Open Access Article
Open Access ArticleDevelopment of a mitochondrial sirtuin 4 FRET assay based on its activity for removing 3-hydroxy-3-methylglutaryl (HMG) modification†
Yan
Li
a,
Yonglong
Zhao
a,
Zhuoxian
Cao
a,
Jie
Wang
a,
Ting
Liu
a,
Yongjun
Li
a,
Yuanyuan
Wang
*b and
Bin
He
 *a
*a
aState Key Laboratory of Functions and Applications of Medicinal Plants, Engineering Research Center for the Development and Application of Ethnic Medicine and TCM (Ministry of Education), Guizhou Provincial Key Laboratory of Pharmaceutics, School of Pharmacy, School of Basic Medicine, Guizhou Medical University, Guiyang 550004, China. E-mail: binhe@gmc.edu.cn; Fax: +86 851 6908218; Tel: +86 137 65113985
bDepartment of Endocrine and Breast Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, China
First published on 13th January 2021
Abstract
SIRT4 belongs to one of three mitochondrial sirtuins, which plays important roles in regulating many biological processes and has significant implications for treating several human diseases. However, the development of those small compounds that can modulate SIRT4 activities is limited because there is no efficient SIRT4 assay available for screening its modulators. Thanks to recent discoveries of several enzymatic activities and substrates for SIRT4, we have developed a FRET-based assay suitable for screening SIRT4 modulators based on its activity of removing 3-hydroxy-3-methylglutaryl (HMG) lysine modification, which could be further coupled with a secondary FRET-based assay for other sirtuins to identify SIRT4-specific inhibitors or activators.
Sirtuins are nicotinamide adenine dinucleotide (NAD)-dependent deacylases, which play crucial roles in regulating many biological processes such as aging, transcription, metabolism, and thus are related with many human diseases like cancer, neurodegeneration, diabetes, obesity.1,2 There are seven sirtuins in mammals, called SIRT1–SIRT7, respectively.3 They differ in different organelles and functions. SIRT4 is one of three mitochondrial sirtuins while SIRT5 and SIRT6 are the other two.3 Due to the key role of mitochondrial function in metabolism, SIRT4 have been implicated in the emergence and pathogenesis of several human diseases, mainly including diabetes, cancer, ageing, etc., by regulating metabolic processes in mitochondrion.4 For an example, SIRT4 can decrease insulin secretion in pancreatic β-cells stimulated by amino acid or glucose and further be proven the critical role of SIRT4 in type II diabetes by the fact that SIRT4 knockout mice help hyperinsulinemia, glucose intolerance, and insulin resistance.5–12 Additionally, SIRT4 in different cells or organs is highly associated with ageing.13–16 SIRT4 was shown to be upregulated during ageing or UV radiation, which is implicated in skin aging-related phenotypes.15,16 On the other hand, SIRT4 has been considered as a tumor suppressor. SIRT4 expression was found to be downregulated in many human cancers while SIRT4 can also inhibit glucose dehydrogenase (GDH) activity to decrease glutaminolysis and then glycolysis.17–19
Despite the significance of SIRT4 in both physiology and human diseases, there are still lack of SIRT4 modulators including inhibitors and activators for evaluating the potential of SIRT4 as a therapeutic target or for elucidating the better mechanistic understanding of SIRT4 in physiology.20 The major difficulty is to establish high-throughput assays for screening SIRT4 modulators since there is no robust enzymatic activities reported for SIRT4 until recently.20 Thanks to recent discoveries of several enzymatic activities and substrates for SIRT4 (Fig. 1), SIRT4 has been getting more and more understanding in biological and physiological functions.6,21–24 At the very beginning, SIRT4 had been shown very weak deacetylase activity but relatively detectable ADP-ribosylase activity.8 Later on, SIRT4 was found to possess substrate-specific deacetylase activity.21 For an example, SIRT4 can deacetylate malonyl-CoA decarboxylase (MCD) and thus inhibit its activity (Fig. 1). In 2014, a study demonstrated that SIRT4 can remove lipoyl and biotinyl lysine modifications more efficiently than acetyl lysine modification22 (Fig. 1). Finally, SIRT4 was further found to hydrolyse several negative charged modifications of 3-hydroxy-3-methylglutaryl (HMG), 3-methylglutaryl, 3-methylglutaconyl, and glutaryl modifications from lysine residues of substrates both in vitro and in vivo24 (Fig. 1).
