Open Access Article
Open Access ArticleImmunoassay-aptasensor for the determination of tumor-derived exosomes based on the combination of magnetic nanoparticles and hybridization chain reaction†
Hua Zhang‡
a,
Yajuan Zhou‡b,
Dan Luoa,
Jingjian Liua,
E. Yangc,
Guangyi Yangc,
Guangjun Fengc,
Qinhua Chen *c and
Lun Wu*a
*c and
Lun Wu*a
aAffiliated Dongfeng Hospital, Hubei University of Medicine, Shiyan, 442008, Hubei, China. E-mail: wulun0909@163.com
bDepartment of Radiotherapy, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430074, China
cShenzhen Baoan Authentic TCM Therapy Hospital, Shenzhen, Guangdong 518101, China. E-mail: cqh77@163.com; Tel: + 86-0719-8272238
First published on 27th January 2021
Abstract
The detection of tumor-related exosomes is of great significance. In this work, a fluorescence aptasensor was designed for the determination of tumor-related exosomes based on the capture of magnetic nanoparticles (MNPs) and specific recognition of an aptamer. MNPs were used as substrates to capture the exosomes by modifying the CD63 antibody on the MNP surface. Probe 1 consists of PDL-1 aptamer sequence and a section of other sequences. PDL-1 expression was observed on the surface of exosomes; the aptamer of PDL-1 could combine with PDL-1 with high affinity. Thus, the immunoassay-type compounds of “MNPs–exosomes–probe 1” were formed. The other section of probe 1 triggered the HCR with probe 2 and probe 3 and formed the super-long dsDNA. The addition of GelRed resulted in the generation of an amplified fluorescence signal. The proposed design demonstrated a good linearity with the exosome concentration ranging from 300 to 107 particles per mL and with a low detection limit of 100 particles per mL. This aptasensor also exhibited high specificity for tumor-related exosomes, and was successfully applied in biological samples.
1. Introduction
The worldwide morbidity and mortality of cancer are increasing every year. Cancer is a serious threat to human life and health.1 Tumor markers play an important role in the early diagnosis and treatment of cancers.2 As effective cancer monitors, exosomes have shown promising advantages in liquid biopsies because of their noninvasive measurement and predictability of the disease.3 Exosomes are membrane-coated extracellular vesicles with a diameter of about 30–200 nm,4 and are secreted by many types of eukaryotic cells into the extracellular space.5 They are detectable in various bodily fluids, including blood, urine, saliva, breast milk and amniotic fluid.6 Exosomes carry large amounts of proteins, lipids, DNA, RNA, and other biological molecules, which can provide information about the originating tumor.7 For tumors, especially for malignant tumors, exosomes participate in the composition and regulation of the local microenvironment, which affect the growth and development of the tumor.8 Tumor-derived exosomes play crucial roles in cancer occurrence and progression.9 They have been recognized as promising biomarkers for an early noninvasive diagnosis of cancers.10However, the small size and low buoyant density of exosomes pose significant challenges for their separation and quantification from the complex biological fluids.11 At present, the recognized standard methods widely used for exosome quantification and analysis include transmission electron microscopy (TEM), dynamic light scattering (DLS), nanoparticle tracking analysis (NTA), atomic force microscopy (AFM), western blot (WB), enzyme-linked immunosorbent assay (ELISA), tunable resistive pulse sensing (RPS), flow cytometry and mass spectrometry (MS).11–14 All of the aforementioned methods are used to analyze one characteristic aspect of exosomes, e.g. TEM is usually used to characterize the morphological characteristics, DLS and NTA are used to analyse the size and concentration of exosomes. WB and ELISA are the methods used to analyze the marker proteins of exosomes.15 However, there are several drawbacks that limit the application scope of the above methods. First, all of the above-mentioned methods require state-of-the-art instruments, and these instruments are very expensive and thus increase the cost. Second, the selectivity for some specific exosomes is poor. Most importantly, the accuracy of exosome determination is affected by unknowable nanoparticle counting, which formed by proteins aggregate and lipoprotein particles, and even some exosomes less than a certain size being ignored.12 Fortunately, with in-depth research, some advanced technologies with recognition element for exosome assay have been developed.
