Open Access Article
Open Access ArticleSingle nucleotide detection using bilayer MoS2 nanopores with high efficiency†
Payel Sen a and
Manisha Gupta
a and
Manisha Gupta *b
*b
aDepartment of Chemical and Materials Engineering, University of Alberta, Edmonton, Canada. E-mail: payel@ualberta.ca
bDepartment of Electrical and Computer Engineering, University of Alberta, Edmonton, Canada. E-mail: mgupta1@ualberta.ca
First published on 3rd February 2021
Abstract
Single nucleotide detection is important for early detection of diseases and for DNA sequencing. Monolayer (ML) MoS2 nanopores have been used to identify and distinguish single nucleotides with good signal-to-noise ratio in the recent past. Here, we use a bilayer (BL) MoS2 nanopore (∼1.3 nm thick) to detect distinct single nucleotides with high spatial resolution and longer dwell time. In this study, the performance of similar sized (<3 nm) ML and BL MoS2 nanopores for detection of a single nucleotide has been compared. Both single nucleotide and single stranded DNA translocations through them are studied. For single nucleotide detection, we observe that BL MoS2 nanopores demonstrate twice the dwell time as compared to ML MoS2 nanopores with 95% confidence. Single nucleotide detection rate for BL MoS2 nanopores (50–60 nucleotides per s) is five-fold higher as compared to ML MoS2 nanopores (10–15 nucleotides per s) in 10 pM analyte concentration. For single stranded DNA, we observe 89% (for 60 DNA molecules detected) single nucleotide detection efficiency with BL MoS2 nanopores as compared to 85% for ML MoS2. The DNA sequencing efficiency through BL MoS2 nanopores is also found to be 8–10% better than through ML MoS2 nanopores, irrespective of DNA sequencing orientation. Thus, owing to improved analyte/nanopore charge interaction BL MoS2 nanopores can be used for single nucleotide detection with high resolution due to longer dwell time, detection rate and efficiency. This study demonstrates the improved ability of BL MoS2 nanopores in sequencing DNA with 8–10% higher efficiency, two-times temporally resolved single-nucleotide current signatures and five-times higher detection rate, compared to ML MoS2 nanopores.
Introduction
DNA sequencing data can be used for detecting hereditary diseases based on genetic information and also determine the health effects of a microbial strain. DNA sequencing performed with nanopores is an important technique as it is label free and can be conducted in real-time with relatively low-cost and long-read without amplification.1 Solid-state nanopores are used extensively for DNA sequencing due to nanopore stability and ease of fabrication.2–6 However, the sensing efficiency and resolution depend upon a lot of factors including the solid-state membrane thickness, nanopore size and material properties. A variety of materials like silicon,7 silicon oxide,8 silicon nitride,9 aluminium dioxide,10 and hafnium oxide11 have been used for sequencing DNA. In spite of being mechanically stable, sensing suffers from poor spatial resolution making them incapable of properly distinguishing four DNA bases. 2D material nanopores are rapidly emerging as a solution to improve spatial resolution due to their ultra-thinness for identifying single nucleobases.12–27 Molybdenum disulphide (MoS2) is a type of 2D metal dichalcogenide that has been used in the last decade for nanopore fabrication. These MoS2 nanopores have demonstrated good DNA sequencing with low sticking of the DNA and high signal-to-noise ratio.12,16,20,21 High molybdenum (Mo) concentration around the MoS2 nanopores is favorable for the analyte/nanopore wall surface charge interactions as this minimizes the noise. Thus, this is a promising material for nanopore fabrication.van der Waals force separate the layers of 2D materials like MoS2. Thus, the number of layers of MoS2 separated can be controlled, which allows tunability of the membrane thickness. Single nucleotide and polynucleotide translocation through monolayer MoS2 nanopores has been previously demonstrated experimentally.13 However, MoS2 nanopores also suffer from high translocation velocity similar to other solid-state nanopores. Although different techniques have been employed to slow down DNA translocation through solid state nanopore,28–31 the control of translocation speed is still challenging. The thickness of monolayer (ML) MoS2 (0.65 nm) is less than a single nucleotide (1.6–1.8 nm). Thus, the charge interaction along DNA/nanopore interface for ML MoS2 gets compromised making DNA translocation fast. Molecular translocation through ML MoS2 nanopore has been slowed down by using transmembrane viscosity gradient for producing temporally resolved current blockades capable of distinguishing different types of DNA nucleotides.13
In this work, we experimentally investigated the role of 2D material membrane properties on the translocation speed of DNA. It is found that the behavior of monolayer (ML) and bilayer (BL) MoS2 is different under a vertical electrical field applied using the cis and trans electrodes.32 The strong interlayer coupling in the BL MoS2 leads to a gradation in potential throughout the thickness as each layer will experience a different electric potential.32 Due to this potential gradient the phosphate groups are trapped and detrapped by the Molybdenum atoms in the membrane. An additional pull is created because of the increased negative (analyte DNA)–positive (Mo atoms lining the nanopore surface) charge interaction. This can help immobilize molecules at the nanopore for longer time producing highly resolved signals. Molecular dynamics simulation study conducted previously concluded that BL MoS2 can slow down peptide translocation producing stepwise distinguishable current blockades for single nucleotide sensing.22 Thus, we have conducted detailed single and poly nucleotide translocation experimental studies through ML and BL MoS2 nanopores. The translocations were conducted using picomolar concentration and the dwell times were then extracted from these measurements for further analysis.