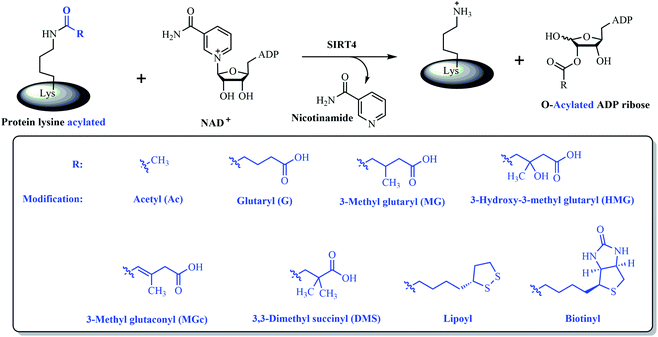 | ||
| Fig. 1 Some reported enzymatic activities for SIRT4 including removal of various lysine acyl modifications. | ||
Through the comparison of the reported catalytic efficiencies of SIRT4 on different acyl peptides in vitro, we can conclude that the catalytic efficiency of SIRT4 deacetylation, even for its sequence dependent deacetylation, is quite poor.20 Compared to deacetylation, SIRT4 showed slightly higher activities for hydrolysing biotinyl and lipoyl modifications with increasing several folds. Strikingly, although there are similar Kcat values for the modifications of HMG, lipoyl, and acetyl, their Km values are totally different.24 The best catalytic efficiency of SIRT4 was achieved by using carbamoyl phosphate synthetase 1, CPS1(524–531) K527HMG as a substrate although this is still lower than other sirtuin activities such as the deacetylation of SIRT2, desuccinylation of SIRT5 and demyristoylation of SIRT6.20 This discovery has enabled us to design a high-throughput assay to screen SIRT4 modulators.
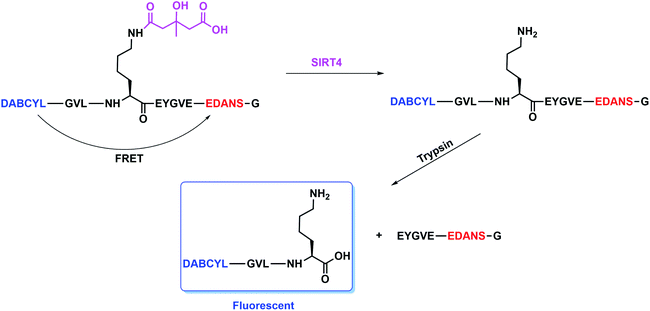
According to our previous strategy for the development of SIRT5 and SIRT6 assays,25,26 we would like to report a fluorescence resonance energy transfer (FRET)-based assay where a donor dye and a quencher dye are connected to a 3-hydroxy-3-methylglutaryl (HMG) peptide substrate for screen SIRT4 modulators. Furthermore, we also shown that combined with a secondary screen of other sirtuins (SIRT2/SIRT5/SIRT6) will identify whether those modulators are selective or not (Fig. 2).
The design is to assemble an HMG peptide with a pair of donor dye and quencher dye at the end of each terminal, respectively (Fig. 2). At the absence of SIRT4, there exists a FRET effect in this HMG peptide that the fluorescence of a donor dye would be quenched by a quencher dye. The FRET will be disrupted and the fluorescence from a donor dye will be released once the removal of HMG group by SIRT4 and then the digestion by trypsin sequentially happens. According to our previous work, we chose a FRET pair of 4-(dimethylaminoazo)benzene-4-carboxylic acid (DABCYL) and 5-(2-aminoethyl)aminonaphthalene-1-sulfonic acid (EDANS) as a pair of donor dye and quencher dye, which exhibits better sensitivity at Ex/Em (maximum excitation and emission) = 340 nm/490 nm (structures shown in Fig. S1 in the ESI†).27 According to the standard procedure of solid phase peptide synthesis shown in the ESI,† we made two peptides. There are a tryptophan containing peptide and a FRET peptide both with an HMG modification on the lysine residue (K527) of CPS1(524–531), whose sequences are GVLK(HMG)EYGVW and (DABCYL)GVLK(HMG)EYGVE(EDANS)G (Fig. 2), respectively.