Recently, considering the importance of exosomes in the liquid biopsy of cancer, many studies have explored new detection techniques over the past few decades.16 A series of novel technologies have been developed for the determination of exosomes, including fluorescence detection,17 electrochemical biosensors,5,14,18,19 immunosensors,13,20 surface plasmon resonance (SPR),21–23 microfluidic devices,21 electrochemiluminescence,24 aptasensors,25 and spectroscopic analysis.26 However, these established exosome assays usually involve labour-intensive steps and require extended quantitative protocols and expensive laboratory infrastructures. They suffer from low sensitivity and high-cost and thus are impractical for wide use. In addition, most of these strategies are unable to distinguish cancer cell-derived exosomes from the normal exosomes, which limits their application in clinical cancer diagnosis. Based on this, we designed a novel biosensor that could with high specificity determine the concentration of tumor-related exosomes by measuring the fluorescence intensity, which combined GelRed with magnetic nanoparticles (MNPs).
In this design, aptamer plays an extremely important role. Aptamers are single-stranded oligonucleotides folding into specific tertiary structures, which bind to their molecular targets, including small molecules, peptides, proteins and cells with high specificity and affinity.27,28 Aptamers have been termed “chemical antibodies” and predicted to replace antibodies in the near future.29 Therefore, the aptamer of PDL-1 can bind with PDL-1 specifically in this research. For signal amplification, hybridization chain reaction (HCR) is one of the most popular techniques to improve the detection sensitivity.30 Thus, this signal amplification method can significantly improve the sensitivity for the determination of tumor-related exosomes. In this work, we used the exosomes triggering the HCR and obtained the super-long dsDNA. The GelRed, being a stable, sensitive and environmentally friendly fluorescent nucleic acid staining dye,31,32 could chimerize into the dsDNA structure and generate a strong fluorescence signal.33 Thus, we could realize the detection of tumor-related exosomes by measuring the final fluorescence signal.
2. Material and methods
2.1 Reagents
All synthetic DNA sequences and aptamer were purchased from Sangon Biotechnology Co. Ltd. (Shanghai, China) and are listed in Table 1. 10% fetal bovine serum was purchased from Sigma-Aldrich (St. Louis, MO, USA). Anti-CD63 antibody was purchased from Abcam (Cambridge, MA, UK), and carboxylated MNPs were bought from XFNANO Co., Ltd. (Nanjing, China). The GelRed (1 × 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was bought from Solarbio (Beijing, China). A549 non-small cell lung cancer (NSCLC) cells and BEAS-2B normal human bronchial epithelial cells were obtained from Procell (Wuhan, China). Ultrapure water prepared from a Millipore water purification system (a resistivity of 18 MΩ cm, Milli-Q Direct 8) was used in all the runs.
000) was bought from Solarbio (Beijing, China). A549 non-small cell lung cancer (NSCLC) cells and BEAS-2B normal human bronchial epithelial cells were obtained from Procell (Wuhan, China). Ultrapure water prepared from a Millipore water purification system (a resistivity of 18 MΩ cm, Milli-Q Direct 8) was used in all the runs.
| Name | Sequences |
|---|---|
| Aptamer of PDL-1 | 5′-ACG GGC CAC ATC AAC TCA TTG ATA GAC AAT GCG TCC ACT GCC CGT-3′ |
| Probe 1 | 5′-AGC TTT AAT TAA CAT GTC CGA CTTT ACG GGC CAC ATC AAC TCA TTG ATA GAC AAT GCG TCC ACT GCC CGT-3′ |
| Probe 2 | 5′-GTC GGA CAT GTT AAT TAA AGC TTA GCA TCG ACT AGC TTT AAT TA-3′ |
| Probe 3 | 5′-AGC TTT AAT TAA CAT GTC CGA CTA ATT AAA GCT AGT CGA TGC TA-3′ |
| Probe 4 | 5′-AGC TTT AAT TAA CAT GTC CGA C ATC ATC GTA TTA CCG AAC TCG GTA CAT-3′ |
2.2 Cell culture
All cell culture media were supplemented with 10% fetal bovine serum, 1% 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin at 37 °C in a humidified 5% CO2 incubator. The conditioned medium was collected after 48 h following serum-free defined medium. Exosomes were isolated using the standard ultracentrifugation method with slight modifications. A549 non-small cell lung cancer (NSCLC) cells and BEAS-2B normal human bronchial epithelial cells were cultured in RPMI 1640 and DMEM, respectively, with 10% FBS, at 5% CO2 in culture bottles.2.3 Extraction of exosomes
The supernatant was collected from the conditioned medium after A549 and BEAS-2B cells were cultured in a serum-free defined medium for 48 h. Then, standard ultracentrifugation with slight modifications was employed to extract exosomes from the supernatant. First, the medium was centrifuged at 300g at 4 °C for 10 min to remove the residual cells. Second, the cell debris and apoptotic vesicles were removed by centrifugation at 2000g for 20 min. The supernatant was collected and filtered through a filter with a pore size of 0.22 μm (Millipore). The filtrate was further centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min to remove the cell vesicles and biological macromolecules. Subsequently, the supernatant was centrifuged at 100
000g for 30 min to remove the cell vesicles and biological macromolecules. Subsequently, the supernatant was centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 70 min, and the sediment was resuspended in 20 mL sterile phosphate-buffered saline (PBS). Then, the resuspension was pelleted again by ultracentrifugation for 2 h at 100
000g at 4 °C for 70 min, and the sediment was resuspended in 20 mL sterile phosphate-buffered saline (PBS). Then, the resuspension was pelleted again by ultracentrifugation for 2 h at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C. Finally, the supernatant was discarded, and exosomes were obtained as residual sediments. The extracted exosomes were resuspended in 200 μL PBS and was ready for use.