Experimental
Standard microfabrication techniques were used to fabricate free-standing solid-state silicon nitride (SiNx) membranes on silicon support. Electron Beam Lithography (EBL) was used along with Reactive Ion Etching (RIE) to fabricate a 50 nm pore at the centre of the previously fabricated SiNx membrane. Then, monolayer (ML) and bilayer (BL) MoS2 flakes were transferred on the pore via exfoliation using scotch tape. A dual-stage microscope was used to ensure a properly aligned and centred MoS2 transfer. ESI Fig. S1a† presents the detailed fabrication flowsheet of MoS2 nanopores. 20 μm sized MoS2 flakes were obtained repeatedly by this method. The membranes were first characterized to determine and ensure the number of MoS2 layers using JEOL JEM-ARM200CF Scanning/Transmission Electron Microscope (STEM). The nanopore was STEM drilled on the free standing MoS2 membrane. Nanopores with diameters between 2.5–3 nm were obtained repeatedly and were suitable for our application. To prevent nanopore expansion or material redeposition due to prolonged beam exposure, intense imaging on the same nanopore was avoided.300 mM filtered KCl solution at pH = 7 was prepared by adding 3 mM Tris–HCl buffer. The solution was then degassed in vacuum for 90 minutes and used for the experiments after attaining room temperature. 0.5 mM solutions of single nucleotides (deoxyadenosine triphosphate: dA, deoxythymidine triphosphate: dT, deoxycytidine triphosphate: dC and deoxyguanosine triphosphate: dG) were procured from Integrated DNA Technologies for nanopore translocation and dwell-time analysis thereof. 5 mM solution of single stranded (ss) customized DNA oligos (3′-ATCGATCGATCGATCGATCGATCGATCG-5′) were procured from Integrated DNA Technologies to evaluate the efficiency of ML and BL MoS2 nanopores for detection of single nucleotides from a DNA strand. Analyte solutions at 10 pM concentration were then prepared by dilution of the purchased single nucleotide and ssDNA solutions.
Custom-made Teflon half-cells were used to contain the electrolyte (KCl). As Teflon or poly-tetra-fluoro-ethylene are both hydrophobic polymers, they help in maintaining a clean environment. They also have high chemical resistance which ensures a well-insulated sensing environment. The half cells were sealed with polydimethylsiloxane (PDMS) to ensure that the electrolytic exchange between the chambers is only via the nanopore. PDMS, also reduces capacitive noise which helps in obtaining improved signals. The half-cells were thoroughly cleaned by ultra-sonication before each experiment. The nanopores/membranes were dipped in acetone and kept in vacuum for 30 min. The nanopore was cleaned by gradually pulling the acetone through it. The cleaning steps include three acetone-cleaning repeats followed by four IPA washes in vacuum (20 min each) to remove acetone residue. Fresh solvent is used for every cycle of acetone and IPA treatment. The membrane with the embedded nanopore was then mounted between the half cells. Two Ag/AgCl electrodes were cleaned using ethanol and DI water and re-chlorinated by treating them with bleach for an hour. The electrodes were then dipped in each half-cell to apply a voltage across the nanopore bearing membrane. The entire cell-assembly was enclosed in a Faraday cage (Warner instruments) to prevent electrical interference. Filtered and buffered 300 mM KCl solution (previously prepared) was introduced in both chambers and the ionic current at varying voltage (−200 mV to 200 mV) was recorded to evaluate the channel conductance. The nanopore was electrically conditioned at a constant 100 mV bias to obtain a stable baseline. The analytes were then added to the trans chamber and pulled electrophoretically to the cis chamber through the nanopore under a constant bias of 200 mV. Single channel blockade recordings obtained from translocating nucleotides and DNA were then filtered (20 kHz 8-pole Bessel filter), amplified (Axon MultiClamp 700B) and finally digitized (Axon Digidata 1550B) for further analysis. ESI Fig. S1b† presents schematic of the sensing assembly.
A customized MATLAB algorithm was used to identify translocation peaks. The raw traces were first loaded as an .abf file. The peaks were identified as change points and the blockade current was calculated by subtracting the mean blockade current from the mean baseline current. The mean was calculated for the 100 samples before and after the peak to account for shift in baseline during the translocation. The dwell time was calculated from the difference of the initial and final changepoint indices multiplied by the sampling frequency. The mean and median of the dwell time were then compared to test whether the distribution is normal or not. We then assumed a null hypothesis that the dwell times for ML and BL MoS2 nanopore is the same and then tested the hypothesis by conducting T-test at 95% confidence on the dwell time data. All statistical analysis was performed for a population of 300 nucleotides. For the mixed DNA, a MATLAB algorithm was built to first truncate the data at each molecular entry and exit. Then the dwell time and blockade current at each individual current drop steps within a molecular translocation event were obtained. The individual nucleotides were then identified by comparing the blockades with that obtained from single nucleotide translocation performed earlier through the same nanopore.
