Open Access Article
Open Access ArticlePhysiological effect of colloidal carbon quantum dots on Bursaphelenchus xylophilus†
Yi Han,
Yaqian Han,
Guicai Du,
Tingting Zhang,
Qunqun Guo,
Hong Yang,
Ronggui Li * and
Yuanhong Xu
* and
Yuanhong Xu *
*
College of Life Sciences, Qingdao University, Qingdao 266071, China. E-mail: lrg@qdu.edu.cn; yhxu@qdu.edu.cn
First published on 3rd February 2021
Abstract
Bursaphelenchus xylophilus (B. xylophilus) is a dangerous plant pest which could result in Pine Wild Disease (PWD). To investigate the physiological activity of B. xylophilus and utilize the fluorescent properties of quantum dots, carbon quantum dots (CQDs) using glucose as precursor were synthesized through a hydrothermal reaction. The properties of the CQDs strongly depended on the reaction temperature and reaction time. The transcriptome analysis was implemented to study the molecular toxicology mechanism of CQDs on B. xylophilus. Based on the analysis results, it can be concluded that CQDs have the potential to stimulate the detoxification process and fatty acid degradation mechanism of B. xylophilus. Through observing the biodistribution of CQDs, lifespan, locomotion and egg-laying behavior of B. xylophilus, the intestine was the main target organ of the CQDs, and the CQDs could affect the locomotion and reproduction activities of B. xylophilus.
1. Introduction
Pinewood nematode (PWN), Bursaphelenchus xylophilus (B. xylophilus) is a well-known and dangerous plant pest, which causes pine wild disease (PWD) in East Asia and Europe.1 B. xylophilus is usually 1 mm long, with insect vectors in the genus Monochamus. Similar to other nematodes, B. xylophilus has four juvenile stages and feeds on fungi and can be easily cultured and propagated. Due to the strong pathogenicity and rapid transmission, it leads to high-infection rates in pinewood. Only a few years from the onset of the disease could see the death of an entire pine forest, which is a great threat to natural resources and the environment. Many insecticides such as organophosphates, carbamates, emamectin benzonate, milbemectin and abamectin have been developed to restrain PWD.2,3 However, most current chemical insecticides are not environmentally friendly, and contaminate the soil, affect other biology and are a hazard to human life. Nanomaterials have been suggested as promising eco-friendly and harmless insecticides. There are many nanoparticles used as insecticides and nematicides against Meloidogyne incognita,4,5 which is a biotrophic parasite of crops. It is important to research the dependence of the physiological activity of B. xylophilus on nanomaterials.Colloidal quantum dots (QD) are semiconducting nanocrystals with size typically less than 20 nm, which have been widely used as building blocks in many applications, such as biosensing, bioimaging, biomedicine delivery system, chemical sensing, antimicrobial activity, and electrocatalysis, etc.6–8 Depending on their size/shape/composition and surface functional groups, colloidal QDs exhibit fresh luminescent properties, which provides an effective way to track the distribution of the QDs in vivo. Among QDs, carbon QDs (CQDs) have attracted a lot of attention due to their abundance, high quantum yield, low cost, and eco-friendly composition.9–14 Thus it is supposed that the CQDs could be used as efficient fluorescent probe for understanding the physiological activity of B. xylophilus through the CQDs bio-distribution.
On the other hand, some studies revealed that the toxicity of CQDs on cancer cells,15–17 mice,18 zebrafish,19 and Caenorhabditis elegans (C. elegans)20,21 was relatively low compared to inorganic QDs. While, there were also some studies reporting that CQDs had potential toxicity.22–24 C. pyrenoidosa was used to compare the toxicity between CQDs and metal-based QDs (MQDs), and the results suggested that the toxicity of the CQDs derived from biomass was less compared to the MQDs, but higher than the toxicity of CuInS2/ZnS QDs.22 Transcriptome analysis of zebrafish exposed to graphene QDs (GQDs) showed that potential bleeding risks and detoxifying processes were stimulated.23 In addition, zebrafish embryonic development could be disrupted by GQDs at high concentrations.24 Similarly, GQDs synthesized from electrochemical exfoliation were found to cause the death of U251 human glioma cells through oxidative stress.25 The mortality and immobility of D. magna zooplankton (Daphnia magna) were induced by CQDs, suggesting that its nervous system function was affected.26
Although many researches have used the CQDs for bio-application, there is still no available detailed study on the effect CQDs derived from glucose affect the physiological activity of B. xylophilus. In this work, we studied systematically the toxicity of the CQDs by choosing B. xylophilus as a simple model and concluded that CQDs could affect the locomotion and reproduction activities of B. xylophilus. CQDs were synthesized using glucose as precursor through hydrothermal reaction.27,28 The structure of the as-produced CQDs were characterized by Transmission electron microscopy (TEM) and Fourier transform infrared (FTIR) spectroscopy. The optical properties of the CQDs synthesized at different temperature and time were measured by fluorescence spectroscopy. Moreover, the bio-distribution of the as-obtained CQDs was studied based on the intrinsic fluorescence properties. In addition, the physiological effects of the CQDs on the B. xylophilus have been carried out through transcriptome analysis, lifespan, locomotion and egg-laying behaviour observation of B. xylophilus.