000g at 4 °C. Finally, the supernatant was discarded, and exosomes were obtained as residual sediments. The extracted exosomes were resuspended in 200 μL PBS and was ready for use.
2.4 Characterization of exosomes
2.5 Fluorescent aptasensor construction and exosome determination
The strategy of exosome determination was carried out by following the procedure below: the carboxylated MNPs (20 μg mL−1) were activated with a mixture of 200 mM EDC and 50 mM NHS for 30 min. The anti-CD63 antibody (10 μg mL−1) was modified on the above prepared MNPs for 2 h. Then, the MNPs were blocked with BSA solution (100 μg mL−1) for 2 h. Consecutively, 100 μL of NTA-calibrated exosome solution was added and incubated at 37 °C for 30 min. The marker protein CD63 on the exosome surface can specifically bind to the CD63 antibody on the surface of the MNPs, which made the MNPs capture the exosomes. Then, 20 nM probe 1 was added and incubated for 30 min. Probe 1 (DNA sequence) consisting of an aptamer of PDL-1 and an initiator of HCR can bind to exosomes and form the complex of “MNPs–exosomes–probe 1”. A mixture of probe 2 (40 nM) and probe 3 (40 nM) strands was heated at 95 °C for 5 min, followed by slow cooling to room temperature. Then, the mixture was incubated with the above obtained MNPs for 30 min at 37 °C. Probe 2 and probe 3 hybridized with the HCR initiator and extended the hybridization chain. Then, GelRed (1 × 600) was added to the mixture. All reaction solutions were PBS solution and the volume was 1 mL. The modified MNPs were obtained through magnetic separation in each step and washed with PBS for 3 times. The content of exosomes was determined by detecting the final fluorescence intensity.2.6 Fluorescence measurements
The fluorescence measurement was carried out at room temperature on a Hitachi F-4600 spectrophotometer (Hitachi Co. Ltd., Japan, https://www.hitachi.co.jp) equipped with a xenon lamp as the excitation source. The excitation wavelength was set at 518 nm. The fluorescence spectra were collected at an emission wavelength of 580–750 nm. The optimum experimental parameters were studied by using the fluorescence intensity at 606 nm. The performance of this designed detection system was also evaluated. The excitation and emission intervals were both set at 5 nm. Except for the special groups, the fluorescence intensity of each group was measured three times and the standard deviation was calculated. The quantitative determination of exosomes was expressed by the final fluorescence intensity.2.7 Application in clinical blood samples
The blood samples collected from 8 normal human subjects and 8 lung cancer patients were added with an appropriate amount of thrombin and were centrifuged at 3000g at 4 °C for 5 min. Afterward, the supernatant samples were further filtered through a 0.8 μm pore filter. Finally, the exosomes were collected by centrifuging the obtained filtrate at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 60 min. All experiments in this study were conducted in strict accordance with the institutional regulations and relevant laws (National Health Commission of the People's Republic of China, Ethical Review of Biomedical Research Involving Human Beings). The plasma of patients with lung cancer and the healthy human plasma were acquired from Affiliated Dongfeng General Hospital, Hubei University of Medicine. This study was approved by the Dongfeng General Hospital's Ethics Committee (Shiyan, China). Informed consent was obtained for any experiment performed on patients with lung cancer and healthy human subjects.
000g at 4 °C for 60 min. All experiments in this study were conducted in strict accordance with the institutional regulations and relevant laws (National Health Commission of the People's Republic of China, Ethical Review of Biomedical Research Involving Human Beings). The plasma of patients with lung cancer and the healthy human plasma were acquired from Affiliated Dongfeng General Hospital, Hubei University of Medicine. This study was approved by the Dongfeng General Hospital's Ethics Committee (Shiyan, China). Informed consent was obtained for any experiment performed on patients with lung cancer and healthy human subjects.