Results
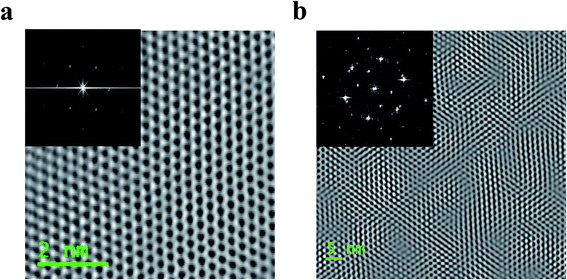
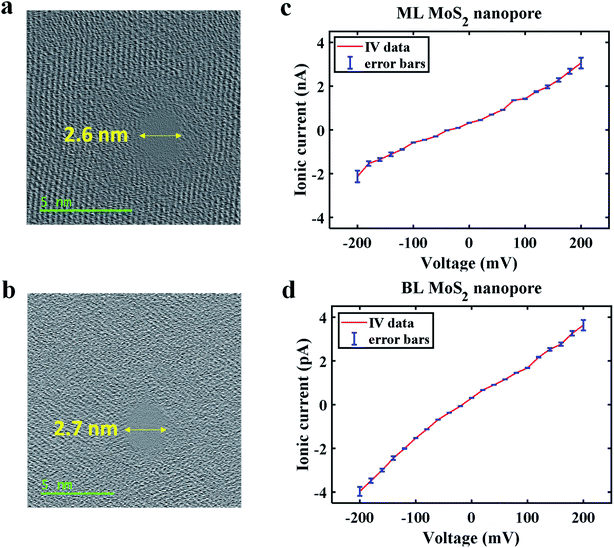
The free-standing ML and BL MoS2 membranes were first characterized by High Resolution TEM (HRTEM) microscopy to confirm number of layers. Fig. 1a shows STEM image of molecular arrangement of a ML MoS2 membrane. Fig. 1b shows STEM image of an AA′ stacked BL MoS2 membrane showing Moiré pattern of two overlapped domains rotated by 60°. The TEM structure obtained for ML MoS2 and Moiré patterns observed for BL MoS2 are in agreement with the literature.33–35 All the HRTEM images are low pass filtered using Gatan Microscope software to remove noise for prominent visualization of the molecular structure. No additional artifacts were produced from the simple filtering. ESI Fig. S2a and b† shows the profilometric studies of ML and BL MoS2 on SiNx support showing the corresponding membrane thicknesses. ESI Fig. S2c and d† shows additional STEM images of the MoS2 membranes.The ionic conductance of the fabricated ML and BL MoS2 nanopores were measured following the procedure described in the methods section to ensure properly cleaned and conducting nanopores. Fig. 2a and b shows MoS2 nanopores with diameters of 2.6 nm for ML and 2.7 nm for BL respectively. Ionic current through each nanopore was measured by applying a trans bias in the range of −200 mV to 200 mV in 300 mM KCl. 4 consecutive ionic conductance measurements were conducted through the nanopore along with intermediate cleaning steps. Here, we have reported the average conductance for each pore. Fig. 2c and d shows the IV plot for both the nanopores. The mean of ionic current recorded at each bias is displayed as scatter plot, with the error bars showing the current variation between the 4 measurements at each voltage. The mean nanopore conductance (calculated as the average of the ratio of ionic current and voltage at each bias value) was found to be 10.5 ± 0.016 nS for 2.6 nm ML MoS2 nanopore and 17.2 ± 0.023 nS for 2.7 nm BL MoS2 nanopore.
To study the effect of ML and BL MoS2 on single nucleotide translocation and sensing, 10 pM concentration of dA, dT, dC and dG were electrophoretically pulled through the above-mentioned ML and BL MoS2 nanopores. For these nucleotide translocations 200 mV bias was applied and the translocation traces were recorded. The ESI Fig. S3a–h† shows the recorded data for single nucleotide (for each of dA, dT, dC and dG) translocation for 5 s and 1.5 s through ML MoS2 and BL MoS2 nanopores respectively. We also observe a detection rate of 10–15 nucleotides per s and 50–60 nucleotides per s for ML and BL MoS2 nanopores respectively (see Fig. S3a–h†). The five-fold higher detection rate obtained for BL nanopores may be due to faster capture of the translocating nucleotides at the nanopore due to stronger electrophoretic pull towards the layered BL MoS2.
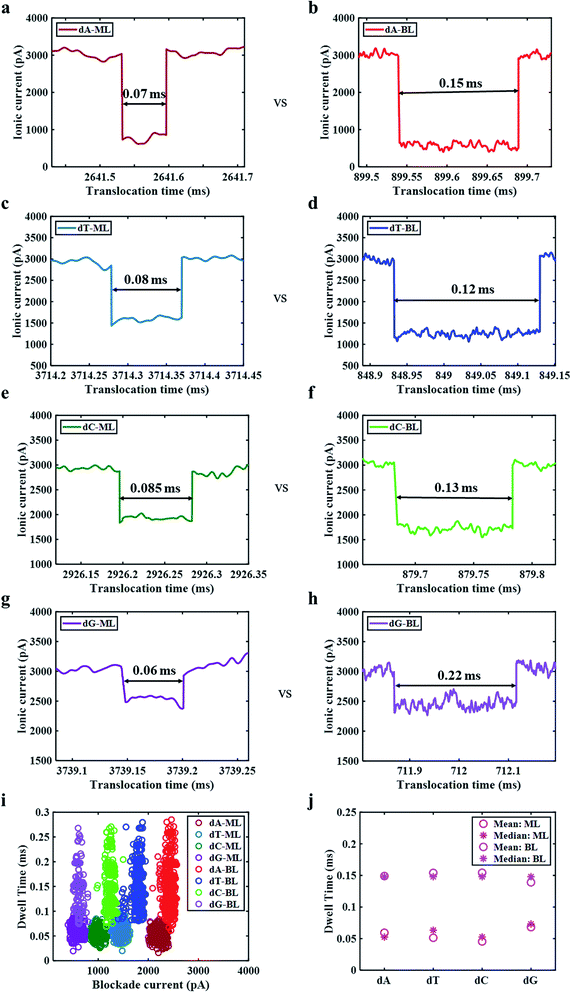
Fig. 3a–h presents truncated single nucleotide peaks obtained for ML and BL MoS2 nanopores for a direct comparison of dwell times. It is observed that the dwell times are higher for BL as compared for the ML MoS2 nanopores for all the different nucleotides. Blockade current is plotted as a function of dwell time for 3000 single nucleotide transport events in Fig. 3i. We observe four distinct blockade current regions for the different nucleotides. Thus, we can conclude that both ML and BL MoS2 nanopores are capable of detecting single nucleotides. The blockade current for the nucleotide translocation is plotted as histograms to observe their distribution (ESI Fig. S4†). We observe normal distribution for all the nucleotides for both ML and BL nanopores. Thus, the mean blockade current values along with their standard deviations can be obtained.