2. Methods
2.1 Preparation and characterization of the CQDs
The CQDs were prepared via a hydrothermal approach using glucose as precursors. Typically, glucose (1 g) was mixed with 10 mL of water. After 10 min sonication, the clear mixture was transferred into Teflon-lined autoclave (25 mL). The mixture was heated at 140 °C and 160 °C for different times (0.5–3 h), respectively, and naturally cooled down to room temperature (20 °C).Then the mixture was transferred into dialysis bags with a molecular weight of 500 Da for 6 days with changing the water every day. The CQDs solution inside the dialysis bag was collected for further experiment. The concentration of CQDs in the fresh-prepared solution was found at 6 mg mL−1 after purification. Absorption spectra were acquired with a UV-2600 UV-Vis spectrophotometer (Shimadzu). Steady state photoluminescence (PL) characterization of the as-prepared CQDs was performed on an Edinburgh FLS980 instrument. The quantum yield of CQDs was measured by using dye as reference. FTIR spectra were acquired with a thermo scientific (Nicolet iS5) spectrophotometer. TEM characterization of the CQDs was carried out using a JEOL 2100F.
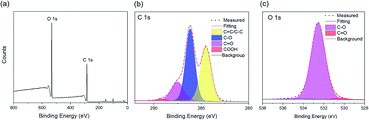
X-ray photoelectron spectroscopy (XPS) was performed in a Thermal Fisher equipped with a hemispherical analyzer recorded for a twin anode X-ray source. All the spectra were corrected to give the adventitious C 1s spectral component (C–C, C–H) a binding energy of 284.6 eV.
2.2 Extraction of B. xylophilus
B. xylophilus strain was isolated from diseased pinewood procured locally, which was cultured on potato dextrose agar plates at 26 °C for seven days. The Baermann funnel method was used to extract B. xylophilus from plates, and the proportion of adult male, adult female, and juvenile was approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, which was similar with the previous report.29 The suspension of nematodes (about 50 nematodes per μl) was collected by centrifugation (3 min, 3000 g) for RNA sequencing, bioimaging and activity observation.
2, which was similar with the previous report.29 The suspension of nematodes (about 50 nematodes per μl) was collected by centrifugation (3 min, 3000 g) for RNA sequencing, bioimaging and activity observation.
2.3 Lifespan, body bending rate and reproduction assay
Lifespan was measured in two ways to investigate the effect of different concentrations and different exposure time of certain concentrations, which was performed on 24 well plates. Twenty five synchronized B. xylophilus were exposed to the CQDs with different concentrations from 0.1 to 6 mg mL−1, 100 mg mL−1 glucose aqueous solution and sterilized distilled water, respectively. Then the lifespan of B. xylophilus in each treatment was recorded every day for five days. For locomotion assay, the number of thrashes within 1 minute was recorded to evaluate the vitality of nematodes. In addition, the number of eggs from female nematodes was counted. The microscope (OLYMPUS) was used for all these observations, which were replicated three times.2.4 Bioimaging
To investigate the possible effects of CQDs on B. xylophilus, three samples of the nematodes immersed in CQDs solution (4 mg mL−1) was considered as the G treatments. The treated nematodes were collected for confocal imaging. Worms were washed three times with sterilized distilled water and fixed with 10% formaldehyde. Subsequently, worms mounted on slides following standard procedures. Leica TCS-SP8 inverted confocal laser microscope was used. Images were viewed and processed with Leica image analysis software. Twenty nematodes were examined, and three replicates were performed.2.5 RNA extraction, cDNA library construction and sequencing and quality control
Six identical nematodes suspensions (1 mL each) were used for RNA extraction and sequencing. Of them, three suspensions were treated with CQDs (4 mg mL−1) as G treatments and the other three ones were exposed to the sterilized distilled water as CK. All the samples were cultured in the dark for 72 h, and further freezed by liquid nitrogen after centrifugation.Total RNA of six samples were qualified using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions. DNase I (TaKara, Dalian, China) was used to remove Genomic DNA. The RNA concentration and purity were quantified on a NanoDrop2000 (Thermo Fisher Scientific) and the quality was checked on the 2100 BioAnalyzer (Agilent). One microgram of total RNA was used for RNA-seq transcriptome libraries construction following TruSeqTM RNA sample preparation kit from Illumina (San Diego, CA). And Paired-end libraries were sequenced by Illumina NovaSeq 6000 sequencing (150bp*2, Shanghai BIOZERON Co., Ltd), which were trimmed and went through quality control by Trimmomatic with parameters (SLIDINGWINDOW:4:15 MINLEN:75) (version 0.36). The remaining clean reads were aligned to reference genome (PRJEA64437, http://www.wormbase.org) with orientation mode using hisat2 software. Htseq (https://htseq.readthedocs.io/en/release_0.11.1/) was used to count each gene reads.