At first, we used GVLK(HMG)EYGVW (a tryptophan containing peptide) to evaluate the catalytic efficiency of SIRT4 removing the HMG modification by an HPLC assay to detect substrate consumption and product formation at the wavelength of 280 nm. We measured the initial rate velocities as a function of substrate concentration and fit the data to the Michaelis–Menten equation to give the Km and Kcat values as shown in Table 1. The catalytic efficiency (Kcat/Km) for SIRT4 removing HMG modification was 827 M−1 s−1, which is consistent with that of the previously reported.24 Then, we turn to check the kinetic constants of SIRT4 on (DABCYL)GVLK(HMG)EYGVE(EDANS)G, a FRET peptide. Compared to the tryptophan containing peptide, the catalytic efficiency is slightly decreased with a value of 464 M−1 s−1, which is still sufficient to be a right substrate to develop the following SIRT4 assay.
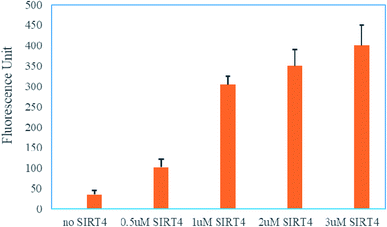
Encouraged by this, we next to test whether the (DABCYL)GVLK(HMG)EYGVE(EDANS)G peptide by coupled the SIRT4 enzymatic reaction and trypsin digestion could be used to read out the activity of SIRT4. In brief, the assay was carried out in one-hour incubation with different concentrations of SIRT4 and (DABCYL)GVLK(HMG)EYGVE(EDANS)G peptide (10 μM) followed by one-hour incubation with 6.25 U of trypsin. As shown in Fig. 3, the fluorescence was increased nearly 10-fold compared with the treatment of 1 μM (or above) SIRT4 and the control of no SIRT4. The fluorescence is also showing a dose-dependent manner against the different concentration of SIRT4. These results demonstrate that the (DABCYL)GVLK(HMG)EYGVE(EDANS)G peptide is a suitable FRET substrate for SIRT4 activity assay.
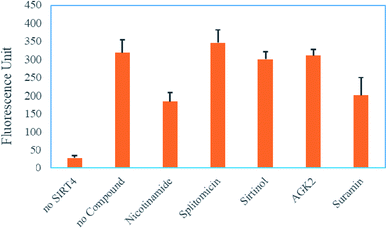
Next, to check whether this FRET-based assay could be utilized to pick up compounds that can modulate SIRT4, we chose several compounds known to be sirtuin inhibitors, including nicotinamide,28 splitomicin,29 sirtinol,30 AGK2 (ref. 31) and suramin32 (structures shown in Fig. S2, ESI†). Setting the concentration at 300 μM, we investigated the capability of these compounds to inhibit SIRT4 activity in this FRET-based assay. Among all the compounds tested, nicotinamide and suramin showed the best inhibition of near 50% at 300 μM. All other compounds showed less than 50% inhibition at 300 μM (Fig. 4).
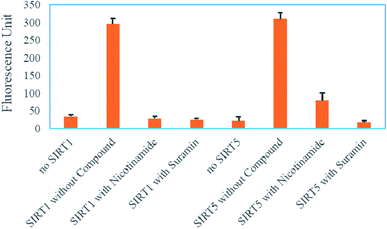
As we known, nicotinamide and suramin are pan sirtuin inhibitors. Therefore, we could perform the correspondingly secondary assay of either SIRT1 or SIRT5 to rule out those compounds that can also inhibit SIRT1 or SIRT5 after the screening of the primary assay of SIRT4. As shown in ESI,† we have utilized the (DABCYL)ISGASEK(Ac)DIVHSE(EDANS)G and (DABCYL)ISGASEK(Su)DIVHSE(EDANS)G peptides (a sequence derived from GDH(497–506) K503) to test whether nicotinamide and suramin also can inhibit the deacetylation of SIRT1 and the desuccinylation of SIRT5, respectively. As shown in Fig. 5, both nicotinamide and suramin at 300 μM also can significantly decrease the fluorescence produced by SIRT1 or SIRT5 to almost background level. The result is consistent with literatures that nicotinamide and suramin are pan inhibitors for almost all sirtuins28,32 and further demonstrates that the SIRT4 assay with this FRET peptide containing HMG lysine coupled with other sirtuin assays can be used to tell whether compounds can selectively modulate SIRT4 activity.