Table 1 lists the dwell time and blockade current values for all single nucleotides sensed using ML and BL MoS2 nanopores. The blockade current magnitudes measured are found to be the dA, dT, dC and dC in decreasing order which agrees with previous study.13 The mean blockade current obtained through BL MoS2 is found to be slightly higher than or comparable to ML MoS2, which may be due to better charge interaction at the van der Waals separated nanopore interface. Based on these numbers we can conclude that the current resolution obtained for ML is preserved in BL MoS2 nanopores. On the other hand, it was observed that the dwell time values of the translocations for BL nanopore are spread over a larger range as compared to the ML nanopore. The events detected through ML nanopores show mean dwell times in range of 0.053–0.061 ms with a 17–20% of deviation. For BL nanopore detection the mean dwell time range is 0.141–0.143 ms with a deviation of 28–30% (see Table 1 and ESI Fig. S5†). The mean and median dwell time values were then evaluated for ML and BL nanopores (see Fig. 3j) and the values were found to be pretty close to each other, supporting an assumption of a normal distribution. Therefore, T-tests were done on the values for a statistical comparison. T-Tests thus performed, indicate that for all the nucleotides, the dwell times for ML and BL nanopores are different at a 95% confidence level (shown in ESI Table T1†). The sample mean of the dwell time for the BL nanopore is approximately twice that of the ML nanopore. A higher dwell time indicates an improvement in sensing.
| Analyte | ML MoS2 nanopore | BL MoS2 nanopore | ||
|---|---|---|---|---|
| Blockade current | Dwell time | Blockade current | Dwell time | |
| dG | 0.595 ± 0.057 nA | 0.0529 ± 0.0092 ms | 0.608 ± 0.036 nA | 0.1409 ± 0.0424 ms |
| dC | 0.987 ± 0.066 nA | 0.0577 ± 0.0102 ms | 1.256 ± 0.048 nA | 0.1430 ± 0.0424 ms |
| dT | 1.454 ± 0.076 nA | 0.0610 ± 0.0110 ms | 1.809 ± 0.07 nA | 0.1422 ± 0.0405 ms |
| dA | 2.245 ± 0.079 nA | 0.0615 ± 0.0122 ms | 2.434 ± 0.06 nA | 0.1412 ± 0.0401 ms |
To demonstrate the single nucleotide detection capability of ML and BL MoS2 nanopores mixed nucleotide translocations through ML and BL MoS2 nanopores were conducted. Fig. S6a and b† show translocation traces for 5 s and 1.5 s through ML and BL MoS2 nanopores respectively. The nucleotide distinction is made based on the current blockades obtained for individual nucleotides sensed separately. The dwell time range for all nucleotides (as observed in the scatter plot in Fig. S6c†) is found to be the same as that when detected separately through ML and BL MoS2 nanopores.
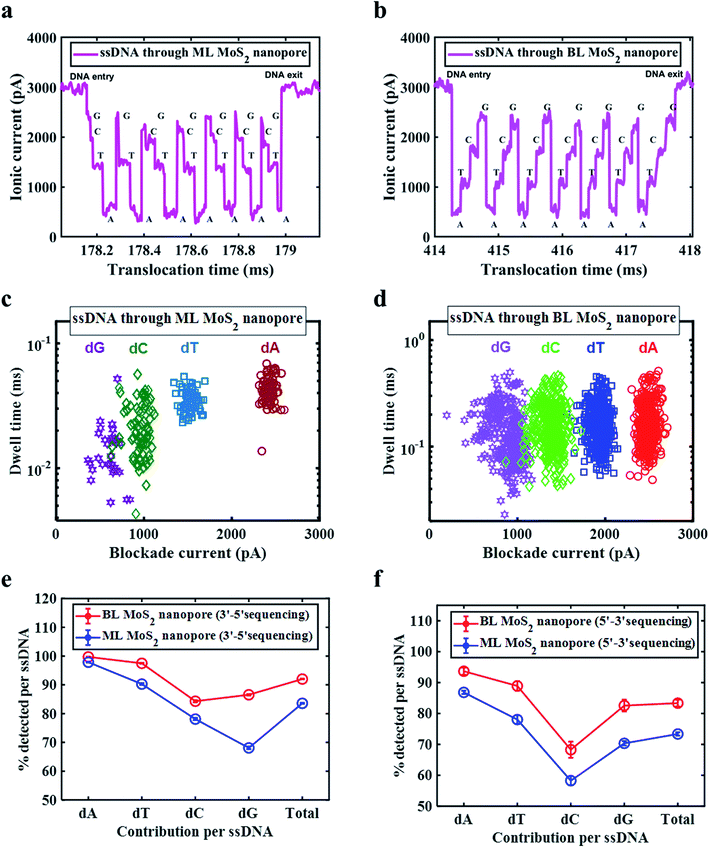
To demonstrate that the BL MoS2 improves the dwell time of the translocation and can detect single nucleotides, we used 10 pM single stranded (ss) DNA oligos with customized mixed sequence. The translocations were conducted using ML and BL nanopores. Fig. 4a and b show the translocation trace for a single ssDNA strand through the ML and BL MoS2 nanopore respectively. For both the nanopores distinct molecular current blockades were observed. From careful inspection of each molecular blockade, it can be clearly understood that the discrete blockade steps correspond to the constituent nucleotides. All the translocations for the ML and BL were analysed using the code developed and dwell time as a function of blockade current for 3 s translocations are plotted in Fig. 4c and d. For BL MoS2 nanopore, we observe four different current levels similar to observed for each nucleotide with dwell times ranging from 0.02–0.6 ms with the highest density obtained around 0.08–0.3 ms which is similar to what we obtained for single nucleotide translocations. On the other hand, for ML MoS2 nanopore, four different nucleotides are also detected. However, the time resolution obtained for dC and dG are particularly low (0.004–0.04 ms for dG and 0.004–0.06 ms for dG) compared to dA and dT, the latter nucleotides showing dwell times in the range of 0.01–0.07 ms, with the mean ∼0.05 ms, which is close to that obtained for single nucleotide triphosphates.
