beclin1, might prevent the dephosphorylation of the proteins within the ensemble and thereby inhibit antitumor activity in cells.39
4 Conclusions
To conclude, we directly show how post-translational modifications, i.e. phosphorylation, change the mechanical properties of vimentin filaments: Vimentin filaments become softer with increasing amount of phosphorylated protein within the filament. These findings may help to understand the relation between the role of phosphorylation in cancer metastasis and the pronounced motility of metastasizing cells. The interaction of phosphorylated vimentin with 14-3-3 enhances this softening effect and may even protect the softer state. We suggest that these changes are induced by reduced electrostatic coupling within the ULF due to additional negative charges introduced by the phosphate groups and support this assumption by a physical model. We thus hypothesize that cells are able to fine-tune and adapt their mechanical properties locally and within seconds by modifications like phosphorylation according to specific external requirements.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Susanne Bauch for preparing the vimentin protein, Anna V. Schepers for providing the comparison of vimentin mechanics measurements in different buffer conditions, Monika Raabe for carrying out the phosphopeptide enrichment and Harald Herrmann for helpful discussions. This work was financially supported by the European Research Council (ERC) under the European Unions Horizon 2020 Research and Innovation Program (Consolidator Grant Agreement No. 724932, to S. K.). Further financial support was received from the Deutsche Forschungsgemeinschaft (DFG) in the framework of SFB 860 (project number B10, to S. K.), SFB 1286 (project number A08, to H. U.) and of Germany's Excellence Strategy – EXC 2067/1-390729940 (MBExC, to S. K. and B. S.). C. L. received a fellowship of the Studienstiftung des deutschen Volkes.References
- F. Huber, A. Boire, M. Preciado López and G. H. Koenderink, Curr. Opin. Cell Biol., 2015, 32, 39–47 CrossRef CAS.
- I. Szeverenyi, A. J. Cassidy, C. W. Chung, B. T. Lee and J. E. Common, Hum. Mutat., 2008, 29, 351–360 CrossRef CAS.
- H. Herrmann, M. Häner, M. Brettel, S. A. Müller, K. N. Goldie, B. Fedtke, A. Lustig, W. W. Franke and U. Aebi, J. Mol. Biol., 1996, 264, 933–953 CrossRef CAS.
- A. A. Chernyatina, D. Guzenko and S. V. Strelkov, Curr. Opin. Cell Biol., 2015, 32, 65–72 CrossRef CAS.
- H. Herrmann and U. Aebi, Cold Spring Harbor Perspect. Biol., 2016, 8, a018242 CrossRef.
- Z. Qin, L. Kreplak and M. Buehler, PLoS One, 2009, 4, 1–14 CrossRef.
- J. Block, H. Witt, A. Candelli, E. J. G. Peterman, G. J. L. Wuite, A. Janshoff and S. Köster, Phys. Rev. Lett., 2017, 118, 1–5 CrossRef.
- C. Lorenz, J. Forsting, A. V. Schepers, J. Kraxner, S. Bauch, H. Witt, S. Klumpp and S. Köster, Phys. Rev. Lett., 2019, 123, 188102 CrossRef CAS.
- J. E. Eriksson, T. Dechat, B. Grin, B. Helfand, M. Mendez, H.-M. Palari and R. D. Goldman, J. Clin. Invest., 2009, 119, 1763–1771 CrossRef CAS.
- L. Kreplak, H. Bär, J. F. Leterrier, H. Herrmann and U. Aebi, J. Mol. Biol., 2005, 354, 569–577 CrossRef CAS.
- L. Kreplak, H. Herrmann and U. Aebi, Biophys. J., 2008, 94, 2790–2799 CrossRef CAS.
- T. Ackbarow and M. Buehler, J. Mater. Sci., 2007, 42, 8771–8787 CrossRef CAS.
- N. Pinto, F.-C. Yang, A. Negishi, M. C. Rheinstädter, T. E. Gillis and D. S. Fudge, Biomacromolecules, 2014, 15, 574–581 CrossRef CAS.
- J. Block, H. Witt, A. Candelli, J. C. Danes, E. J. Peterman, G. J. Wuite, A. Janshoff and S. Köster, Sci. Adv., 2018, 4, eaat1161 CrossRef.
- J. Forsting, J. Kraxner, H. Witt, A. Janshoff and S. Köster, Nano Lett., 2019, 19, 7349–7356 CrossRef CAS.
- E. Fuchs and D. W. Cleveland, Science, 1998, 279, 514–519 CrossRef CAS.