2.6 Differentially expressed genes analysis
The fragments per kilobase of exon per million mapped reads (FRKM) method was used for each transcript to identify differentially expressed genes (DEGs) between the two different groups. And edgeR package in BioConductor was used to filter out DEGs using false discovery rate (FDR) ≤ 0.05 and log2 (fold-change) ≥2. Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) were used for GO functional enrichment and KEGG pathways analysis at Bonferroni-corrected from p < 0.05 to understand the functions of the DEGs.2.7 Gene expression validation via quantitative real-time PCR technique
Twenty-four DEGs (12 up-regulated DEGs, 12 down-regulated DEGs) were selected for gene expression validation quantitative real-time PCR (qRT-PCR) technique. The primers of selected gene were designed as shown in Table S1,† and the actin gene of B. xylophilus was selected as the internal control. Total RNA was extracted through the aforementioned method used in the RNA sequencing. And 10 μL of SYBR Green PCR Master Mix (2X), 0.4 μL of Prime F, 0.4 μL of Prime R, 7.2 μL of ddH2O and 2μL of cDNA samples were mixed into 20 μL of reaction, which was used for qRT-PCR through ABI Stepone plus (Applied Biosystems, Foster City, CA, USA). The procedure was carried out according to follow procedure: 95 °C for 3 minutes, 45 cycles of 95 °C for 5 s, and 60 °C for 30 s. All qRT-PCRs were conducted with three biological replicates performed in triplicate and 2−ΔΔCt method was used to for gene expression levels quantification.3. Results
3.1 Synthesis and structure of the CQDs
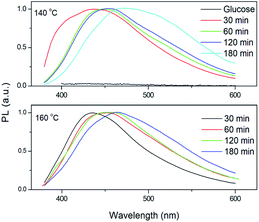
The CQDs were synthesized via a hydrothermal approach using glucose as the precursor. The morphology and structure of as-synthesized CQDs were characterized by TEM and high-resolution TEM (HRTEM), as shown in Fig. 1. The CQDs have a typical irregular-shape with a mean size of less than 10 nm (Fig. 1a). HRTEM image of an individual CQDs reveals the graphene crystalline structure of CQDs with a measured d spacing of around 2.1 Å, corresponding to the (100) plane of hexagonal graphene structure (Fig. 1b). These results indicate the successful synthesis of CQDs. Compared to glucose, the FTIR spectra of CQDs clearly show the peaks at 2919, 1712, 1657 cm−1 (Fig. 2). The peak at 2919 cm−1 indicates the presence of the C–H of the CQDs. The peaks at 1712, 1657 cm−1 correspond to stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. The peaks at 1154 and 1104 cm−1 for glucose almost disappeared because the C–O group is disappeared in the CQDs. The FTIR results revealed that the existence of carboxylate and hydroxyl groups on the surfaces of the CQDs. The quantum yield of CQDs (160 °C, 90 min) was 20%. The excitation-dependent photoluminescence spectra of the as-produced CQDs under 140 °C and 160 °C were provided in Fig. S1.†
O. The peaks at 1154 and 1104 cm−1 for glucose almost disappeared because the C–O group is disappeared in the CQDs. The FTIR results revealed that the existence of carboxylate and hydroxyl groups on the surfaces of the CQDs. The quantum yield of CQDs (160 °C, 90 min) was 20%. The excitation-dependent photoluminescence spectra of the as-produced CQDs under 140 °C and 160 °C were provided in Fig. S1.†
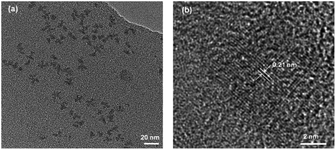 | ||
| Fig. 1 (a) Representative TEM image of CQDs synthesized at 160 °C for 3 h. (b) HRTEM of an individual CQDs. | ||
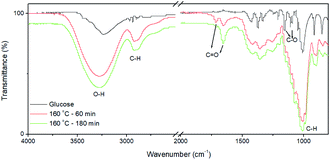 | ||
| Fig. 2 FI-IR spectra of fresh-prepared CQDs without post purification. The as-prepared reaction solution was dried for FI-IR measurement. | ||
3.2 Optical properties of the CQDs
As shown in Fig. 3a and b, the as-synthesized CQDs have an absorption spectrum ranging from 200–330 nm with an absorption peak at 283 nm. The influence of reaction time and temperature on the optical properties of the CQDs have been investigated in detail. With the increase of the reaction time, the absorbance of the CQDs gradually increased (Fig. 3c). There was no obvious difference for the samples synthesized at different temperature when the reaction was lower than 120 minutes. While the absorbance of CQDs synthesized at 160 °C was almost two-time higher than CQDs synthesized at 140 °C with the reaction time of 3 hours, indicating large amount of CQDs was formed at this reaction condition (Fig. 3c). All CQDs samples have similar PL spectra (Fig. 4). With the increasing reaction time, the PL spectrum shows a slightly red shift. As the longer reaction could lead to higher concentration of CQDs, there was no variation in absorption peak position of CQDs synthesized at different conditions (Fig. 3), the red-shift of the PL peaks with higher CQDs concentration may be due to the energy transfer between the CQDs with high concentration.30 Consistently, XPS further confirmed the prevalent chemical composition of the C-dots (160 °C, 180 min) after reaction (Fig. 5a–c). The high-resolution XPS C1s spectra indicate that the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C at 284.5 eV, C–O at 286.1 eV,C
C/C–C at 284.5 eV, C–O at 286.1 eV,C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 287.5 eV and COOH at 289.2 eV (Fig. 5b). We investigated the PL spectrum under initial concentration, 10 times dilution and 20 times dilution (Fig. S2†). Based on the data, we concluded that the red-shift is due to the structure variation of the CQDs, other than the energy transfer as we measured the CQDs in a very diluted solution (more than 10 time).
O at 287.5 eV and COOH at 289.2 eV (Fig. 5b). We investigated the PL spectrum under initial concentration, 10 times dilution and 20 times dilution (Fig. S2†). Based on the data, we concluded that the red-shift is due to the structure variation of the CQDs, other than the energy transfer as we measured the CQDs in a very diluted solution (more than 10 time).