Based on the different dwell times and blockade current four different nucleotides can be distinctly identified. The blockade levels are then matched with the single nucleotide data recorded before to obtain and relate the DNA sequence. Fig. S7a and b† shows the sequence of nucleotides detected and left undetected by ML and BL MoS2 nanopore sensing in a color-coded format, thus representing the single nucleotide efficiency of detection in each case. ML MoS2 nanopore is found to demonstrate a single nucleotide detection efficiency of 84.9% ((total − undetected)/total nucleotides) considering 60 DNA molecules translocation. Similarly, the experimental analysis shows that BL MoS2 nanopore are capable of identifying single nucleotides with about 89.1% efficiency. For both ML and BL nanopores, it is observed that mostly dC and dG (especially at the tailing end or entering end of the DNA) are undetected or detected with poor time resolution. This may be due to the low density of these two nucleotides along with their low blockade current.
In addition, we have extracted the efficiency of ssDNA sequencing for both 3′–5′ and 5′–3′ orientations. BL MoS2 nanopores demonstrate 83 ± 1.07% and 92 ± 0.14% for 5′–3′ and 3′–5′-ssDNA sequencing efficiency respectively. These are 10% better than ML MoS2 for 5′–3′-orientation (73.35 ± 0.55%) and 8.4% better for 3′–5′ sequencing (83.6 ± 0.16%). Fig. S8 and S9† shows sequencing events for 3′–5′ and 5′–3′ ssDNA translocations for ML and BL MoS2 nanopores respectively.
Fig. 4e and f shows the efficiency of detection of individual molecules from ssDNA and ssDNA sequencing as a whole. It is found that the efficiency of BL MoS2 nanopores is better in both cases. The role of BL MoS2 is observed to be most prominent in improving the temporal resolution of the signature for dC and dG. It was observed that the 47/60 ssDNA translocations were in 3′–5′ orientation for ML MoS2 nanopore and 51/60 ssDNA translocations were in 3′–5′ orientation for BL MoS2 nanopore.
Discussions
Short dwell time or high translocation speed in a nanopore prevents precise sequencing of nucleotides. Different techniques have been used to slow down the DNA translocation using trans-membrane viscosity gradient, electric field tuning and surface charge modification of the nanopore/electrolyte interface.28–31 DNA molecule sensing has been demonstrated through sub-5 nm nanopores on 25 nm thick SiNx membranes in 38 pM concentration solution.36 The study demonstrated that unamplified dsDNA molecules (as opposed to single nucleotides) can be sensed by slowing down the translocation using 20-fold salt gradient.36 A ML MoS2 nanopore-based viscosity gradient system has also been used for improved DNA sequencing by slowing down the translocation by two orders of magnitude.13 A simulation study has also been used to demonstrate improved peptide dwell times for BL MoS2 nanopores.22In this study, we experimentally explore the unaided capacity of BL MoS2 nanopores in improving real-time sensing resolution, detection rate and efficiency for ssDNA sequencing. We first analysed the blockade current obtained for single nucleotide using ML and BL MoS2 nanopores. Our ML MoS2 nanopore current signatures are similar to that obtained in previous study.13 However, we observed geometry defying blockade signatures for guanine. This may be due to the increased hydrogen bonding potential of guanine or increased physical adsorption energy of guanine on MoS2 which may increase the pore conductance, thus producing a lower overall ionic blockade event.37 For ssDNA sequencing, we also observed lower time resolution of guanine base of the ssDNA compared to other bases, which may indicate that the effect of surrounding A and C over nanopore blockade are dominant over G. Few studies have shown exceptions to the geometry-determined blockade when larger A and G have produced lower blockades than C and T, for both single nucleotide and polynucleotide sensing.13,20,38
We also obtain higher blockade current levels for BL MoS2 nanopore as compared to ML MoS2 nanopore. In general, for 3D bulk materials like SiNx, the conductance decreases with increase in pore/membrane thickness.39 However, in our study we find that 2D MoS2 nanopores do not demonstrate this behaviour. Unlike 3D bulk material nanopores, in 2D material nanopores interlayer charge storage, electro-activity and field confinement between the van der Waals coupled monolayers increases the capacitance of the pore.40 COMSOL Multiphysics simulation study indicates that the ionic conductance of MoS2 nanopores demonstrates the following trend 2-layers > 1-layer > 4-layers > 3-layers > 6-layers > 5-layers and so on above 2 nm diameter nanopore.41 Few studies on 2D materials nanopores like graphene also demonstrate similar behaviour.18,42,43
We further observe nearly double the dwell time for BL MoS2 as compared to ML MoS2 nanopore, with 95% confidence. To observe the effect of the bias on the blockade current we measured the blockade current for three different voltages. Fig. S11† shows blockade current vs. dwell time scatter plot for single nucleotide translocations through ML and BL MoS2 nanopores for varying transmembrane bias values (100 mV, 150 mV and 200 mV). However, significant change in dwell times is not observed for either of ML or BL MoS2 nanopore sensing. This further proves that the events considered are indeed translocation events and strengthens the inference drawn about improved ability of BL MoS2 nanopores in providing better resolution for single nucleotide sensing.
The effect of the temporal resolution improvement offered by BL MoS2 nanopore on sequencing single-stranded DNA oligos was investigated next. From these experiments, we found that the capability of BL MoS2 nanopores can resolve the nucleotide blockade currents and hence can distinguish single distinct nucleotides. Thus, we observed an overall single nucleotide (for 1800 nucleotides in 60 ssDNA strands) detection efficiency of 89% using BL MoS2 nanopore, a 4% improvement compared to that for ML MoS2 nanopores. We also observed an 8–10% improved ssDNA sequencing efficiency for both 3′–5′ and 5′–3′ ssDNA orientations. The interlayer potential gradient for BL MoS2 offers improved capture of analytes inside the nanopore even at low (picomolar) concentration leading to 5× better detection rate. Previous studies have shown that improved nanopore/DNA interaction can result in faster capture of DNA within the nanopore and also cause longer residence of DNA at the nanopore.44 We believe that the improved capture rate and nanopore dwell time is a result of the enhanced DNA/nanopore interaction experienced in BL MoS2 nanopore as compared to ML MoS2 nanopore.
Moreover, the behaviour of even and odd layers of MoS2 is found to differ under an applied vertical electrical field.45 The electric field confinement within the layers is more for odd-numbered layers than even-numbers, which thus improves the ionic conduction for even layers. Moreover, the BL MoS2 thickness is close to nucleotide size which helps maintain a good spatial resolution. However, >2 layers can hamper the spatial resolution. Additionally, our simulation results also show a reduction in blockade signal amplitude above two layers. All things considered; we believe >2-layers MoS2 may not be a better choice for DNA sequencing. The results conclusively suggest that BL MoS2 nanopores can distinguish four type of nucleotides with significantly higher dwell time. Thus, BL MoS2 nanopores can be a suitable choice for more efficient and faster DNA sequencing.
Conclusion
In summary, we experimentally demonstrate that BL MoS2 nanopores are capable of detecting distinct single nucleotides with twice the dwell time as compared to ML MoS2 nanopores. The van der Waals separated layers of BL MoS2 experience different potentials when a bias is applied across them creating an interlayer potential gradient. This makes each individual molecule experience an increased charge interaction and stronger immobilization at the nanopore, leading to prominently sharp and broad translocation events. The translocation profiles through BL MoS2 nanopores show distinct peaks for each type of DNA nucleobase of 10 pM concentration. Thus, we have been able to detect nucleotides at 89% efficiency using BL MoS2 nanopores for 60 detected DNA molecules. Furthermore, we observed an 8–10% improved sequencing efficiency by using BL MoS2 nanopores. The BL MoS2 nanopore can resolve single-nucleotide signals temporally, while maintaining a good spatial resolution as well. This high detection rate, efficiency and dwell time makes BL MoS2 nanopores promising for high-speed detections in low concentration analyte solutions which is essential for several biosensing applications.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
P. S. conducted all the simulations, fabrication, and experiments on the nanopores. P. S. and M. G. analyzed and wrote the manuscript.Funding sources
This work was funded by Natural Sciences and Engineering Research Council of Canada award # 06096 and Alberta Innovates.Conflicts of interest
The authors declare that there are no competing interests.References
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Characterization of Individual Polynucleotide Molecules Using a Membrane Channel, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13770–13773 CrossRef CAS.
- F. Haque, J. Li, H. C. Wu, X. J. Liang and P. Guo, Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA, Nano Today, 2013, 8, 56–74 CrossRef CAS.
- F. Sawafta, A. T. Carlsen and A. R. Hall, Membrane thickness dependence of Nanopore formation with a focused helium ion beam, Sensors, 2014, 14, 8150–8816 CrossRef.
- Y. H. Lanyon, G. D. Marzi, Y. E. Watson, A. J. Quinn, J. P. Gleeson, G. Redmond and D. W. M. Arrigan, Fabrication of nanopore array electrodes by focused ion beam milling, Anal. Chem., 2007, 79, 3048–3055 CrossRef CAS.
- H. M. Kim, M. H. Lee and K. B. Kim, Theoretical and experimental study of nanopore drilling by a focused electron beam in transmission electron microscopy, Nanotechnology, 2011, 22(27), 275303 CrossRef.
- O. K. Zahid and A. R. Hall, Helium ion microscope fabrication of solid-state nanopore devices for biomolecule analysis, in NanoScience and Technology, ed. G. Hlawacek and A. Gölzhäuser, Springer, Cham, 2016, pp. 447–470 Search PubMed.
- T. Schmidt, M. Zhang, I. Sychugov, N. Roxhed and J. Linnros, Nanopore arrays in a silicon membrane for parallel single-molecule detection: fabrication, Nanotechnology, 2015, 26(31), 314001 CrossRef.
- A. J. Storm, J. H. Chen, H. W. Zandbergen and C. Dekker, Translocation of double-strand DNA through a silicon oxide nanopore, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2005, 71(5), 051903 CrossRef CAS.
- K. Briggs, H. Kwok and V. Tabard-Cossa, Automated fabrication of 2-nm solid-state nanopores for nucleic acid analysis, Small, 2014, 10, 2077–2086 CrossRef CAS.
- B. M. Venkatesan, B. Dorvel, S. Yemenicioglu, N. Watkins, I. Petrov and R. Bashir, Highly sensitive, mechanically stable nanopore sensors for DNA analysis, Adv. Mater., 2009, 21, 2771–2776 CrossRef CAS.
- J. Larkin, R. Henley, D. C. Bell, T. Cohen-Karni, J. K. Rosenstein and M. Wanunu, Slow DNA transport through nanopores in hafnium oxide membranes, ACS Nano, 2013, 7, 10121–10128 CrossRef CAS.
- K. Liu, J. Feng, A. Kis and A. Radenovic, Atomically thin molybdenum disulfide nanopores with high sensitivity for dna translocation, ACS Nano, 2014, 8, 2504–2511 CrossRef CAS.
- J. Feng, K. Liu, R. D. Bulushev, S. Khlybov, D. Dumcenco, A. Kis and A. Radenovic, Identification of single nucleotides in MoS2 nanopores, Nat. Nanotechnol., 2015, 10, 1070–1076 CrossRef CAS.
- A. B. Farimani, K. Min and N. R. Aluru, DNA base detection using a single-layer MoS2, ACS Nano, 2014, 8, 7914–7922 CrossRef CAS.
- H. Chen, L. Li, T. Zhang, Z. Qiao, J. Tang and J. Zhou, Protein Translocation through a MoS2 Nanopore: A Molecular Dynamics Study, J. Phys. Chem. C, 2018, 122, 2070–2080 CrossRef CAS.
- M. Graf, M. Lihter, D. Altus, S. Marion and A. Radenovic, Transverse Detection of DNA in a MoS2 Nanopore, Nano Lett., 2019, 19(12), 9075–9083 CrossRef CAS.
- Y. Deng, Q. Huang, Y. Zhao, D. Zhou, C. Ying and D. Wang, Precise fabrication of a 5 nm graphene nanopore with a helium ion microscope for biomolecule detection, Nanotechnology, 2017, 28(4), 045302 CrossRef.
- C. A. Merchant, K. Healy, M. Wanunu, V. Ray, N. Peterman, J. Bartel, M. D. Fischbein, K. Venta, Z. Luo, A. T. C. Johnson and M. Drndić, DNA translocation through graphene nanopores, Nano Lett., 2010, 10, 2915–2921 CrossRef CAS.
- B. Luan and R. Zhou, Spontaneous Transport of Single-Stranded DNA through Graphene-MoS2 Heterostructure Nanopores, ACS Nano, 2018, 12, 3886–3891 CrossRef CAS.
- A. D. Carral, S. C. Sarap, K. Liu, A. Radenovic and M. Fyta, 2D MoS2 nanopores: ionic current blockade height for clustering DNA events, 2D Mater., 2019, 6(4), 045011 CrossRef CAS.
- J. Shim, S. Banerjee, H. Qiu, K. K. H. Smithe, D. Estrada, J. Bello, E. Pop, K. Schulten and R. Bashir, Detection of methylation on dsDNA using nanopores in a MoS2 membrane, Nanoscale, 2017, 9, 14836–14845 RSC.
- A. Nicolaï, M. D. B. Pérez, P. Delarue, V. Meunier, M. Drndić and P. Senet, Molecular Dynamics Investigation of Polylysine Peptide Translocation through MoS2 Nanopores, J. Phys. Chem. B, 2019, 123, 2342–2353 CrossRef.
- A. Smolyanitsky, B. I. Yakobson, T. A. Wassenaar, E. Paulechka and K. A. Kroenlein, A MoS2-Based Capacitive Displacement Sensor for DNA Sequencing, ACS Nano, 2016, 10, 9009–9016 CrossRef CAS.
- G. Danda, P. M. Das, Y. C. Chou, J. T. Mlack, W. M. Parkin, C. H. Naylor, K. Fujisawa, T. Zhang, L. B. Fulton, M. Terrones, A. T. C. Johnson and M. Drndić, Monolayer WS2 Nanopores for DNA Translocation with Light-Adjustable Sizes, ACS Nano, 2017, 10, 9009–9016 Search PubMed.
- H. Arjmandi-Tash, L. A. Belyaeva and G. F. Schneider, Single molecule detection with graphene and other two-dimensional materials: nanopores and beyond, Chem. Soc. Rev., 2016, 45, 476–493 RSC.
- W. Chen, G. C. Liu, J. Ouyang, M. J. Gao, B. Liu and Y. D. Zhao, Graphene nanopores toward DNA sequencing: a review of experimental aspects, Sci. China: Chem., 2017, 60, 721–729 CrossRef CAS.