- A. V. Schepers, C. Lorenz and S. Köster, Nanoscale, 2020, 12, 15236–15245 RSC.
- C. L. Hyder, H.-M. Pallari, V. Kochin and J. E. Eriksson, FEBS Lett., 2008, 582, 2140–2148 CrossRef CAS.
- N. T. Snider and M. B. Omary, Nat. Rev. Mol. Cell Biol., 2014, 15, 163–177 CrossRef CAS.
- T. Busch, M. Armacki, T. Eiseler, G. Joodi, C. Temme, J. Jansen, G. von Wichert, M. B. Omary, J. Spatz and T. Seufferlein, J. Cell Sci., 2012, 125, 2148–2159 CrossRef CAS.
- J. E. Eriksson, T. He, A. V. Trejo-Skalli, A. S. Harmala-Brasken, J. Hellman, Y.-H. H. Chou, R. D. Goldman, A.-S. Härmälä-Braskén, J. Hellman, Y.-H. H. Chou and R. D. Goldman, J. Cell Sci., 2004, 117, 919–932 CrossRef CAS.
- P. Traub and C. E. Vorgias, J. Cell Sci., 1983, 63, 43–67 CAS.
- G. Tzivion, Z. J. Luo and J. Avruch, J. Biol. Chem., 2000, 275, 29772–29778 CrossRef CAS.
- A. K. Gardino and M. B. Yaffe, Semin. Cell Dev. Biol., 2011, 22, 688–695 CrossRef CAS.
- N.-O. Ku, S. Michie, E. Z. Resurreccion, R. L. Broome and M. B. Omary, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4373–4378 CrossRef CAS.
- L. Miao, J. Teng, J. Lin, X. Liao and J. Chen, J. Cell Sci., 2013, 126, 427–436 CrossRef CAS.
- H. Herrmann and U. Aebi, Annu. Rev. Biochem., 2004, 73, 749–789 CrossRef CAS.
- H. Wu, Y. Shen, D. Wang, H. Herrmann, R. D. Goldman and D. A. Weitz, Biophys. J., 2020, 119, 55–64 CrossRef CAS.
- S. Winheim, A. R. Hieb, M. Silbermann, E.-M. Surmann, T. Wedig, H. Herrmann, J. Langowski and N. Mücke, PLoS One, 2011, 6, e19202 CrossRef CAS.
- R. Janissen, B. A. Berghuis, D. Dulin, M. Wink, T. Van Laar and N. H. Dekker, Nucleic Acids Res., 2014, 42, e137 CrossRef.
- R. M. Evans, FEBS Lett., 1988, 234, 73–78 CrossRef CAS.
- D. A. Parry and P. M. Steinert, Intermediate filament structure, Springer-Verlag, 1995, p. 183 Search PubMed.
- A. J. Smith, J. Daut and B. Schwappach, Physiology, 2011, 26, 181–191 CrossRef CAS.
- L. Barberis, C. Pasquali, D. Bertschy-Meier, A. Cuccurullo, C. Costa, C. Ambrogio, F. Vilbois, R. Chiarle, M. Wymann, F. Altruda, C. Rommel and E. Hirsch, Eur. J. Immunol., 2009, 39, 1136–1146 CrossRef CAS.
- Y.-P. Lim, C. Y. Wong, L. L. Ooi, B. J. Druker and R. J. Epstein, Clin. Cancer Res., 2004, 10, 3980–3987 CrossRef CAS.
- Q. Zhu, K. Rosenblatt, K. Huang, G. Lahat, R. Brobey, S. Bolshakov, T. Nguyen, Z. Ding, R. Belousov, K. Bill, X. Luo, A. Lazar, A. Dicker, G. Mills, M. Hung and D. Lev, Oncogene, 2011, 30, 457–470 CrossRef CAS.
- J. Ivaska, H.-M. Pallari, J. Nevo and J. E. Eriksson, Exp. Cell Res., 2007, 313, 2050–2062 CrossRef CAS.
- R. C. Wang, Y. Wei, Z. An, Z. Zou, G. Xiao, G. Bhagat, M. White, J. Reichelt and B. Levine, Science, 2012, 338, 956–959 CrossRef CAS.
- M. E. Kidd, D. K. Shumaker and K. M. Ridge, Am. J. Respir. Cell Mol. Biol., 2014, 50, 1–6 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed description of the model and of material and methods, individual data sets and ESI figures. See DOI: 10.1039/d0nr07322a |
| This journal is © The Royal Society of Chemistry 2021 |