3. Results and discussion
3.1 Principle of exosome determination
As shown in Fig. 1, the determination mechanism of exosome is based on the recognition of the antibodies and the specific binding of aptamers to the target. CD63 is the marker protein expressed on the surface of exosomes.34 The surface of the carboxylated MNP was modified with CD63 antibody, which could capture exosomes specifically. PDL-1 was expressed on the surface of the tumor-related exosomes,35,36 so that probe 1 (consisting of aptamer of PDL-1 and initiator of HCR) could bind with the complexes of “MNPs–exosomes” and form “MNPs–exosomes–probe 1”. With the addition of probe 2 and probe 3, the initiator of HCR could bind with probe 3 and made the HCR occur in probe 2. With the addition of GelRed and the extension of probe 2/probe 3 hybrid duplex, GelRed formed a soluble complex with nucleic acids and generated fluorescence signals. Thus, the concentration of the tumor-associated exosome could be quantified specifically by the measurement of the final fluorescence intensity.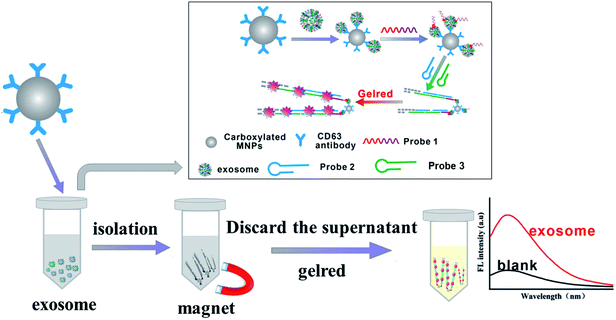 | ||
| Fig. 1 Schematic illustration of the immunoassay-aptasensor for exosome detection based on the combination of MNPs and HCR. | ||
3.2 Characterization of exosomes
NSCLC-specific exosomes were isolated from the culture medium of A549 NSCLC cell lines by ultracentrifugation. In order to confirm the successful extraction of exosomes, the morphology of the exosomes was observed by using TEM. In addition, the concentration and size distribution of the exosome were analyzed by NTA. As shown in Fig. 2a, we can clearly see the presence of exosomes with different sizes, and the average diameter is about 150 nm in TEM. The concentration and diameter of the extracted exosomes can be seen in Fig. 2b. The exosomes were resuspended in PBS and the mother liquor was diluted for 50 times. The detecting concentration of exosomes was 3.7 × 108 particles per mL, and the actual concentration of exosomes was about 1.85 × 1010 particles per mL with a size distribution of 50 to 200 nm. The above characterization results showed that a large amount of exosomes with narrow and uniform size had been extracted successfully.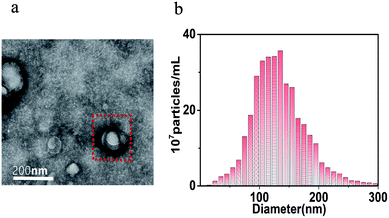 | ||
| Fig. 2 Characterization of exosomes. (a) The TEM image of exosomes and (b) the NTA analysis of exosomes. | ||
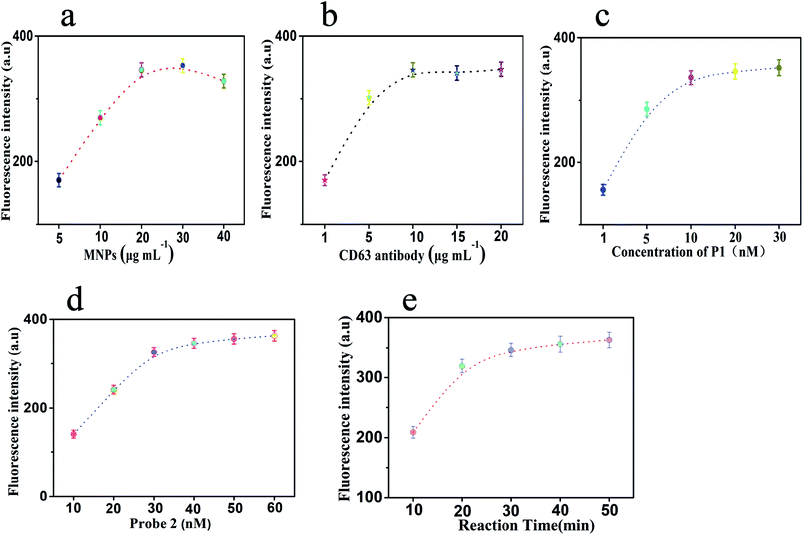
3.3 Experimental condition optimization
During the experimental process, there were five parameters that affected the sensing efficiency. In order to obtain the ideal performance for the determination of exosomes, these conditions were optimized: (a) the concentration of MNPs, (b) the concentration of CD63 antibody, (c) the concentration of probe 1, (d) the concentration of probe 2, and (e) the reaction time of HCR.It was worth noting that as the substrate for capturing exosomes, the concentration of MNPs affects the efficiency of exosome determination. Under different concentrations of MNPs, there were several changes in the final fluorescence intensities. Fig. 3a shows that the fluorescence signal changes with an increase in the concentration of MNPs from 5 to 40 μg mL−1. The fluorescence signal stabilized when the concentration of MNPs was 20 μg mL−1. We chose the point where the fluorescence intensity became stable as the optimal concentration. MNPs were utilized as the substrate to capture exosomes. The exosomes would