- G. F. Schneider, Q. Xu, S. Hage, S. Luik, J. N. H. Spoor, S. Malladi, H. Zandbergen and C. Dekker, Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation, Nat. Commun., 2013, 4(1), 2619 CrossRef.
- B. Luan, G. Stolovitzky and G. Martyna, Slowing and controlling the translocation of DNA in a solid-state nanopore, Nanoscale, 2012, 4, 1068–1077 RSC.
- D. V. Melnikov, J. P. Leburton and M. E. Gracheva, Slowing down and stretching DNA with an electrically tunable nanopore in a p-n semiconductor membrane, Nanotechnology, 2012, 23(25), 255501 CrossRef.
- R. Akahori, T. Haga, T. Hatano, I. Yanagi, T. Ohura, H. Hamamura, T. Iwasaki, T. Yokoi and T. Anazawa, Slowing single-stranded DNA translocation through a solid-state nanopore by decreasing the nanopore diameter, Nanotechnology, 2014, 25(27), 275501 CrossRef.
- M. Waugh, A. Carlsen, D. Sean, G. W. Slater, K. Briggs, H. Kwok and V. Tabard-Cossa, Interfacing solid-state nanopores with gel media to slow DNA translocations, Electrophoresis, 2015, 36, 1759–1767 CrossRef CAS.
- Q. Liu, L. Li, Y. Li, Z. Gao, Z. Chen and J. Lu, Tuning electronic structure of bilayer MoS2 by vertical electric field: a first-principles investigation, J. Phys. Chem. C, 2012, 116, 21556–21562 CrossRef CAS.
- P. Yang, X. Zou, Z. Zhang, M. Hong, J. Shi, S. Chen, J. Shu, L. Zhao, S. Jiang, X. Zhou, Y. Huan, C. Xie, P. Gao, Q. Chen, Q. Zhang, Z. Liu and Y. Zhang, Batch production of 6-inch uniform monolayer molybdenum disulfide catalyzed by sodium in glass, Nat. Commun., 2018, 9, 1–10 CrossRef CAS.
- H. Qiu, T. Xu, Z. Wang, W. Ren, H. Nan, Z. Ni, Q. Chen, S. Yuan, F. Miao, F. Song, G. Long, Y. Shi, L. Sun, J. Wang and X. Wang, Hopping transport through defect-induced localized states in molybdenum disulphide, Nat. Commun., 2013, 4, 2642 CrossRef.
- S. Hussain, J. Singh, D. Vikraman, A. K. Singh, M. Z. Iqbal, M. F. Khan, P. Kumar, D. C. Choi, W. Song, K. S. An, J. Eom, W. G. Lee and J. Jung, Large-area, continuous and high electrical performances of bilayer to few layers MoS2 fabricated by RF sputtering via post-deposition annealing method, Sci. Rep., 2016, 6, 1–13 CrossRef.
- M. Wanunu, W. Morrison, Y. Rabin, A. Y. Grosberg and A. Meller, Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient, Nat. Nanotechnol., 2010, 5(2), 160–165 CrossRef CAS.
- N. M. Luscombe, R. A. Laskowskia and J. M. Thornton, Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level, Nucleic Acids Res., 2001, 29, 2860–2874 CrossRef CAS.
- S. Bhattacharya, J. Yoo and A. Aksimentiev, Water Mediates Recognition of DNA Sequence via Ionic Current Blockade in a Biological Nanopore, ACS Nano, 2016, 10, 4644–4651 CrossRef CAS.
- S. W. Kowalczyk, A. Y. Grosberg, Y. Rabin and C. Dekker, Modeling the conductance and DNA blockade of solid-state nanopores, Nanotechnology, 2011, 22, 315101 CrossRef.
- A. J. Bard and L. R. Faulkner, Electrochemical Methods: Fundamentals and Applications, Department of Chemistry and Biochemistry, University of Texas, Austin, Second edn, 2001 Search PubMed.
- P. Sen, H. Hoi, D. Nandi and M. Gupta, Highly accurate random DNA sequencing using inherent interlayer potential traps of bilayer MoS2 nanopores, bioRxiv, 2020 DOI:10.1101/2020.04.21.053595.
- G. F. Schneider, S. W. Kowalczyk, V. E. Calado, G. Pandraud, H. W. Zandbergen, L. M. K. Vandersypen and C. Dekker, DNA Translocation through Graphene Nanopores, Nano Lett., 2010, 10, 3163–3167 CrossRef CAS.
- W. Lv, M. Chen and R. Wu, The impact of the number of layers of a graphene nanopore on DNA translocation, Soft Matter, 2013, 9, 960–966, 10.1039/C2SM26476E.
- D. Wang, S. Harrer, B. Luan, G. Stolovitzky, H. Peng and A. Afzali, Regulating the Transport of DNA through Biofriendly Nanochannels in a Thin Solid Membrane, Sci. Rep., 2014, 4, 3985, DOI:10.1038/srep03985.
- H. Zhong, R. Quhe, Y. Wang, Z. Ni, M. Ye, Z. Song, Y. Pan, J. Yang, L. Yang, M. Lei, J. Shi and J. Lu, Interfacial Properties of Monolayer and Bilayer MoS2 Contacts with Metals: Beyond the Energy Band Calculations, Sci. Rep., 2016, 6, 21786, DOI:10.1038/srep21786.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10222a |
| This journal is © The Royal Society of Chemistry 2021 |