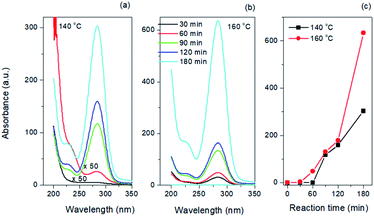 | ||
| Fig. 3 Absorption spectra of CQDs synthesized at (a) 140 °C and (b) 160 °C. (c) Absorbance of CQDs as a function of reaction time and temperature. | ||
3.3 Effects of CQDs exposure on lifespan, locomotion and egg-laying behavior of B. xylophilus
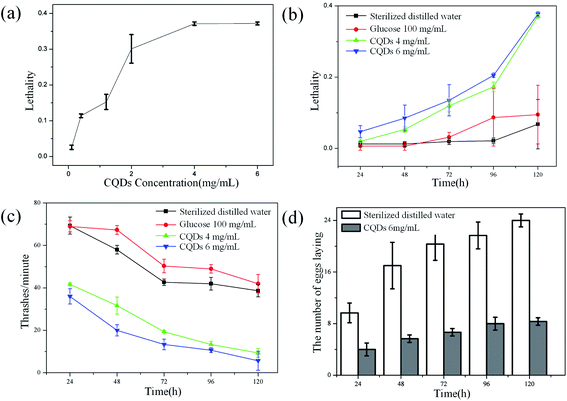
Effects of CQDs exposure on the B. xylophilus was evaluated in detail. To find out the possible effect of CQDs on lifespan, B. xylophilus was exposed to sterilized distilled water with different concentrations of CQDs ranging from 0.1 to 6 mg mL−1 for 1–5 days. The results are shown in Fig. 6a, which indicated that the lethality was relatively higher for CQDs at concentration over 4 mg mL−1 compared with the control. Besides, to find the relationship between the exposure time and the lethality, nematodes were processed with sterilized distilled water, glucose aqueous solution (100 mg mL−1), CQDs suspension at different concentrations of 4 mg mL−1 and 6 mg mL−1, respectively. Fig. 6b showed that the high concentration of CQDs affected the survival rate of nematode. After exposure with 120 h to CQDs with high concentration, the lethality was about 40%. However, the high concentration of glucose aqueous solution has no obvious impact on lifespan of B. xylophilus. We chose the concentration of 100 mg mL−1 for comparison of glucose since this was the initial concentration of precursors for CQDs synthesis. Similarly, the lethality of sterilized distilled water as control was lower than 1%, suggesting that the CQDs with high concentration (4 mg mL−1) have potential toxicity of altering the longevity of B. xylophilus. It was reported that 0.1 to 0.3 mg mL−1 of CdTe,31–33 which was defined as high concentration of semiconductor QDs, could affect the lifespan of C. elegan. However, the CQDs with concentration of 1.5 mg mL−1 also showed negligible toxicity.20 In this work, our results indicated that CQDs at higher concentration (over 4 mg mL−1) could induce serious influence of B. xylophilus.Besides, the effect of locomotion was evaluated. Under the same treatment as aforementioned, the number of thrashes was observed every minute, and the results are shown in Fig. 6c. The results were consistent with that of lifespan study. The frequency of thrashes of CQDs-processed nematode was relatively lower than that of control groups, although frequency of all four groups dropped down. As shown in Fig. 6d, the number eggs of CQDs-processed B. xylophilus were much lower than that of control group. All these observations suggested that the CQDs altered the locomotion and reproductive function of the B. xylophilus.
3.4 Whole body distribution of CQDs in B. xylophilus
Through observations of different nematodes, the distribution of CQDs was determined as shown in Fig. 7. The CQDs covered almost the whole body of the B. xylophilus in different amount (Fig. 7a and b), and the intestine was the main target organ since the CQDs distributed all over the intestine. The distribution in gonad was relatively low (Fig. 7d). Moreover, it was found that CQDs also accumulated in the head in certain amount (Fig. 7e).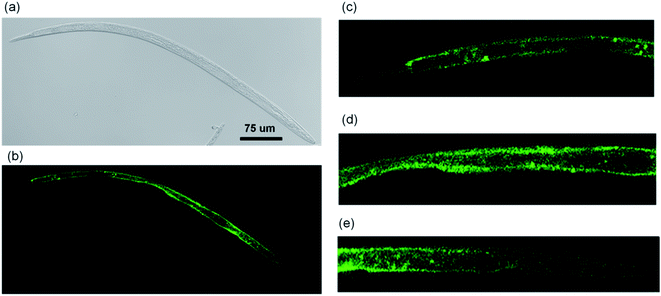 | ||
| Fig. 7 Whole body distribution of CQDs in B. xylophilus (a) DIC of the whole body. (b) Fluorescent image of the whole body. Fluorescent image of (c) tail part, (d) middle part and (e) head part. | ||
3.5 RNA sequencing
The RNA sequencing was implemented and more than 40.45 Mb clean-reads were obtained after quality control for each sample (Table 1). The Q20 values of all samples were over 98.10%. When mapping to the reference genome data set, the proportion of total reads ranged from 90.54% to 91.87%. After removing all transcripts with FPKM = 0, there were more than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 genes of each samples for analysis. Around 1680 genes were identified as DEGs between the CK and G treatments. Among these genes, 523 genes were up-regulated, and 1157 genes were down-regulated by the CQDs. DEGs related to the detoxification, fatty acid, locomotion and reproduction were focused in this study.