aggregate at a high concentration of MNPs. Moreover, at low concentrations of MNPs, less exosomes were captured. About 20 μg mL−1 was found to be the optimum concentration of MNPs. The concentration of CD63 antibody played a significant role in the sensing performance of this sensor. The concentration of CD63 antibody affected the formation of the antibody modified MNPs and whether this method can detect exosomes. The results shown in Fig. 3b display significant changes in the fluorescence intensities at a series of CD63 antibody concentrations. The relatively steady fluorescence intensity was observed when the concentration of CD63 antibody was 10 μg mL−1. When the concentration of CD63 antibody was decreased gradually, the fluorescence intensity decreased dramatically. Furthermore, when the concentration of CD63 antibody was increased to 15 μg mL−1, the change in fluorescence was almost negligible and tended to be stable. Therefore, the optimal concentration of CD63 antibody was 10 μg mL−1, which could be used in the subsequent experiments.
At the same time, the concentration of probe 1 would affect the connection between the MNPs and the exosomes. It would also affect whether this method can detect tumor-related exosomes. Thus, we chose different concentrations of probe 1 to optimize. Fig. 3c shows the results. With an increase in the concentration of probe 1 from 1 to 30 nM, the fluorescence intensity increased gradually and then tended to be steady. The changes in the fluorescence intensity with respect to the concentration of probe 1 at 20 nM could be neglected. A relatively stable fluorescence intensity was observed when the concentration of probe 1 was 20 nM. Therefore, 20 nM was considered as the best concentration of probe 1. The concentration of probe 2 affects the formation and extension of the hybrid duplexes (probe 2/probe 3) and further affects the dyeing efficacy of GelRed with probe 2/probe 3 to generate fluorescence signals. As shown in Fig. 3d, with an increase in the concentration of probe 2 from 10 to60 nM, the fluorescence intensity increases gradually and tends to be stable. When the concentration of probe 2 was 40 nM, the fluorescence intensity reached a relatively stable state. When the concentration was increased to 50 nM or 60 nM, there was a slight change in fluorescence. To obtain a better performance, we chose 40 nM as the optimal concentration.
Finally, the reaction time of HCR was another significant parameter that affected the fluorescence intensity. As shown in Fig. 3e, with an increase in the reaction time, the fluorescence intensity increases first and then becomes steady. A relatively stable fluorescence intensity was observed when the reaction time was 30 min. After 30 minutes, the signal continued to increase when prolonging the reaction time. However, with the extension of reaction time, the background signal will also increase. An increase in the background signal was not conducive to reducing the minimum detection limit of the method. Considering the influence, when the reaction time was 30 min, better experimental results could be obtained in a shorter time. On the other hand, MNPs will be inactivated if the reaction time was too long. In addition, the shorter reaction time will lead to an incomplete dyeing of GelRed and probe 2/probe 3 duplexes. Therefore, the reaction time of 30 min was regarded as the optimum reaction time for the rest of the experiments.