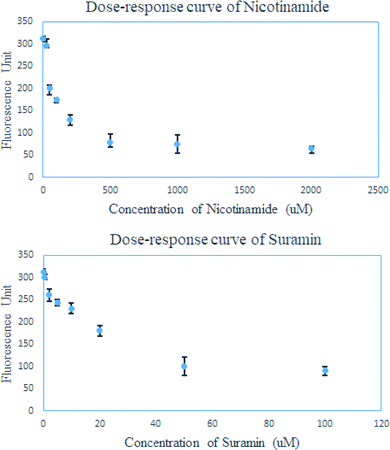
At last, we tested whether this FRET-based assay can evaluate the IC50 of SIRT4 inhibitors. Accordingly, the dose–response curve for nicotinamide and suramin were obtained using this FRET-based assay (Fig. 6). As shown in the dose–response curve, SIRT4 activity for the removal of HMG modification decreased in a dose-dependent manner at presence of either nicotinamide or suramin. The dose–response curve of nicotinamide in a concentration range of 25 μM to 2000 μM gave the estimated IC50 value of about 200 μM, while that of suramin in a concentration range of 0.5 μM to 100 μM gave the estimated IC50 value of about 40 μM (Fig. 6).
Conclusions
Among three mitochondrial sirtuins, SIRT3 and SIRT5 assays have been developed while SIRT4 assay is still lack.27 To solve this problem, we have developed a FRET-based assay for SIRT4, using an HMG lysine peptide containing a FRET pair of DABCYL/EDANS that can be used to screen for compounds that can modulate SIRT4 activity. Coupled with a secondary FRET-based assay for other sirtuins, inhibitors selective for SIRT4 could be identified. This assay is a “mix-and-measure” type of assay and should be easily miniaturized and automated for a high-throughput screening to identify SIRT4-specific inhibitors or activators.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
We thank the following for the financial support to this work: National Natural Science Foundation of China (No. 21662010 and 81960628), Guiyang Municipal Science and Technology Project (No. [2017]30-38 and [2017]30-18), Department of Science and Technology of Guizhou Province (No. [2019]2760, [2020]1Y395 and [2020]4Y206).References
- G. Blander and L. Guarente, The Sir2 family of protein deacetylases, Annu. Rev. Biochem., 2004, 73, 417–435 CrossRef CAS PubMed.
- P. Bheda, H. Jing, C. Wolberger and H. Lin, The Substrate Specificity of Sirtuins, Annu. Rev. Biochem., 2016, 85, 405–429 CrossRef CAS PubMed.
- R. A. Fyre, Phlogenetic classification of prokaryotic and eukaryotic Sir2-like proteins, Biochem. Biophys. Res. Commun., 2000, 273, 793–798 CrossRef PubMed.
- Y. Han, S. Zhou, S. Coetzee and A. Chen, SIRT4 and Its Roles in Energy and Redox Metabolism in Health, Disease and During Exercise, Front. Physiol., 2019, 10, 1006 CrossRef PubMed.
- C. Argmann and J. Auwerx, Insulin Secretion: SIRT4 Gets in on the Act, Cell, 2006, 126, 837–839 CrossRef CAS PubMed.
- K. A. Anderson, F. K. Huynh and K. Fisher-Wellman, et al., SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion, Cell Metab., 2017, 25, 838–855 CrossRef CAS PubMed.
- M. C. Haigis, R. Mostoslavsky and K. M. Haigis, et al., SIRT4 Inhibits Glutamate Dehydrogenase and Opposes the Effects of Calorie Restriction in Pancreatic β Cells, Cell, 2006, 126, 941–954 CrossRef CAS PubMed.
- N. Ahuja, B. Schwer and S. Carobbio, et al., Regulation of Insulin Secretion by SIRT4, a Mitochondrial ADP-ribosyltransferase, J. Biol. Chem., 2007, 282, 33583–33592 CrossRef CAS PubMed.
- J. X. Shi, Q. J. Wang, H. Li and Q. Huang, SIRT4 overexpression protects against diabetic nephropathy by inhibiting podocyte apoptosis, Exp. Ther. Med., 2017, 13, 342–348 CrossRef CAS PubMed.
- L. Ho, A. S. Titus, K. K. Banerjee and S. George, et al., SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK, Aging, 2013, 5, 835–849 CrossRef CAS PubMed.
- R. Song, W. Xu, Y. Chen, Z. Li, Y. Zeng and Y. Fu, The expression of sirtuins 1 and 4 in peripheral blood leukocytes from patients with type 2 diabetes, Eur. J. Histochem., 2011, 55, 55–59 CAS.
- E. Pirinen, G. Lo Sasso and J. Auwerx, Mitochondrial sirtuins and metabolic homeostasis, Best Pract. Res., Clin. Endocrinol. Metab., 2012, 26, 759–770 CrossRef CAS PubMed.
- Y. X. Luo, X. Tang and X. Z. An, et al., SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity, Eur. Heart J., 2017, 38, 1389–1398 CrossRef CAS PubMed.
- A. Lang, S. Grether-Beck and M. Singh, et al., MicroRNA-15b regulates mitochondrial ROS production and the senescence-associated secretory phenotype through sirtuin 4/SIRT4, Aging, 2016, 8, 484–505 CrossRef CAS PubMed.
- D. W. Wong, T. Soga and I. S. Parhar, Aging and chronic administration of serotonin-selective reuptake inhibitor citalopram upregulate Sirt4 gene expression in the preoptic area of male mice, Front. Genet., 2015, 6, 1–9 Search PubMed.
- K. Dong, E. Pelle, D. B. Yarosh and N. Pernodet, Sirtuin 4 identification in normal human epidermal keratinocytes and its relation to sirtuin 3 and energy metabolism under normal conditions and UVB-induced stress, Exp. Dermatol., 2012, 21, 231–233 CrossRef CAS PubMed.
- M. Miyo, H. Yamamoto and M. Konno, et al., Tumour-suppressive function of SIRT4 in human colorectal cancer, Br. J. Cancer, 2015, 113, 492–499 CrossRef CAS PubMed.
- S. M. Jeong, A. Lee, J. Lee and M. C. Haigis, SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma, J. Biol. Chem., 2014, 289, 4135–4144 CrossRef CAS PubMed.
- S. M. Jeong, C. Xiao and L. W. S. Finley, et al., SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism, Cancer Cell, 2013, 23, 450–463 CrossRef CAS PubMed.
- Y. Li, Y. Zhou and F. Wang, et al., SIRT4 is the last puzzle of mitochondrial sirtuins, Bioorg. Med. Chem., 2018, 26, 3861–3865 CrossRef CAS PubMed.
- G. Laurent, N. J. German and A. K. Saha, et al., SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase, Mol. Cell, 2013, 50, 686–698 CrossRef CAS PubMed.
- R. A. Mathias, T. M. Greco and A. Oberstein, et al., Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity, Cell, 2014, 159, 1615–1625 CrossRef CAS PubMed.
- K. A. Anderson, M. F. Green and F. K. Huynh, et al., SnapShot: Mammalian Sirtuins, Cell, 2014, 159, 956 CrossRef CAS PubMed.
- M. Pannek, Z. Simic and M. Fuszard, et al., Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features, Nat. Commun., 2017, 8, 1513 CrossRef PubMed.
- Y. Li, L. You and W. Huang, et al., A FRET-based assay for screening SIRT6 modulators, Eur. J. Med. Chem., 2015, 96, 245–249 CrossRef CAS PubMed.
- Y. Li, W. Huang and L. You, et al., A FRET-based assay for screening SIRT5 specific modulators, Bioorg. Med. Chem. Lett., 2015, 25, 1671–1674 CrossRef CAS PubMed.
- Y. Li, T. Liu and S. Liao, et al., A mini-review on Sirtuin activity assays, Biochem. Biophys. Res. Commun., 2015, 467, 459–466 CrossRef CAS PubMed.
- K. J. Bitterman, R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves and D. A. Sinclair, Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1, J. Biol. Chem., 2002, 277, 45099 CrossRef CAS PubMed.
- A. Bedalov, T. Gatbonton and W. P. Irvine, et al., Identification of a small molecule inhibitor of Sir2p, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 15113 CrossRef CAS PubMed.
- C. M. Grozinger, E. D. Chao and H. E. Blackwell, et al., Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening, J. Biol. Chem., 2001, 276, 38837 CrossRef CAS PubMed.
- T. F. Outeiro, et al., Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease, Science, 2007, 317, 516 CrossRef CAS PubMed.
- A. Schuetz, J. Min and T. Antoshenko, et al., Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin, Structure, 2007, 15, 377 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09424b |
| This journal is © The Royal Society of Chemistry 2021 |