100 genes of each samples for analysis. Around 1680 genes were identified as DEGs between the CK and G treatments. Among these genes, 523 genes were up-regulated, and 1157 genes were down-regulated by the CQDs. DEGs related to the detoxification, fatty acid, locomotion and reproduction were focused in this study.
| Samples | Clean reads | Mapped reads | Mapped-rate (%) | Q20 | Q30 | FPKM > 0 |
|---|---|---|---|---|---|---|
| CK-1 | 45474946 | 41273060 | 90.76 | 98.37 | 94.64 | 16362 |
| CK-2 | 46042292 | 41686691 | 90.54 | 98.10 | 93.95 | 16386 |
| CK-3 | 40453300 | 36715415 | 90.76 | 98.52 | 95.03 | 16288 |
| G-1 | 49589808 | 45528402 | 91.81 | 98.27 | 94.4 | 16279 |
| G-2 | 45149436 | 41478786 | 91.87 | 98.51 | 95.02 | 16339 |
| G-3 | 40688886 | 36925164 | 90.75 | 98.47 | 94.89 | 16162 |
3.6 GO classification for DEGs of the B. xylophilus
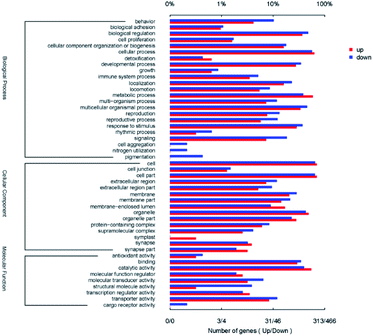
Gene Ontology (GO) analysis was utilized for functional annotation and classification of DEGs of nematode (Fig. 8). All DEGs were classified into 47 classes in the three categories of GO, including molecular function (9 classes), cellular component (15 classes) and biological process (23 classes). For the biological process, the up-regulated DEGs mostly participated in the cellular process (GO: 0009987, 62.9%), the metabolic process (GO: 0008152, 58.8%) and biological regulation (GO: 0065007, 36.7%). The down-regulated DEGs mainly focused on cellular process (GO: 0009987, 56.9%), biological regulation (GO: 0065007, 47.9%) and multicellular organismal process (GO: 0032501, 45.7%) in biological process category. In the cellular component categories, the three most prominent classes containing up-regulated DEGs and the down-regulated ones were cell (GO: 0005623, 70.3% and 65.0%), cell part (GO: 0044464, 70.3% and 64.4%) and organelle (GO: 0043226, 48.6% and 42.9%) successively. For the molecular function category, both the up-regulated and the down-regulated DEGs mostly belonged to the classes of catalytic activity (GO: 0003824, 55.0% and 40.0%) and binding (GO: 0005488, 30.0% and 34.8%). Among all DEGs, the gene BXY_0116800 which encodes propionyl-CoA carboxylase beta chain and participated in eight GO classes was up-regulated with the minimum Q value of 2.66 × 10−288. The gene BXY_1574200 was down-regulated with the minimum Q value of 2.39 × 10−164, which encodes alcohol dehydrogenase and participated in three GO classes. It was shown in Fig. 8 that reproduction (7.7%, 13.3%) and locomotion (5.4%, 8.6%) were also enriched with up-regulated and the down-regulated DEGs. All of these results indicated that the CQDs affected the viability of B. xylophilus, and changed the expression of the genes which related to the detoxification process.3.7 Pathways related to the detected genes
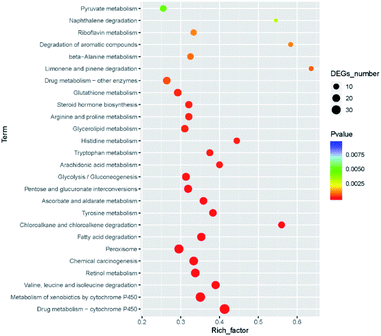
There were 250 KEGG pathways were annotated. After filtering the unimportant pathways, 26 significant pathways were selected (FDR < 0.05), including drug metabolism (38 DEGs), metabolism of xenobiotics by cytochrome P450 (34 DEGs). Among all the 26 pathways, there were five pathways participating in the xenobiotics biodegradation and metabolism, and five pathways participating in the amino acid metabolism. All 26 pathways and their P-values are shown in Fig. 9. The role of xenobiotics biodegradation and metabolism is to protect the B. xylophilus from the toxic substances, which indicated that the nematode response to CQDs toxicity.3.8 qRT-PCR validation of transcriptome data
We selected 24 DEGs to validate differential expression profiles by qRT-PCR technique. The actin expression was selected as the internal reference to normalize the transcriptional levels of these DEGs. The expression patterns of selected genes were consistent with RNA-seq data (Fig. S3†), which further confirmed the reliability and accuracy of the RNA-seq data.3.9 DEGs related to the detoxification process
Through the analysis of KEGG pathway and GO function, it was found that the most enriched pathways or classes were related to the detoxification process. For the KEGG pathways, drug metabolism, cytochrome P450 and metabolism of xenobiotics by cytochrome P450 were related to the detoxification process. In the GO analysis, the transferase activity and glutathione transferase activity were also enriched with DEGs. All detoxification-related DEGs (|log2(FC)| ≥ 2) are listed in Table S2,† including 10 DEGs of cytochrome P450 (CYP), 7 DEGs of UDP-glucuronosyl transferase (UGT) and 10 DEGs of glutathione S-transferase (GST), 1 DEG of carboxylesterase. Specifically, the expression level of BXY_1188700 (CYP) and BXY_1345300 (CYP) were up-regulated over 4.4 fold. The expression level of BXY_1673100 (UGT) was up-regulated by 5 fold. And the expression level of BXY_1297200 (GST) and BXY_0629000 (GST) were up-regulated 4.8 fold and 6.2 fold, respectively. All these remarkably up-regulated DEGs indicated that the CQDs activated the detoxification process.3.10 DEGs related to the fatty acid degradation
Moreover, the fatty acid degradation pathway (ko00071, 24 DEGs), fatty acid beta-oxidation activity (GO: 0006635, 26 DEGs) and long-chain fatty acid metabolic process (GO: 0001676, 35 DEGs) were significantly enriched with DEGs (Table S3†). Two Acyl-CoA synthetase family member DEGs (BXY_0104400, log2(FC): 7.31; BXY_0104500, log2(FC): 4.99), two propionyl-CoA carboxylase DEGs (BXY_0116800, log2(FC): 5.02; BXY_0419500, log2(FC): 4.49), and one acetyl-CoA acyl transferase DEG (BXY_0670900, log2(FC): 2.70) were up-regulated suggesting that the fatty acid β-oxidation process was stimulated.3.11 DEGs related to locomotion and reproduction