3.4 Analytical performance
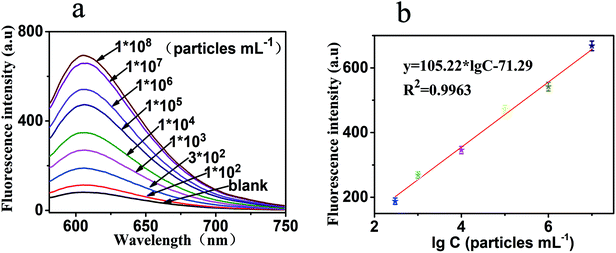
Under the above optimal parameters, the analytical performance of this sensor was evaluated at different concentrations of exosomes. Each experiment was repeated three times, and the result was the mean value of three measurements. There were numerous PDL-1 on the surface of the exosomes, and one exosome can combine with multiple probe 1 to form the 1:N signal amplification mode. Secondly, one probe 1 can also induce HCR to obtain a super long double-stranded DNA, which formed exponential amplification of the signal. Therefore, an increase in the concentration of exosomes could increase the signal exponentially. As shown in Fig. 4a, of the 9 different samples we tested, 6 samples show a linear relationship. The fluorescence intensity increased gradually as the concentration of exosomes was increased from 0 to 1 × 108 particles per mL. Furthermore, a good linearity between the fluorescence intensity and the logarithm to the base 10 of exosome concentration was observed. The linear regression equation was y = 105.22 × lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C − 71.29 (R2 = 0.9963), where y and lg
C − 71.29 (R2 = 0.9963), where y and lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C, respectively, represented the fluorescence intensity and the logarithm of exosome concentration. These data can be seen in Fig. 4b. The detection limit of this method for exosome determination was calculated to be as low as 100 particles per mL, and the linear range was 300 to 107 particles per mL. As shown in Table 2, the analytical performance of this strategy was compared with the other recently reported literature.
C, respectively, represented the fluorescence intensity and the logarithm of exosome concentration. These data can be seen in Fig. 4b. The detection limit of this method for exosome determination was calculated to be as low as 100 particles per mL, and the linear range was 300 to 107 particles per mL. As shown in Table 2, the analytical performance of this strategy was compared with the other recently reported literature.
| Analytical method | Detection limit | Linear range | Ref. |
|---|---|---|---|
| Fluorescence detection | 3.12 × 103 particles per μL | 1.0 × 104 to 5.0 × 105 particles per μL | 12 |
| Fluorescence detection | 1.0 × 105 particles per μL | 1.0 × 105 to 1.0 × 109 particles per μL | 17 |
| Electrochemical biosensor | 1.72 × 104 particles per mL | 1.0 × 105 to 5.0 × 107particles per mL | 5 |
| Electrochemical biosensor | 7.83 × 103 particles per μL | 9.5 × 103 to 1.9 × 107 particles per μL | 14 |
| Electrochemical biosensor | 1.44 × 103 particles per mL | 2.38 × 104 to 2.38 × 109 particles per mL | 19 |
| Immunosensor | 1.4 × 108 particles per mL | — | 13 |
| Immunosensor | 96 particles per μL | 1.12 × 102 to 1.12 × 108 particles per μL | 20 |
| Surface plasmon resonance | 3.86 × 108 particles per mL | — | 21 |
| Surface plasmon resonance | 5 × 103particles per mL | — | 23 |
| Microfluidic devices | 2.18 × 109 particles per mL | — | 21 |
| Electrochemiluminescence | 37.0 particles per μL | 1.1 × 102 to 1.1 × 107 particles per μL | 24 |
| Aptasensor | 3.94 × 105 particles per mL | 8.3 × 105 to 5.3 × 107 particles per mL | 25 |
| Immunoassay-aptasensor | 100 particles per mL | 300 to 107 particles per mL | This sensor |
In order to verify the specificity of this biosensor for exosome determination, control experiments were carried out by using cTnI, probe 4, normal cells and the blank group. We used cTnI instead of CD63 to modify MNPs, probe 4 instead of probe 1, and the BEAS-2B normal human bronchial epithelial cells replaced A549 NSCLC cells; results are shown in Fig. 5. Testing these five different groups under the same conditions exhibited the specificity of this method for exosome determination. Compared with the blank group, there were weak and negligible fluorescence peaks observed in the other three groups (including cTnI, probe 4, and normal cells). In the experimental group of exosomes from A549 NSCLC cells, there was a strong and obvious fluorescence peak. All of these results suggested that this sensor could only achieve the specific determination of tumor-related exosomes, and neither exosomes secreted by the normal cells nor other reagents replacing the originals. The excellent specificity of our strategy was mainly attributed to the high affinity and specific recognition between the aptamers (or antibodies) and the targets.
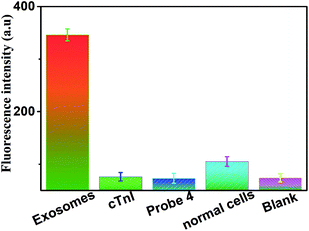 | ||
| Fig. 5 Fluorescence intensity (at an emission wavelength of 606 nm) of the sensor in the presence of exosomes, cTnI, probe 4, normal cells and blank, respectively. Error bars: SD, n = 3. | ||
3.5 Application in biological samples
In order to investigate the feasibility and practical application of this sensor in real biological samples, we obtained plasma from patients with lung cancer (LC) and healthy human subjects. We extracted exosomes from 8 normal human subject plasma and 8 LC patient plasma; other conditions remained identical. The exosomes of the two groups were treated as described previously, added into the mixtures and measured the final fluorescence intensity. The results are shown in Fig. 6, and the fluorescence intensities of the samples from LC patients are stronger than that of the group of healthy human subjects. Furthermore, the difference in the fluorescence intensity between the two groups was statistically significant (P < 0.0001). These results indicated that the sensor could be applied for the analysis of exosomes from biological samples.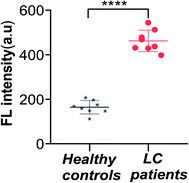 | ||
| Fig. 6 Fluorescence intensity of the sensor for the detection of exosomes from LC patients and healthy controls (****P < 0.0001). Error bars: SD, n = 3. | ||
4. Conclusion
A fluorescent aptasensor was developed for the determination of exosomes. Based on the specific recognition of immunoassay, high specificity and sensitivity, the determination of LC-related exosomes was completed with a wide linearity range (300 to 107 particles per mL) and a low detection limit of 100 particles per mL. The proposed immunoassay-aptasensor could also be applied for the analysis of clinical samples, which has a practical application value and potential significance. Considering the wide presence of exosomes in types of tumor-derived biological fluids, by binding with the different aptamers of exosome marker protein, this strategy has broad application prospects in early diagnosis, treatment evaluation, as well as monitoring and prognosis of various cancers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
These works were supported by the National Natural Science Foundation of China (81872509 and 81803065), the Hubei Province Health and Family Planning Scientific Research Project (WJ2019M054), 2020 Baoan District Medical and Health Basic Research Project (2020JD491 and 2020JD072), Bao'an TCM Development Foundation (2020KJCX-KTYJ-200), and the Internal Research Project of Shenzhen Baoan Authentic TCM Therapy Hospital (BCZY2021003).References
- J. Tang, Y. Wang, Y. Luo, J. Fu, Y. Zhang, Y. Li, Z. Xiao, Y. Lou, Y. Qiu and F. Zhu, Comput. Struct. Biotechnol. J., 2020, 18, 2012–2025 CrossRef CAS.
- A. Bobrie and C. Théry, Biochem. Soc. Trans., 2013, 41, 263–267 CrossRef CAS.
- M. L. Gao, F. He, B. C. Yin and B. C. Ye, Analyst, 2019, 144, 1995–2002 RSC.
- K. Kalimuthu, W. Y. Kwon and K. S. Park, J. Biol. Eng., 2019, 13, 31–38 CrossRef.
- Y. Cao, L. Li, B. Han, Y. Wang, Y. Dai and J. Zhao, Biosens. Bioelectron., 2019, 141, 111397 CrossRef CAS.
- H. Zhang, B. Qiao, Q. Guo, J. Jiang, C. Cai and J. Shen, Analyst, 2020, 145, 3557–3563 RSC.
- L. Huang, D. Wang, N. Singh, F. Yang, N. Gu and X. Zhang, Nanoscale, 2018, 10, 20289–20295 RSC.
- C. Wang, D. Jin, Y. Yu, L. Tang, Y. Sun, Z. Sun and G. Zhang, Sens. Actuators, B, 2020, 314, 128056 CrossRef CAS.
- R. Huang, L. He, Y. Xia, H. Xu, C. Liu, H. Xie, S. Wang, L. Peng, Y. Liu, Y. Liu, N. He and Z. Li, Small, 2019, 15, e1900735 CrossRef.
- Z. Yan, S. Dutta, Z. Liu, X. Yu, N. Mesgarzadeh, F. Ji, G. Bitan and Y. Xie, ACS Sens., 2019, 4, 488–497 CrossRef CAS.
- H. Xu, C. Liao, P. Zuo, Z. Liu and B. Ye, Anal. Chem., 2018, 90, 13451–13458 CrossRef CAS.
- Y. Xia, T. Chen, G. Chen, Y. Weng, L. Zeng, Y. Liao, W. Chen, J. Lan, J. Zhang and J. Chen, Talanta, 2020, 214, 120851 CrossRef CAS.
- J. Suthar, E. S. Parsons, B. W. Hoogenboom, G. R. Williams and S. Guldin, Anal. Chem., 2020, 92, 4082–4093 CrossRef CAS.
- Z. Sun, L. Wang, S. Wu, Y. Pan, Y. Dong, S. Zhu, J. Yang, Y. Yin and G. Li, Anal. Chem., 2020, 92, 3819–3826 CrossRef CAS.
- Y. Sun, H. Jin, X. Jiang and R. Gui, Anal. Chem., 2020, 92, 2866–2875 CrossRef CAS.