It was found that the CQDs could affect the function of locomotion and reproduction of the B. xylophilus. DEGs belonging to locomotion (GO: 0040011, 17 up-regulated DEGs, 40 down-regulated DEGs) and reproduction (GO: 0000003, 24 up-regulated DEGs, 62 down-regulated DEGs) were also enriched. All DEGs belonging to these two classes with |log2(FC)| ≥ 3 are listed (Tables S4 and S5†). All these DEGs were consistent with the results of locomotion and egg-laying observation.4. Discussion
B. xylophilus is a dangerous plant pest affecting the environment and economic. To our best knowledge, there isn't any report about the physiological effect of CQDs on B. xylophilus. To research on the potential toxicity of QDs on B. xylophilus, CQDs were utilized in this paper. Moreover, the transcriptome analysis was implemented to study the molecular toxicology mechanism of CQDs on B. xylophilus.Colloidal QDs are very attractive for bio applications, while it is extremely important to select the QD system with excellent performance but low toxicity. To date, various types of QDs have been synthesized and investigated in vitro or in vivo for its potential toxicity to human beings. For example, CdTe QDs were firstly found to be toxic to C. elegans under prolonged exposure at the concentration of 2.5 mg L−1, which was demonstrated through the reactive oxygen species (ROS) production.34 Although carbon based QDs do not have very toxic heavy-metals, researchers found that GQDs showed potential bleeding risks and detoxifying processes on zebrafish larvae through transcriptomic analysis.23 Rare minnow was used to evaluate the toxicity of CQDs and the results showed that spontaneous movements, body length, heart rate, etc. were all affected by the CQDs.35 Our results also showed that detoxification was the main affected process of the nematode after exposed to CQDs. 5 DEGs encoding cytochrome P450 (CYP), 6 DEGs encoding UDP-glucuronosyl transferase and 7 DEGs encoding glutathione S-transferase (GST) were up-regulated. The detoxification process of B. xylophilus contains three phases.36 In the first phase CYPs are used to generate enzyme substrates. There were 5 up-regulated DEGs encoding cytochrome P450 genes, which indicated that the detoxification process was activated. Also, P450 was important for nematode life cycle, and CYP33C9 gene affected the reproduction and pathogenicity of nematode.37 Some studies found that the P450 gene was up-regulated in responses to other toxic material.38,39 These P450 genes including CYP33C2, CYP33C4 and CYP33cC9 were associated with the detoxification mechanism of B. xylophilus.40 Both GSTs and UGTs are important and essential enzymes at the second phase of detoxification. Two UGT DEGs and three GST DEGs were up-regulated at least four-fold. The GSTs were also demonstrated to be involved in host defense and participated in the detoxification.41 All these results indicated that CQDs activated and affected the detoxification process of B. xylophilus.
Two DEGs encoding acyl-CoA and one DEG encoding acetyl-CoA were up-regulated. Acyl-CoA or acetyl-CoA are reported to be related to the fatty acid metabolism.42 Accordingly, we think that CQDs could stimulate the fatty acid metabolism of B. xylophilus. Moreover, the propionyl-CoA also will be generated in addition to acetyl-CoA,42 and two propionyl-CoA carboxylase DEGs were also up-regulated, which coincided with the results of the transcriptomic analysis. The graphene oxide QD (GOQD) was found to down regulate the genes related to unsaturated fatty acid biosynthesis of Chlorella vulgaris through both metabolomics and transcriptomics analysis.43
Because of the fluorescent feature of CQDs, the distribution of the CQDs in the B. xylophilus could be observed directly. The intestine was the primary targeted organ for toxicants which accumulated a large amount of CQDs. Similar observations were reported in C. elegans. The distribution of the CQDs generated from beetroot was mainly accumulated in the intestine of C. elegans,20 nitrogen-doped GQDs were also found distributed in the intestines of C. elegans.44 On the contrast, the nitrogen-doped CQDs were uniformly distributed within the whole body of C. elegans, indicating that the nitrogen-doped CQDs could stain proteins.45
Through the behavior observation of B. xylophilus, the CQDs were tested to affect the lifespan. In 72 hours, the survival rate didn't decrease obviously. When nematodes were exposed to the CQDs for more than 72 hours, the lethality reached 40% with high concentration of CQDs. This result suggested that the toxicity caused by CQDs may be accumulated to shorten the longevity of the B. xylophilus for long-term exposure. There is a similar finding that CdTe QDs could shorten the lifespan of C. elegans after 72 h exposure.46
Moreover, locomotion and egg deposition were also affected by the CQDs, which was consistent with the transcriptome data on the locomotion class and reproduction class. In the GO class of reproduction, 17b-hydroxysteroid dehydrogenases (17b-HSDs, BXY_0499000) was down-regulated over 3 fold, which played an important role in regulating androgen and estrogen concentrations.47 With exposure time increasing, the locomotion and egg deposition behavior were affected obviously. Similarly, the frequency of body bends decreased with CdTe QDs with concentration of 10 or 20 mg L−1.48 The CdTe QDs also induced egg-laying defects.49 However, the adverse effects of egg laying and locomotion were not detected in C. elegans exposed to nitrogen-dope GQDs44 and other CQDs.
5. Conclusions
In this paper, the CQDs were synthesized through hydrothermal reaction, and the quantity of which was highly depends on the temperature of reaction. The toxicity of the CQDs in B. xylophilus was evaluated through activity observation, bioimaging and transcriptome data analysis. The intestine of B. xylophilus was accumulated a large amount of CQDs. The lifespan, locomotion and reproduction system of B. xylophilus were altered by CQDs of high concentration after 72 h, which was consistent with the transcriptome data. The genes related to detoxification, fatty acid metabolism, locomotion and reproduction were overexpressed, suggesting that CQDs were chronic toxic and could stimulate the detoxification process. Because the method CQDs of synthesis is relatively easy, and eco-friendly, it could be used as a new nematicide.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Applied Basic Research Special Fund for Young Talents of Original Innovation Project of Qingdao (18-2-2-54-jch), Postdoctoral Applied Research Project of Qingdao (Grant Number 2015142) and National Natural Science Foundation of China (31901313).References
- J. T. Jones, A. Haegeman, E. G. J. Danchin, H. S. Gaur, J. Helder, M. G. K. Jones, T. Kikuchi, R. Manzanilla-López, J. E. Palomares-Rius, W. M. L. Wesemael and others, Mol. Plant Pathol., 2013, 14, 946–961 CrossRef.
- J. S. Kang, Y. S. Moon and S. H. Lee, Pestic. Biochem. Physiol., 2012, 104, 157–162 CrossRef CAS.
- S. K. Rajasekharan, J.-H. Lee, V. Ravichandran and J. Lee, Sci. Rep., 2017, 7, 1–13 CrossRef CAS.
- E. A. Mohamed, S. F. Elsharabasy and D. Abdulsamad, J. Parasitol., 2019, 2, 1–6 Search PubMed.
- M. A. Abbassy, M. A. Abdel-Rasoul, A. M. K. Nassar and B. S. M. Soliman, Arch. Phytopathol. Plant Prot., 2017, 50, 909–926 CrossRef CAS.
- X. He and N. Ma, Colloids Surf., B, 2014, 124, 118–131 CrossRef CAS.
- S. Sadhu and A. Patra, ChemPhysChem, 2013, 14, 2641–2653 CrossRef CAS.
- H. Song, Y. Su, L. Zhang and Y. Lv, Luminescence, 2019, 34, 530–543 CrossRef.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- Y. Wang and A. Hu, J. Mater. Chem. C, 2014, 2, 6921–6939 RSC.
- L. Cao, S.-T. Yang, X. Wang, P. G. Luo, J.-H. Liu, S. Sahu, Y. Liu and Y.-P. Sun, Theranostics, 2012, 2, 295 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, S. Perumal, N. Muthuchamy and Y. R. Lee, Fuel, 2020, 275, 117821 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, K. R. Aseer, S. Perumal and Y. R. Lee, Colloids Surf., B, 2018, 169, 321–328 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, K. R. Aseer, S. Perumal, N. Karthik and Y. R. Lee, Biosens. Bioelectron., 2018, 99, 303–311 CrossRef CAS.
- N. Md, K. Zehedina, H. K. Moo, P. Sung Young, L. D. Yun, C. Kwang Jae and L. Yong-Kyu, ACS Nano, 2013, 7, 6858–6867 CrossRef.
- R. Atchudan, T. N. J. I. Edison, S. Perumal, R. Vinodh and Y. R. Lee, J. Mol. Liq., 2019, 296, 111817 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, M. G. Sethuraman and Y. R. Lee, Appl. Surf. Sci., 2016, 384, 432–441 CrossRef CAS.
- Y. Chong, Y. Ma, H. Shen, X. Tu, X. Zhou, J. Xu, J. Dai, S. Fan and Z. Zhang, Biomaterials, 2014, 35, 5041–5048 CrossRef CAS.
- Y.-F. Kang, Y.-H. Li, Y.-W. Fang, Y. Xu, X.-M. Wei and X.-B. Yin, Sci. Rep., 2015, 5, 1–12 CrossRef.
- V. Singh, K. S. Rawat, S. Mishra, T. Baghel, S. Fatima, A. A. John, N. Kalleti, D. Singh, A. Nazir, S. K. Rath and others, J. Mater. Chem. B, 2018, 6, 3366–3371 RSC.
- R. Atchudan, T. N. J. I. Edison, S. Perumal, R. Vinodh and Y. R. Lee, Mater. Lett., 2020, 261, 127153 CrossRef CAS.
- A. Xiao, C. Wang, J. Chen, R. Guo, Z. Yan and J. Chen, Ecotoxicol. Environ. Saf., 2016, 133, 211–217 CrossRef CAS.
- S. Deng, P.-P. Jia, J.-H. Zhang, M. Junaid, A. Niu, Y.-B. Ma, A. Fu and D.-S. Pei, J. Hazard. Mater., 2018, 357, 146–158 CrossRef CAS.
- Z. G. Wang, Z. Rong, D. Jiang, E. S. Jing, X. U. Qian, S. I. Jing, Y. P. Chen, Z. Xin, G. A. N. Lu, J. Z. Li and others, Biomed. Environ. Sci., 2015, 28, 341–351 Search PubMed.
- Z. M. Markovic, B. Z. Ristic, K. M. Arsikin, D. G. Klisic, L. M. Harhaji-Trajkovic, B. M. Todorovic-Markovic, D. P. Kepic, T. K. Kravic-Stevovic, S. P. Jovanovic, M. M. Milenkovic and others, Biomaterials, 2012, 33, 7084–7092 CrossRef CAS.
- K. Yao, X. Lv, G. Zheng, Z. Chen, Y. Jiang, X. Zhu, Z. Wang and Z. Cai, Environ. Sci. Technol., 2018, 52, 14445–14451 CrossRef CAS.
- M. Shehab, S. Ebrahim and M. Soliman, J. Lumin., 2017, 184, 110–116 CrossRef CAS.
- T. Van Tam, S. G. Kang, K. F. Babu, E.-S. Oh, S. G. Lee and W. M. Choi, J. Mater. Chem. A, 2017, 5, 10537–10543 RSC.
- Q.-Q. Guo, G.-C. Du, T.-T. Zhang, M.-J. Wang, C. Wang, H. T. Qi and R. G. Li, J. Nematol., 2020, 52, 1–14 CrossRef.
- H. Zhao, G. Liu, S. You, F. V. A. Camargo, M. Z. Rossi, X. Wang, C. Sun, B. Liu, Y. Zhang, G. Han, A. Vomiero and X. Gong, Energy Environ. Sci., 2021, 14, 396–406 RSC.
- M. Qu, Y. Qiu, R. Lv, Y. Yue, R. Liu, F. Yang, D. Wang and Y. Li, Ecotoxicol. Environ. Saf., 2019, 173, 54–62 CrossRef CAS.
- E. Q. Contreras, M. Cho, H. Zhu, H. L. Puppala, G. Escalera, W. Zhong and V. L. Colvin, Environ. Sci. Technol., 2013, 47, 1148–1154 CrossRef CAS.
- S. Srivastava, A. Pant, S. Trivedi and R. Pandey, Environ. Toxicol. Pharmacol., 2016, 42, 55–62 CrossRef CAS.
- Y. Zhao, X. Yu, R. Jia, R. Yang, Q. Rui and D. Wang, Sci. Rep., 2015, 5, 17233 CrossRef CAS.
- Y.-Y. Xiao, L. Liu, Y. Chen, Y.-L. Zeng, M.-Z. Liu and L. Jin, BioMed Res. Int., 2016, 2016, 4016402 Search PubMed.
- T. Kikuchi, J. A. Cotton, J. J. Dalzell, K. Hasegawa, N. Kanzaki, P. McVeigh, T. Takanashi, I. J. Tsai, S. A. Assefa and P. J. A Cock, PLoS Pathog., 2011, 7(9), e1002219 CrossRef CAS.
- X. Qiu, L. Yang, J. Ye, W. Wang, T. Zhao, H. Hu and G. Zhou, Int. J. Mol. Sci., 2019, 20, 4520 CrossRef CAS.
- L.-X. He, X.-Q. Wu, Q. Xue and X.-W. Qiu, Int. J. Mol. Sci., 2016, 17, 778 CrossRef.
- X. Qiu, X. Wu, L. Huang, M. Tian and J. Ye, PLoS One, 8, 10, e78063 Search PubMed.
- Y. Li, F. Meng, X. Deng, X. Wang, Y. Feng, W. Zhang, L. Pan and X. Zhang, Int. J. Mol. Sci., 2019, 20, 911 CrossRef CAS.
- M. Espada, J. T. Jones and M. Mota, Nematology, 2016, 18, 697–709 CAS.
- S. M. Houten, S. Violante, F. V Ventura and R. J. A. Wanders, Annu. Rev. Physiol., 2016, 78, 23–44 CrossRef CAS.
- W. Kang, X. Li, A. Sun, F. Yu and X. Hu, Environ. Sci. Technol., 2019, 53, 3791–3801 CrossRef CAS.
- Y. Zhao, Q. Liu, S. Shakoor, J. R. Gong and D. Wang, Toxicol. Res., 2015, 4, 270–280 CrossRef CAS.
- R. Atchudan, T. N. J. I. Edison, S. Perumal, N. C. S. Selvam and Y. R. Lee, J. Photochem. Photobiol., A, 2019, 372, 99–107 CrossRef CAS.
- Q. Wang, Y. Zhou, B. Song, Y. Zhong, S. Wu, R. Cui, H. Cong, Y. Su, H. Zhang and Y. He, Small, 2016, 12, 3143–3154 CrossRef CAS.
- M. E. Baker, Mol. Cell. Endocrinol., 2001, 171, 211–215 CrossRef CAS.
- Z. Liu, X. Zhou, Q. Wu, Y. Zhao and D. Wang, RSC Adv., 2015, 5, 94257–94266 RSC.
- Y. Qu, W. Li, Y. Zhou, X. Liu, L. Zhang, L. Wang, Y. Li, A. Iida, Z. Tang, Y. Zhao and others, Nano Lett., 2011, 11, 3174–3183 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10144c |
| This journal is © The Royal Society of Chemistry 2021 |