- R. Huang, L. He, S. Li, H. Liu, L. Jin, Z. Chen, Y. Zhao, Z. Li, Y. Deng and N. He, Nanoscale, 2020, 12, 2445–2451 RSC.
- X. Yu, L. He, M. Pentok, H. Yang, Y. Yang, Z. Li, N. He, Y. Deng, S. Li, T. Liu, X. Chen and H. Luo, Nanoscale, 2019, 11, 15589–15595 RSC.
- L. Luo, L. Wang, L. Zeng, Y. Wang, Y. Weng, Y. Liao, T. Chen, Y. Xia, J. Zhang and J. Chen, Talanta, 2020, 207, 120298 CrossRef CAS.
- Y. Zhu, Y. An, R. Li, F. Zhang, Q. Wang and P. He, J. Electroanal. Chem., 2020, 862, 113969 CrossRef CAS.
- K. H. Kholafazad and M. Hasanzadeh, Anal. Methods, 2020, 12, 2795–2811 RSC.
- M. Gaillard, A. Thuaire, G. Nonglaton, V. Agache, Y. Roupioz and C. Raillon, Analyst, 2020, 145, 1997–2013 RSC.
- G. Qiu, A. Thakur, C. Xu, S. Ng, Y. Lee and C. L. Wu, Adv. Funct. Mater., 2019, 29, 1806761 CrossRef.
- Q. Wang, L. Zou, X. Yang, X. Liu, W. Nie, Y. Zheng, Q. Cheng and K. Wang, Biosens. Bioelectron., 2019, 135, 129–136 CrossRef CAS.
- D. Fang, D. Zhao, S. Zhang, Y. Huang, H. Dai and Y. Lin, Sens. Actuators, B, 2020, 305, 127544 CrossRef CAS.
- Y. Zhou, H. Xu, H. Wang and B. C. Ye, Analyst, 2019, 145, 107–114 RSC.
- H. Shin, S. Oh, S. Hong, M. Kang, D. Kang, Y. Ji, B. H. Choi, K. Kang, H. Jeong, Y. Park, S. Hong, H. K. Kim and Y. Choi, ACS Nano, 2020, 14, 5435–5444 CrossRef CAS.
- L. Shen, K. Jia, T. Bing, Z. Zhang, X. Zhen, X. Liu, N. Zhang and D. Shangguan, Anal. Chem., 2020, 92, 5370–5378 CrossRef CAS.
- Y. Zhu, Z. Xu, J. Gao, W. Ji and J. Zhang, Biosens. Bioelectron., 2020, 160, 112210 CrossRef CAS.
- S. Arshavsky-Graham, K. Urmann, R. Salama, N. Massad-Ivanir, J. G. Walter, T. Scheper and E. Segal, Analyst, 2020, 145, 4991–5003 RSC.
- Z. Liu, S. Lei, L. Zou, G. Li, L. Xu and B. Ye, Biosens. Bioelectron., 2019, 131, 113–118 CrossRef CAS.
- M. Anjomshoa and M. Torkzadeh-Mahani, J. Fluoresc., 2016, 26, 1505–1510 CrossRef CAS.
- F. A. P. Crisafuli, E. B. Ramos and M. S. Rocha, Eur. Biophys. J., 2015, 44, 1–7 CrossRef CAS.
- J. Q. Chen, S. F. Xue, Z. H. Chen, S. Zhang, G. Shi and M. Zhang, Biosens. Bioelectron., 2018, 100, 526–532 CrossRef CAS.
- N. Cheng, D. Du, X. Wang, D. Liu, W. Xu, Y. Luo and Y. Lin, Trends Biotechnol., 2019, 37, 1236–1254 CrossRef CAS.
- D. Daassi, K. M. Mahoney and G. J. Freeman, Nat. Rev. Immunol., 2020, 20, 209–215 CrossRef CAS.
- G. Chen, A. C. Huang, W. Zhang, G. Zhang, M. Wu, W. Xu, Z. Yu, J. Yang, B. Wang, H. Sun, H. Xia, Q. Man, W. Zhong, L. F. Antelo, B. Wu, X. Xiong, X. Liu, L. Guan, T. Li, S. Liu, R. Yang, Y. Lu, L. Dong, S. McGettigan, R. Somasundaram, R. Radhakrishnan, G. Mills, Y. Lu, J. Kim, Y. H. Chen, H. Dong, Y. Zhao, G. C. Karakousis, T. C. Mitchell, L. M. Schuchter, M. Herlyn, E. J. Wherry, X. Xu and W. Guo, Nature, 2018, 560, 382–386 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10159a |
| ‡ Hua Zhang and Yajuan Zhou are the Co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |