DOI:
10.1039/D1RA00472G
(Paper)
RSC Adv., 2021,
11, 9528-9541
Structure-based design of 5′-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors†
Received
19th January 2021
, Accepted 25th February 2021
First published on 3rd March 2021
Abstract
Resistant viruses containing mutant neuraminidases (NAs) with diminished drug affinity continue to emerge, and new anti-influenza agents are urgently required. Several potent inhibitors targeting the hydrophobic 150-cavity of viral NAs have been developed by modifying the antiviral drugs, oseltamivir carboxylate (OSC) and zanamivir, with hydrophobic groups. Here, we describe a different strategy for exploring novel and efficient NA inhibitors by targeting the charged amino acid residues around the entrance to the 150-cavity. We synthesized a C5-substituted OSC derivative (1e) with a 4′-phenyl-1,2,3-triazolyl group capable of entering the 150-cavity, and solved the crystal structure of 1e in complex with influenza A virus N5 NA. Using the resulting structural information, we next designed and synthesized two series of OSC derivatives carrying various polar substituents at the triazolyl group of 1e and 2e, with 2e being a 5′-phenyl-1,2,3-triazole regioisomer of 1e. The NA inhibition assays demonstrated that the 2 series (2e–n) generally had superior activity compared with the 1 series (1e–n). Compound 2j, bearing a 3-phenylamino group on the triazole ring, was the most potent inhibitor of all tested NAs including an N2 NA containing the E119V OSC-resistant mutation. Moreover, 2j potently inhibited viral replication in vitro, and molecular docking studies revealed that its phenylamino group can form an additional strong hydrogen bond with residue D151 near the entrance of the 150-cavity. The design method described in this study provides useful insights into the development of novel NA inhibitors. Compound 2j warrants further structural optimization to obtain a candidate for clinical use.
Introduction
Despite the availability of vaccines and antiviral drugs, influenza viruses continue to pose a major threat to public health because of their ability to rapidly mutate.1 Over the past 20 years, the emergence of human-infective H5N1 and H7N9 avian influenza viruses has received extensive public attention.2,3 Influenza neuraminidase (NA) is an enzyme on the viral envelope that hydrolyzes sialic acids in glycan receptors on the infected host cells to enable the release and spread of progeny virus from these cells.4 Several neuraminidase inhibitors (NAIs), including oseltamivir,5 zanamivir,6,7 laninamivir octanoate,8 and peramivir,9 have been successfully developed and approved for clinical use. However, the rapid emergence of viruses resistant to these drugs significantly increased the urgent need for the next generation of NAIs.10–13
On the basis of phylogenetic and X-ray crystallographic studies of various NAs, this viral enzyme is classified into two subtypes.14 In group 1 NAs (N1, N4, N5, and N8), there is a 150-cavity adjacent to the enzyme active site (i.e., drug-binding site) with an entrance loop (known as the 150-loop, amino acid residues 147–152) that usually adopts an open conformation (Fig. 1). In group 2 NAs (N2, N3, N6, N7, and N9), the 150-loop is normally closed.15–17 However, when the enzyme is bound to appropriate inhibitors, this flexible loop becomes open and the formerly covered 150-cavity is exposed.18–22 Novel NAIs that target both the active site and the 150-cavity can potentially have more extensive interactions and higher affinities toward both group 1 and 2 NAs compared with conventional inhibitors.23–36
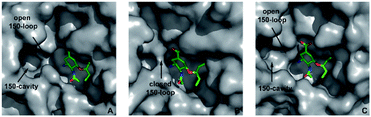 |
| | Fig. 1 Comparison of the crystal structures of representative group 1 and group 2 neuraminidases. (A) N1 virus with an open 150-loop (PDB code: 6HP0);25 (B) N2 virus with a closed 150-loop (PDB code: 4GZP);26 (C) N2 virus with an open 150-loop induced by oseltamivir carboxylate (PDB code: 4K1K).18 | |
Chemical derivatization of conventional NAIs via a group substitution oriented toward the 150-cavity could generate such potent inhibitors. The NAIs reported thus far were mainly designed based on the hydrophobic properties of the 150-cavity and were generally obtained by attaching a hydrophobic group to the structure of oseltamivir carboxylate (OSC) or zanamivir (ZNV).24–36 Herein, we report a different strategy for exploring novel and efficient NA inhibitors by targeting the charged amino acid residues around the entrance to the 150-cavity. We envisioned that the designed compounds, which could simultaneously bind the active site of NA and interact with the charged residues of the 150-cavity, possess high inhibitory potency toward various NA subtypes. Therefore, two series of C5-substituted OSC derivatives carrying various polar substituents at the triazolyl group were synthesized and evaluated for their biological activity in vitro.
Results and discussion
Design of target NA inhibitors
Among the approved NA inhibitors, oseltamivir (OSV), with its superior pharmacokinetics and bioavailability, is the only orally available drug. Therefore, it was chosen as the template molecule for this study. Moreover, the C5 amino group of OSV is well exposed toward the open 150-cavity and can serve as a potential modification site. It has been reported that for some OSC analogs featuring a 1,2,3-triazole at the C5 position, the triazole-extended portion can enter the 150-cavity.24,25 Thus, our strategy involved using a triazole ring with various functional groups as the linker between the OSC scaffold and the group entering the 150-cavity. Furthermore, polar substituents were introduced into the triazole-extended portion to explore the potential for strong interactions (hydrogen bonds or salt bridges) with charged residues around the entrance to the 150-cavity (Fig. 2A).
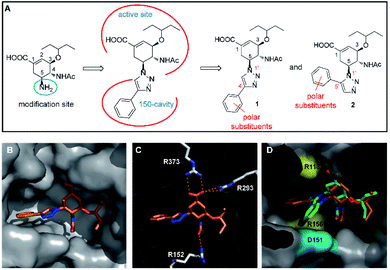 |
| | Fig. 2 Design approach and structural basis of the target neuraminidase (NA) inhibitors. (A) Design of the 1,2,3-triazolylated oseltamivir carboxylate (OSC) derivatives. (B) The X-ray crystal structure of compound 1e in complex with the N5 NA. The 4′-phenyl-1,2,3-triazolyl group can enter the 150-cavity. (C) Key interactions (hydrogen bond interactions) of 1e in complex with the N5 NA. The main structure (OSC scaffold) interacts with the active site in a manner similar to that of the binding pattern of OSC but without the additional strong interaction involving the phenyl group. Polar interactions are represented by dashed lines. Other interactions are not indicated. The residues involved in direct hydrogen bonds with the ligand are represented with sticks. (D) Comparison of the conformation of 1e (orange) and 2e (green) within the N5 NA crystal structure based on molecular docking experiments. Compound 2e could maintain a conformation similar to that of 1e and its phenyl group was closer to the acidic residues (cyan) and basic residues (yellow) near the entrance of the 150-cavity. The oxygen and nitrogen atoms of the compounds are colored red and blue, respectively. | |
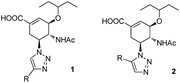
Given these points, we first designed five C5-substituted OSC derivatives (1a–e) with a 4′-aryl-1,2,3-triazolyl group as these derivatives were easy for modification, and expected to be capable of entering the 150-cavity. These compounds were synthesized by copper-catalyzed azide–alkyne cycloaddition (CuAAC),37 and preliminary enzymatic assays of their inhibition of N2 (H3N2) and N5 (H12N5) were performed (Table 1). Although 1c–e exhibited moderate inhibition potency at a similar level, compound 1e bearing a phenyl group on the triazole ring was preferred as the most promising parent molecule because of its simple structure and capacity for easy chemical modification. Subsequently, the X-ray crystal structure of compound 1e in complex with the N5 NA were determined and can be guided for more effective modifications (Fig. 2B and C). Crystals of the complex exhibited that 1e was indeed occupying both the catalytic site and the 150-cavity simultaneously, and its main structure (OSC scaffold) interacted with the active site in a manner similar to that of the binding pattern of OSC.38 However, no additional interactions of the phenyl group were observed, except for some hydrophobic interactions, resulting in a substantial decrease in inhibitory potency. It appeared that a rational modification of 1e would include the introduction of polar substituents such as amino, carboxyl, or alkyl-substituted N-containing groups into the benzene ring, so as to explore additional interactions with charged amino acids near the entrance of the 150-cavity (R118, E119, D151, and R156). However, with the substituent at the 4′ position of the triazole, the phenyl group faced toward the base of the cavity rather than toward the entrance. Therefore, a better approach was to force the orientation of the substituents on the triazole ring toward the charged residues near the entrance of the 150-cavity, using ruthenium-catalyzed 1,3-dipolar cycloaddition (RuAAC).39 This afforded the 5′-substituted isomer (2e) instead of a 4′-substituted triazole. Encouragingly, molecular docking studies showed that 2e maintained a conformation similar to that of 1e in the crystal structure of the N5 NA, allowing it to serve as the precursor molecule for the next round of inhibitor design (Fig. 2D).
Table 1 IC50 values (μM) of the target compounds based on the neuraminidase inhibition assay
Thus, in this study, a series of 5′-substituted 1,2,3-triazolylated OSC derivatives (2f–n) were synthesized and evaluated for their inhibitory potency toward NAs in vitro. Meanwhile, all the corresponding 4′-isomers (1f–n) were also tested to verify the validity of using 5′-substituted triazoles in our strategy.
Chemistry
As shown in Scheme 1, the 4′- and 5′-substituted 1,2,3-triazolylated OSC derivatives were prepared via coupling with azido oseltamivir (3), which can be obtained by the azidation of commercially available oseltamivir phosphate with imidazole-1-sulfonyl azide hydrochloride,25 and the corresponding alkynes (4) using either CuAAC37 or RuAAC,39 followed by hydrolysis or deprotection. Remarkably, although some alkynylamines require N-Boc protection prior to the RuAAc reaction to improve the reaction yield, no protection is required in the CuAAC reaction. All resulting triazolylated OSC derivatives were fully characterized by high-performance liquid chromatography (purity > 95%), nuclear magnetic resonance (NMR), and mass spectroscopy (MS). In most cases, the 1H chemical shift of the triazole H for 4′-substituted 1,2,3-triazolylated OSC derivatives was over 0.4 ppm larger than the corresponding 5′-isomers, and could be served as an identifying feature of the two series.
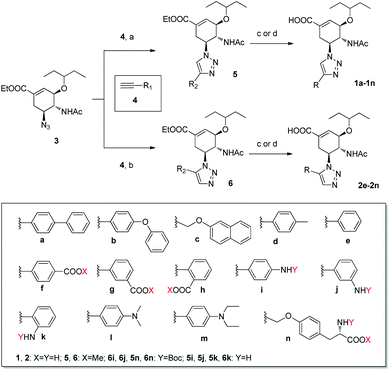 |
| | Scheme 1 Synthesis of target compounds. Reagents and conditions: (a) sodium ascorbate, aq CuSO4, THF/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), 45 °C, 6–12 h, 68–98%; (b) Cp*RuCl(PPh3)2, THF, 60 °C, 12–24 h, 29–89%; (c) aq NaOH, MeOH, rt, 6–12 h, 55–81%; (d) (i) aq NaOH, MeOH, rt, 6–12 h; (ii) TFA/DCM (1 1), 45 °C, 6–12 h, 68–98%; (b) Cp*RuCl(PPh3)2, THF, 60 °C, 12–24 h, 29–89%; (c) aq NaOH, MeOH, rt, 6–12 h, 55–81%; (d) (i) aq NaOH, MeOH, rt, 6–12 h; (ii) TFA/DCM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), rt, 3–4 h, 72–79%. 1), rt, 3–4 h, 72–79%. | |
NA inhibition assay
All the 5′-substituted 1,2,3-triazolylated OSC derivatives, along with their corresponding 4′-isomers, were evaluated for their NA inhibition of N5 (H12N5) from group 1 and N2 (H3N2) from group 2, using 4-methylumbelliferyl-N-acetylneuraminic acid (MUNANA) as a fluorescent substrate.40 To verify their inhibitory activity toward drug-resistant influenza viruses, an N2 (H3N2) NA with the E119V mutation, which confers resistance to OSV,41 was also employed in the assays. OSC was run in parallel as a control.
As shown in Table 1, all the 5′-substituted 1,2,3-triazolylated OSC derivatives exhibited higher IC50 values than their corresponding 4′-isomers and the parent compound (2e), suggesting that the 5′-substituted triazole-extended portions were well exposed toward the residues near the entrance of the 150-cavity, making it easier for these derivatives to gain additional interactions. However, all the tested compounds displayed weaker NA inhibitory activities than that of OSC, demonstrating that the new interactions could not compensate for the hydrogen bonds that were compromised by the insertion of the less basic triazole ring in place of the amino group at C5 of OSC. Generally, the tested compounds exhibited equal inhibition toward both of the NA subtypes and did not display a marked degree of selectivity. Furthermore, the decreased inhibitory activities toward mutN2 compared with wild N2 for most of the compounds suggested that interactions between the E119 residue and these compounds remained.
Structure–activity relationship analysis indicated that the substituent positions on the phenyl ring played an important role in the potency of the compounds. The compound with an electropositive substituent at the meta-position of the phenyl group (2j) showed considerably increased potency compared with its para- and ortho-substituted isomers, whereas the compound with an electronegative substituent at the ortho-position (2h) exhibited higher inhibition activity. This phenomenon is likely largely dependent on the hydrogen-bonding strengths between polar substituents on the benzene ring of the inhibitors and the charged residues near the entrance of the 150-cavity. Furthermore, although compound 2l, with a dimethyl-substituted derivative on the amino group of 2i, showed slightly increased inhibition, 2m had almost no activity. One possibility is that the conformation of 2m, which bears a large substituent on the amino group, may be distorted within the NAs, resulting in unfavorable interactions with the binding pockets of these enzymes.
Remarkably, compound 2j exhibited significantly higher binding potency toward N2 and N5 NA than that of other compounds, with IC50 values of 19 nM and 45 nM, respectively. Importantly, 2j also showed moderate inhibition toward mutN2 in the sub-micromolar range.
Anti-influenza virus activity and cytotoxicity of 2j and OSC in MDCK cells
Next, the anti-influenza virus potency of the most promising compound (2j) was evaluated in Madin–Darby canine kidney (MDCK) cells, and OSC was selected as a reference compound for testing in parallel. Three viruses, comprising H1N1 (A/Puerto Rico/8/34), H3N2 (A/Moscow/10/99), and an OSV-resistant strain of the latter containing the E119V mutation, were rescued with reverse genetics41,42 and used for the assays (Table 2). In agreement with its IC50 against the purified enzymes, compound 2j showed lower activity than OSC against H1N1 and H3N2 (EC50 = 11.53 μM and 7.65 μM, respectively). As for the OSV-resistant influenza A virus (H3N2 E119V), both 2j and OSC exhibited an EC50 value larger than 100 μM, and at this concentration, the inhibition rates were 24.6% and 44.1%, respectively. Notably, 2j exhibited no appreciable cytotoxicity at the highest tested concentrations (CC50 > 500 μM). The cell-based assay demonstrated that compound 2j, with moderate inhibition toward influenza A viruses and low cytotoxicity in vitro, deserves further optimization.
Table 2 Anti-influenza virus activity and cytotoxicity of 2j and oseltamivir carboxylate (OSC)
| Compounds |
EC50 (μM)a |
CC50b |
| H1N1c |
H3N2c |
H3N2 E119Vc |
| EC50 refers to the concentration required to achieve 50% protection against virus-induced cytopathic effect. All the assays were performed in quadruplicate and the results are reported as the mean ± SD. CC50 refers to the concentration of 50% cellular toxicity. A/Puerto Rico/8/1934; H3N2, A/Moscow/10/99; H3N2 E119V, and A/Moscow/10/99H3N2 background containing an E119V mutation in the neuraminidase gene. At 100 μM, the inhibition rates were 24.6% for 2j and 44.1% for OSC, respectively. |
| 2j |
11.53 ± 2.69 |
7.65 ± 0.70 |
>100d |
>500 |
| OSC |
0.25 ± 0.081 |
0.43 ± 0.080 |
>100d |
>500 |
Molecular docking study
Unfortunately, the crystal structure of NA in complex with 2j could not be obtained to present the mechanism of action more intuitively. To further investigate the discrepancy in activity between the 5′-substituted 1,2,3-triazolylated OSC derivatives and their corresponding 4′-isomers within the 150-cavity of the NA structure, molecular docking studies of two representative compounds (2h and 2j) and their 4′-isomers (1h and 1j) with the crystal structure of N5 NA extracted from the complex with 1e was conducted with GOLD software (Genetic Optimization of Ligand Docking),43 and PyMOL44 was used to visualize the results.
As shown in Fig. 3A, the triazole-extended portions of all four compounds occupied the 150-cavity while the other structural regions of the compounds interacted with the active site in a manner similar to that of the binding pattern of OSC. The difference was that the 2 series (2h and 2j) exhibited several key interactions within the 150-cavity that were not observed for the 1 series (1h and 1j). As we had speculated, hydrogen-bonding interactions were clearly observed between the amino group of 2j and the anionic residue D151, whereas the carboxyl group of 2h interacted with the basic amino acids R152 and R156 (Fig. 3B). The main reason for this is that the 5′-substituted triazole-extended portion formed by RuAAC was closer to the charged residues near the entrance of the 150 cavity, which resulted in improved enzyme inhibition.
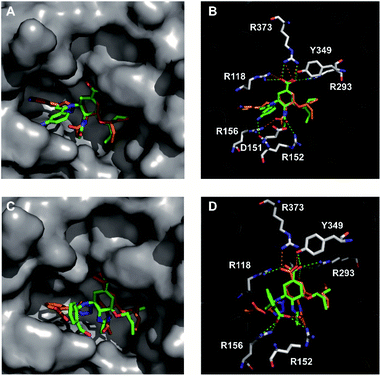 |
| | Fig. 3 Comparison of the binding modes and interactions of 5′-substituted 1,2,3-triazolylated OSC derivatives (green) and their 4′-isomers (orange) with N5 neuraminidase. (A and B) Overlay of N5-1j with N5-2j; (C and D) Overlay of N5-1h and N5-2h. Polar interactions are represented by dashed lines (green for the 2 series and orange for the 1 series). Other interactions are not shown. The residues involved in direct hydrogen bonds with the ligand are represented with sticks. The oxygen and nitrogen atoms of the compounds are colored red and blue, respectively. | |
Taken together, the molecular docking experiments were in agreement with the results of the NA inhibition assays, especially for the OSC analog 2j, which bears a 3-aminophenyl group on the 5′-substituted triazole ring that can simultaneously bind the active site of NA and interact with the residues in the 150-cavity, allowing high inhibitory potency toward both NA subtypes. These findings explain the mechanism behind the remarkable ability of compound 2j to bind and inhibit NA enzymes.
Conclusions
Since the first report of the 150-cavity in 2006, tremendous progress has been made in the development of NA inhibitors specifically targeted toward this hydrophobic cavity. In this study, we described a different strategy that designing two series of 1,2,3-triazolylated OSC derivatives by targeting the charged amino acid residues around the entrance to the 150-cavity. The NA inhibition assays demonstrated that the 2 series (2e–n) generally had better activity than the 1 series (1e–n), whereas the compound 2j, bearing a 3-phenylamino group on the triazole ring, was the most potent inhibitor of all the tested NAs, including an N2 NA that contains an E119V OSC-resistant mutation. Moreover, 2j exhibited acceptable potency and low cytotoxicity in cellular assays. Molecular docking studies revealed that the phenylamino group of 2j can form an additional hydrogen bond with the D151 residue near the entrance of the NA 150-cavity. The design method described in this study provides useful insights into the development of novel NA inhibitors. The compound 2j warrants further structural optimization to obtain a candidate for clinical use.
Experimental
General chemistry information
All chemical regents and solvents, including 4 and OSC, were obtained from commercial source and used as purchased. Compound 3 was synthesized as described previously.25 Silica gel (100–200 mesh) for column chromatography was purchased from Qingdao Haiyang Chemical Co., Ltd (China). Sephadex G-10 was purchased from GE Healthcare (China). IR data were recorded using a Nicolet IS5 FTIR spectrophotometer (Waltham, MA, USA). 1H NMR (500 MHz) and 13C NMR (125 MHz) spectra were measured on a Bruker Advance spectrometer. ESIMS and HR-ESIMS spectra were recorded on a Bruker Apex IV FTMS spectrometer. The purity of final compounds is ≥95%, which was determined by an HPLC system (Agilent Technologies 1200) equipped with a UV detector using conditions as follows: Agilent Eclipse XDB-C18 columns (250 × 9.4 mm, 5 μm), 95 → 20% deionized water/acetonitrile gradient at 2 mL min−1 over 30 min, UV detection at 254 nm wavelength.
NA inhibition assay
The N5 (H12N5, A/Duck/Alberta/60/1976), N2 (H3N2, A/Moscow/10/1999) and its OSV-resistant virus with E119V mutation were obtained from Sino Biological Inc. NA enzyme activity and competitive inhibition by inhibitor (OSC, 1, or 2) were tested using MUNANA (4-methylumbelliferyl-N-acetylneuraminic acid) as a fluorescent substrate, according to published protocols.40 In brief, NA activity was predetermined in an appropriate range (normally in 10–50 nM, in PBS) for fluorescence detection in the presence of MUNANA (167 μM, in PBS). For inhibition assays, the enzyme (10 μL) was mixed with serial 3-fold dilutions of inhibitor (10 μL, initial concentration at 1 mM) in PBS in a 96-well microplate and incubated at 37 °C for 30 min. MUNANA (30 μL, 167 μM, in PBS) was then added to the mixture and fluorescence was measured on a SpectrMax M5 microplate reader (molecular devices), at excitation and emission wavelengths of 355 and 460 nm, respectively. Each assay was performed in no less than triplicates. IC50 values were determined with GraphPad Prism 8.
X-ray crystallography
The N5 enzyme for X-ray crystallography was cloned, expressed, and purified as previously described.45 The N5-1e complex was prepared by soaking N5 crystals in crystallization solution (0.1 M HEPES, pH 7.5, containing 12% v/v polyethylene glycol 3350) supplemented with 1e (20 mM) at 18 °C for 3 h. The crystals were then flash-cooled at 100 K with cryoprotection. Diffraction data were collected on the beamline BL17U at Shanghai Synchrotron Radiation Facility. The data were processed and scaled with HKL2000.46 The structure of N5 in complex with 1e was determined by molecular replacement in Phaser using the CCP4 program suite,47 with the known N5 structure (PDB code 3SAN)45 as a search model. Model building and refinement were performed as described previously45 using COOT,48 REFMAC5,49 and the PHENIX package.50 The stereochemical quality of the final models was assessed with the program PROCHECK.51 All structural figures were generated using PyMOL software (http://www.pymol.org). Statistical support for the structures obtained is presented in ESI Table S1.† Atomic coordinates and experimental structure factors have been deposited in the PDB database under identification code 7E6Q.
Anti-influenza virus activity and cytotoxicity of 2j and OSC in MDCK cells
The anti-influenza activities of compounds were measured by the EC50 values, which were the concentrations of tested molecules for 50% protection of the influenza virus infection-mediated cytopathic effects (CPE).29 Serial 3-fold dilutions of inhibitor (initial concentration at 200 μM) in DMEM containing 2 mg mL−1 TPCK-trypsin and 2% bovine serum albumin (BSA) were prepared and mixed with an equal volume of virus (200 TCID50) in the same medium. The mixtures (100 μL) were used to infect MDCK cells, which were grown on 96-well microplates (104 cells per well). After 72 h of incubation at 37 °C in a humidified 5% CO2 incubator, 50 μL per well of Cell Counting Kit-8 (CCK-8, Dojindo Laboratories) reagent solution (5 μL CCK-8 and 45 μL media) was added. After incubation at 37 °C for additional 2 h, the absorbance was measured at 450 nm using a SpectraMax M5 fluorescence spectrophotometer (molecular devices). The EC50 values were calculated by nonlinear regressions using GraphPad Prism 8. The 50% cytotoxic concentration (CC50) values of compounds to MDCK cells were determined by the procedures analogous to the EC50 determination but without virus infection.
Molecular docking study
GOLD (version 5.8.1, Cambridge Crystallographic Data Centre, Cambridge, UK) is a powerful docking software which docks compounds in the active region to generate a good bonding pose depending on different scoring function and template.52–54 Protein N2 (PDB code: 4GZP) and N5 (extracted from N5-1e complex) structure was prepared using GOLD program and all hydrogens were added. The structure of native ligand present in NA proteins was removed, and the 3D structures of the studied compounds were generated using Chembio3D Ultra 14.0 software and energy minimization using molecular mechanics (MM2). Binding site of the enzyme for docking was defined automatically by setting 6 Å around the coordinates of the native ligand. All available scoring functions in GOLD, namely CHEMPLP, ChemScore, ASP, and GoldScore were tried to re-dock compounds inside the NA protein and then RMSD measure was calculated to validate and find the most appropriate scoring function for docking analyses. For each compound, ten docked solutions were generated with their corresponding GOLD fitness scores. The selection of best ligand pose was done based on their interactions with amino acid residues of the target protein showing least clashes and having highest fitness score. The images were generated using PyMOL (http://www.pymol.org).
Synthetic protocols
General procedure for the synthesis of 5a–5n via CuAAC. To the mixture of 3 (135 mg, 0.4 mmol, 1.0 eq.) and alkyne 4 (0.5 mmol, 1.25 eq.) in 16 mL of THF/H2O (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added freshly prepared 1 M solution of CuSO4·5H2O (24 μL, 0.06 equiv.) in water, followed by the addition of sodium ascorbate (8 mg, 0.04 mmol, 0.1 eq.). The heterogeneous reaction mixture was stirred at room temperature for 6–12 h and monitored by TLC. After the completion, the reaction was evaporated to dryness. The crude product was purified by column chromatography, eluted with 25–75% EtOAc in petroleum ether, yielded the 4′-substituted-1,2,3-triazolylated OSC derivatives 5.
1) was added freshly prepared 1 M solution of CuSO4·5H2O (24 μL, 0.06 equiv.) in water, followed by the addition of sodium ascorbate (8 mg, 0.04 mmol, 0.1 eq.). The heterogeneous reaction mixture was stirred at room temperature for 6–12 h and monitored by TLC. After the completion, the reaction was evaporated to dryness. The crude product was purified by column chromatography, eluted with 25–75% EtOAc in petroleum ether, yielded the 4′-substituted-1,2,3-triazolylated OSC derivatives 5.
General procedure for the synthesis of 6e–6n via RuAAC. To the mixture of 3 (135 mg, 0.4 mmol, 1.0 eq.) and alkyne 4 (0.6 mmol, 1.5 eq.) in 10 mL of dry THF was added Cp*RuCl(PPh3)2 (16 mg, 0.02 mmol, 0.05 eq.) and stirred at 60 °C for 12–24 h under a nitrogen atmosphere. The reaction mixture was monitored by TLC, and after the completion, evaporated to dryness. The crude product was purified by column chromatography, eluted with 25–75% EtOAc in petroleum ether, yielded the 5′-substituted-1,2,3-triazolylated OSC derivatives 6.
General procedure A for the synthesis of 1 or 2 (reaction condition c in Scheme 1). To a solution of compound 5 or 6 (0.2 mmol) in MeOH (10 mL) was added an aqueous solution of NaOH (1 M, 5 mL). The mixture was stirred at rt for 6–12 h, neutralized with Dowex 50 (H+), filtered, and concentrated. The residue was dissolved in deionized water (0.5 mL) and filtered (Millipore Express 0.22 μM filter). Purification with Sephadex G-10 (eluent: 0.1% TFA in H2O) yielded 1 or 2 as a white foam.
General procedure B for the synthesis of 1 or 2 (reaction condition d in Scheme 1). To a solution of compound 5 or 6 (0.2 mmol) in MeOH (10 mL) was added an aqueous solution of NaOH (1 M, 5 mL). The mixture was stirred at rt for 6–12 h, neutralized with Dowex 50 (H+), filtered, and concentrated. The residue was dissolved in CH2Cl2/TFA (10 mL, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The mixture was stirred at rt for 3–4 h and concentrated. The resulting residue was dissolved in deionized water (0.5 mL) and filtered (Millipore Express 0.22 μM filter). Purification with Sephadex G-10 (eluent: 0.1% TFA in H2O) yielded 1 or 2 as a white foam.
1, v/v). The mixture was stirred at rt for 3–4 h and concentrated. The resulting residue was dissolved in deionized water (0.5 mL) and filtered (Millipore Express 0.22 μM filter). Purification with Sephadex G-10 (eluent: 0.1% TFA in H2O) yielded 1 or 2 as a white foam.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(1,1′-biphenyl-4-yl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5a). Using the general procedure for CuAAC, compound 5a was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-biphenylylacetylene (89 mg, 0.5 mmol, 1.25 eq.) as a white foam (188 mg, 91%).1H NMR (500 MHz, CDCl3) δ 7.99 (s, 1H), 7.91–7.35 (m, 9H), 6.92 (s, 1H), 6.16 (s, 1H), 5.50 (dd, J = 17.4, 11.1 Hz, 1H), 4.77 (d, J = 8.6 Hz, 1H), 4.25–4.19 (m, 2H), 4.09–4.07 (m, 1H), 3.43–3.39 (m, 1H), 3.13–3.03 (m, 2H), 1.79 (s, 3H), 1.60–1.50 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.96 (t, J = 7.4 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H). ESIMS calcd for C30H37N4O4 (M + H)+: 517.3. Found: 517.1.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-[4-(4-phenoxyphenyl)-1H-1,2,3-triazol-1-yl]cyclohex-1-ene-1-carboxylate (5b). Using the general procedure for CuAAC, compound 5b was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 1-(4-ethynylphenoxy)benzene (97 mg, 0.5 mmol, 1.25 eq.) as a white foam (183 mg, 86%). 1H NMR (500 MHz, CDCl3) δ 7.84 (s, 1H), 7.77 (d, J = 8.7 Hz, 1H), 7.36 (dd, J = 8.5, 7.5 Hz, 2H), 7.13 (t, J = 7.4 Hz, 3H), 7.06–7.04 (m, 3H), 6.91 (s, 1H), 6.07 (s, 1H), 5.45 (td, J = 11.1, 6.1 Hz, 1H), 4.74 (d, J = 8.8 Hz, 1H), 4.23 (q, J = 7.1 Hz, 2H), 3.42–3.37 (m, 1H), 3.11–3.00 (m, 2H), 1.77 (s, 3H), 1.58–1.49 (m, 4H), 1.30 (t, J = 7.1 Hz, 3H), 0.94 (t, J = 7.4 Hz, 3H), 0.89 (t, J = 7.4 Hz, 3H). ESIMS calcd for C30H37N4O5 (M + H)+: 533.3. Found: 533.2.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(2-naphthyloxy)methyl-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5c). Using the general procedure for CuAAC, compound 5c was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and naphth-2-yloxymethylacetylene (92 mg, 0.5 mmol, 1.25 eq.) as a white foam (183 mg, 88%). 1H NMR (500 MHz, CDCl3) δ 7.79 (s, 1H), 7.76–7.72 (m, 2H), 7.71 (d, J = 3.3 Hz, 1H), 7.43 (t, J = 7.2 Hz, 1H), 7.33 (t, J = 7.2 Hz, 1H), 7.23 (d, J = 1.8 Hz, 1H), 7.14 (dd, J = 8.9, 2.3 Hz, 1H), 6.86 (s, 1H), 6.49 (d, J = 6.7 Hz, 1H), 5.41 (dd, J = 17.3, 10.9 Hz, 1H), 5.32–5.22 (m, 2H), 4.70 (d, J = 8.4 Hz, 1H), 4.19 (q, J = 7.1 Hz, 2H), 3.99 (dd, J = 19.5, 8.7 Hz, 1H), 3.35 (p, J = 5.6 Hz, 1H), 3.09–2.94 (m, 2H), 1.63 (s, 3H), 1.54–1.45 (m, 4H), 1.27 (t, J = 7.1 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C29H37N4O5 (M + H)+: 521.2759. Found: 521.2770.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(4-methylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5d). Using the general procedure for CuAAC, compound 5d was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynyltoluene (58 mg, 0.5 mmol, 1.25 eq.) as a white foam (169 mg, 93%). 1H NMR (500 MHz, CDCl3) δ 7.95 (s, 1H), 7.64 (d, J = 7.7 Hz, 2H), 7.16 (d, J = 7.6 Hz, 2H), 7.02 (d, J = 6.5 Hz, 1H), 6.89 (s, 1H), 5.34 (t, J = 13.5 Hz, 1H), 4.65 (d, J = 7.6 Hz, 1H), 4.24–4.19 (m, 3H), 3.46–3.36 (m, 1H), 3.10–2.96 (m, 2H), 2.34 (s, 3H), 1.66 (s, 3H), 1.57–1.51 (m, 4H), 1.28 (t, J = 7.1 Hz, 3H), 0.93 (t, J = 7.3 Hz, 3H), 0.88 (t, J = 7.3 Hz, 3H). HRESIMS calcd for C25H35N4O4 (M + H)+: 455.2653. Found: 455.2651.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-(4-phenyl-1H-1,2,3-triazol-1-yl)cyclohex-1-ene-1-carboxylate (5e). Using the general procedure for CuAAC, compound 5e was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and phenylacetylene (52 mg, 0.5 mmol, 1.25 eq.) as a white foam (173 mg, 98%). 1H NMR (500 MHz, CDCl3) δ 7.96 (s, 1H), 7.78 (d, J = 7.4 Hz), 7.38 (t, J = 7.5 Hz, 2H), 7.31 (t, J = 7.3 Hz, 1H), 6.90 (s, 1H), 6.72 (d, J = 7.1 Hz, 1H), 5.42–5.33 (m, 1H), 4.68 (d, J = 8.4 Hz, 1H), 4.22 (dd, J = 14.2, 7.1 Hz, 2H), 4.19–4.11 (m, 1H), 3.41 (p, J = 5.5 Hz, 1H), 3.11–2.97 (m, 2H), 1.69 (s, 3H), 1.56–1.52 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.94 (d, J = 7.4 Hz, 3H), 0.88 (d, J = 7.4 Hz, 3H). HRESIMS calcd for C24H33N4O4 (M + H)+: 441.2496. Found: 441.2493.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(4-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5f). Using the general procedure for CuAAC, compound 5f was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynylbenzoic acid methyl ester (80 mg, 0.5 mmol, 1.25 eq.) as a white foam (171 mg, 86%). 1H NMR (500 MHz, CDCl3) δ 8.06 (d, J = 8.1 Hz, 2H), 8.02 (s, 1H), 7.86 (d, J = 8.2 Hz, 2H), 6.91 (s, 1H), 6.35 (d, J = 4.5 Hz, 1H), 5.45 (dd, J = 16.9, 11.1 Hz, 1H), 4.71 (d, J = 8.3 Hz, 1H), 4.23 (q, J = 7.1 Hz, 2H), 4.13–4.07 (m, 1H), 3.92 (s, 3H), 3.43–3.37 (m, 1H), 3.12–2.99 (m, 2H), 1.73 (s, 3H), 1.55–1.51 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.93 (t, J = 7.4 Hz, 3H), 0.88 (t, J = 7.3 Hz, 3H). HRESIMS calcd for C26H35N4O6 (M + H)+: 499.2551. Found: 499.2544.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(3-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5g). Using the general procedure for CuAAC, compound 5g was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 3-ethynylbenzoic acid methyl ester (80 mg, 0.5 mmol, 1.25 eq.) as a white foam (157 mg, 79%). 1H NMR (500 MHz, CDCl3) δ 8.38 (s, 1H), 8.11 (s, 1H), 7.96 (t, J = 7.7 Hz, 2H), 7.41 (t, J = 7.8 Hz, 1H), 6.96 (d, J = 8.0 Hz, 1H), 6.89 (s, 1H), 5.39 (td, J = 11.1, 6.1 Hz, 1H), 4.65 (d, J = 8.3 Hz, 1H), 4.29–4.17 (m, 3H), 3.86 (s, 3H), 3.41 (t, J = 8.4 Hz, 1H), 3.14–2.97 (m, 2H), 1.69 (s, 3H), 1.56–1.48 (m, 4H), 1.27 (t, J = 7.1 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H35N4O6 (M + H)+: 499.2551. Found: 499.2551.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(2-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5h). Using the general procedure for CuAAC, compound 5h was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 2-ethynylbenzoic acid methyl ester (80 mg, 0.5 mmol, 1.25 eq.) as a white foam (149 mg, 75%). 1H NMR (500 MHz, CDCl3) δ 7.87–7.83 (m, 2H), 7.75 (d, J = 7.6 Hz, 1H), 7.57 (t, J = 7.7 Hz, 1H), 7.44 (t, J = 7.5 Hz, 1H), 6.91 (s, 1H), 5.83 (s, 1H), 5.54–5.49 (m, 1H), 4.82 (d, J = 8.4 Hz, 1H), 4.24 (q, J = 7.1 Hz, 2H), 3.98–3.90 (m, 1H), 3.41–3.34 (m, 1H), 3.11 (d, J = 7.1 Hz, 2H), 1.84 (s, 3H), 1.58–1.49 (m, 4H), 1.31 (t, J = 7.1 Hz, 3H), 1.29 (t, J = 7.1 Hz, 3H), 0.94 (t, J = 7.4 Hz, 3H), 0.88 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H34N4NaO6 (M + Na)+: 521.2376. Found: 521.2376.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(4-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy) cyclohex-1-ene-1-carboxylate (5i). Using the general procedure for CuAAC, compound 5i was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynylaniline (58 mg, 0.5 mmol, 1.25 eq.) as a white foam (131 mg, 72%). 1H NMR (500 MHz, CDCl3) δ 7.84 (s, 1H), 7.53 (d, J = 8.4 Hz, 2H), 6.98 (s, 1H), 6.75 (d, J = 8.4 Hz, 2H), 6.17 (d, J = 8.0 Hz, 1H), 5.41 (td, J = 11.2, 6.4 Hz, 1H), 4.75 (d, J = 8.1 Hz, 1H), 4.23 (q, J = 7.2 Hz, 2H), 4.08–4.03 (m, 1H), 3.42–3.40 (m, 1H), 3.07–3.01 (m, 2H), 1.74 (s, 3H), 1.59–1.53 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.96 (t, J = 7.4 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H). ESIMS calcd for C24H34N5O4 (M + H)+: 456.3. Found: 456.2.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy) cyclohex-1-ene-1-carboxylate (5j). Using the general procedure for CuAAC, compound 5j was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 3-ethynylaniline (58 mg, 0.5 mmol, 1.25 eq.) as a white foam (124 mg, 68%). 1H NMR (500 MHz, CDCl3) δ 7.85 (s, 1H), 7.21–7.12 (m, 3H), 6.90 (s, 1H), 6.65 (d, J = 7.3 Hz, 1H), 6.36 (d, J = 6.3 Hz, 1H), 5.38 (td, J = 11.1, 6.1 Hz, 1H), 4.70 (d, J = 8.3 Hz, 1H), 4.22 (q, J = 7.1 Hz, 2H), 4.08 (d, J = 6.7 Hz, 1H), 3.41–3.34 (m, 1H), 3.09–2.98 (m, 2H), 1.71 (s, 3H), 1.57–1.49 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.93 (d, J = 7.4 Hz, 3H), 0.88 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C24H34N5O4 (M + H)+: 456.2605. Found: 456.2603.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(2-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy) cyclohex-1-ene-1-carboxylate (5k). Using the general procedure for CuAAC, compound 5k was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 2-ethynylaniline (59 mg, 0.5 mmol, 1.25 eq.) as a white foam (138 mg, 76%). 1H NMR (500 MHz, CDCl3) δ 7.98 (s, 1H), 7.44 (d, J = 7.5 Hz, 1H), 7.19 (t, J = 7.5 Hz, 1H), 7.09 (s, 1H), 6.94–6.91 (m, 2H), 5.49 (dd, J = 16.3, 10.2 Hz, 1H), 4.75 (d, J = 8.0 Hz, 1H), 4.23 (q, J = 7.1 Hz, 2H), 4.05 (s, 1H), 3.39 (dd, J = 11.3, 5.6 Hz, 1H), 3.09–3.01 (m, 2H), 1.76 (s, 3H), 1.56–1.49 (m, 4H), 1.30 (t, J = 7.1 Hz, 3H), 0.94 (d, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C24H34N5O4 (M + H)+: 456.2611. Found: 456.2609.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(4-dimethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5l). Using the general procedure for CuAAC, compound 5l was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynyl-N,N-dimethylaniline (72 mg, 0.5 mmol, 1.25 eq.) as a white foam (164 mg, 85%). 1H NMR (500 MHz, CDCl3) δ 7.74 (s, 1H), 7.68 (d, J = 8.3 Hz, 2H), 6.90–6.82 (m, 3H), 6.15 (d, J = 7.6 Hz, 1H), 5.40 (td, J = 11.1, 6.1 Hz, 1H), 4.74 (d, J = 8.6 Hz, 1H), 4.23 (q, J = 7.1 Hz, 2H), 4.04 (dd, J = 19.8, 8.7 Hz, 1H), 3.46–3.37 (m, 1H), 3.14–2.93 (m, 8H), 1.73 (s, 3H), 1.58–1.51 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.95 (t, J = 7.4 Hz, 3H), 0.89 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H38N5O4 (M + H)+: 484.3. Found: 484.2.
Ethyl (3R,4R,5S)-4-acetamido-5-[4-(4-diethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (5m). Using the general procedure for CuAAC, compound 5m was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynyl-N,N-diethylaniline (87 mg, 0.5 mmol, 1.25 eq.) as a white foam (162 mg, 79%).1H NMR (500 MHz, CDCl3) δ 7.69 (s, 1H), 7.61 (d, J = 8.4 Hz, 2H), 6.91 (s, 1H), 6.73 (d, J = 8.4 Hz, 2H), 5.67 (d, J = 7.4 Hz, 1H), 5.43 (dd, J = 18.6, 9.9 Hz, 1H), 4.77 (d, J = 7.5 Hz, 1H), 4.34–4.16 (m, 4H), 4.03–3.92 (m, 2H), 3.39–3.36 (m, 1H), 3.07–3.05 (m, 2H), 1.75 (s, 3H), 1.56–1.49 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.98–0.87 (m, 12H). HRESIMS calcd for C28H42N5O4 (M + H)+: 512.3. Found: 512.2.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-{4-{[4-((S)-2-tert-butoxycarbonylamino-3-methoxy-3-oxopropyl)phenoxy]methyl}-1H-1,2,3-triazol-1-yl}cyclohex-1-ene-1-carboxylate (5n). Using the general procedure for CuAAC, compound 5n was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and methyl(S)-2-[(tert-butoxycarbonyl)amino]-3-[4-(prop-2-yn-1-yloxy)phenyl]propanoate (167 mg, 0.5 mmol, 1.25 eq.) as a white foam (207 mg, 77%). 1H NMR (500 MHz, CDCl3) δ 7.68 (s, 1H), 7.02 (d, J = 8.1 Hz, 2H), 6.89 (d, J = 8.3 Hz, 3H), 5.96 (s, 1H), 5.42 (d, J = 8.3 Hz, 1H), 5.20–5.11 (m, 2H), 4.98 (d, J = 7.7 Hz, 1H), 4.72 (d, J = 8.5 Hz, 1H), 4.52 (d, J = 6.8 Hz, 1H), 4.21 (q, J = 7.1 Hz, 2H), 3.91 (d, J = 7.5 Hz, 1H), 3.70 (s, 3H), 3.39–3.31 (m, 1H), 3.09–2.94 (m, 4H), 1.68 (s, 3H), 1.55–1.45 (m, 4H), 1.41 (s, 9H), 1.28 (t, J = 7.1 Hz, 3H), 0.91 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C34H49N5NaO9 (M + Na)+: 694.3423. Found: 694.3431.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-(5-phenyl-1H-1,2,3-triazol-1-yl)cyclohex-1-ene-1-carboxylate (6e). Using the general procedure for RuAAC, compound 6e was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and phenylacetylene (62 mg, 0.6 mmol, 1.5 eq.) as a white foam (157 mg, 89%). 1H NMR (500 MHz, CDCl3) δ 7.67 (s, 1H), 7.49–7.37 (m, 5H), 6.84 (s, 1H), 6.08 (d, J = 5.5 Hz, 1H), 5.57 (dd, J = 19.7, 8.3 Hz, 1H), 5.01 (d, J = 10.6 Hz, 1H), 4.18 (qd, J = 7.1, 2.9 Hz, 2H), 3.97 (dd, J = 16.9, 10.3 Hz, 1H), 3.33–3.27 (m, 1H), 2.84 (d, J = 8.1 Hz, 2H), 1.75 (s, 3H), 1.53–1.43 (m, 4H), 1.26 (t, J = 7.1 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C24H33N4O4 (M + H)+: 441.2496. Found: 441.2494.
Ethyl (3R,4R,5S)-4-acetamido-5-[5-(4-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (6f). Using the general procedure for RuAAC, compound 6f was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynylbenzoic acid methyl ester (96 mg, 0.6 mmol, 1.5 eq.) as a white foam (106 mg, 53%).1H NMR (500 MHz, CDCl3) δ 8.05 (d, J = 8.2 Hz, 2H), 8.01 (s, 1H), 7.86 (d, J = 8.2 Hz, 2H), 6.91 (s, 1H), 6.36 (d, J = 5.8 Hz, 1H), 5.44 (td, J = 11.1, 5.9 Hz, 1H), 4.70 (d, J = 8.3 Hz, 1H), 4.22 (q, J = 7.1 Hz, 2H), 4.11 (dd, J = 19.5, 9.0 Hz, 1H), 3.92 (s, 3H), 3.43–3.36 (m, 1H), 3.12–2.99 (m, 2H), 1.72 (s, 3H), 1.58–1.50 (m, 4H), 1.29 (t, J = 7.1 Hz, 3H), 0.93 (t, J = 7.4 Hz, 3H), 0.88 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H35N4O6 (M + H)+: 499.2551. Found: 499.2537.
Ethyl (3R,4R,5S)-4-acetamido-5-[5-(3-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (6g). Using the general procedure for RuAAC, compound 6g was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 3-ethynylbenzoic acid methyl ester (96 mg, 0.6 mmol, 1.5 eq.) as a white foam (76 mg, 38%). 1H NMR (500 MHz, CDCl3) δ 8.14 (t, J = 4.4 Hz, 1H), 8.01 (s, 1H), 7.72 (s, 1H), 7.60 (d, J = 4.9 Hz, 2H), 6.84 (s, 1H), 5.89 (d, J = 6.0 Hz, 1H), 5.52 (dt, J = 17.3, 8.5 Hz, 1H), 4.96 (d, J = 8.1 Hz, 1H), 4.19 (q, J = 6.5 Hz, 2H), 4.08 (d, J = 6.7 Hz, 1H), 3.94 (s, 3H), 3.29 (p, J = 5.7 Hz, 1H), 2.96–2.85 (m, 2H), 1.74 (s, 3H), 1.52–1.42 (m, 4H), 1.27 (t, J = 7.1 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H35N4O6 (M + H)+: 499.2551. Found: 499.2557.
Ethyl (3R,4R,5S)-4-acetamido-5-[5-(2-methoxycarbonylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (6h). Using the general procedure for RuAAC, compound 6h was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 2-ethynylbenzoic acid methyl ester (96 mg, 0.6 mmol, 1.5 eq.) as a white foam (58 mg, 29%). 1H NMR (500 MHz, CDCl3) δ 7.79–7.77 (m, 2H), 7.44 (t, J = 7.4 Hz, 1H), 7.11 (d, J = 8.4 Hz, 1H), 7.00 (t, J = 7.4 Hz, 1H), 6.88 (s, 1H), 5.67 (d, J = 5.0 Hz, 1H), 5.47 (dd, J = 18.2, 9.0 Hz, 1H), 4.80 (d, J = 8.4 Hz, 1H), 4.22 (q, J = 7.2 Hz, 2H), 3.86 (dd, J = 18.2, 8.4 Hz, 1H), 3.37–3.34 (m, 1H), 3.14 (d, J = 13.5 Hz, 2H), 1.69 (s, 3H), 1.56–1.44 (m, 4H), 1.36 (t, J = 7.2 Hz, 3H), 0.92 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C26H35N4O6 (M + H)+: 499.2557. Found: 499.2555.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-[5-(4-tert-butoxycarbonylaminophenyl)-1H-1,2,3-triazol-1-yl]cyclohex-1-ene-1-carboxylate (6i). Using the general procedure for RuAAC, compound 6i was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and tert-butyl (4-ethynylphenyl)carbamate (130 mg, 0.6 mmol, 1.5 eq.) as a white foam (136 mg, 61%). 1H NMR (500 MHz, CDCl3) δ 7.66 (s, 1H), 7.50 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 8.4 Hz, 2H), 6.84 (s, 1H), 6.63 (s, 1H), 5.76 (d, J = 6.4 Hz, 1H), 5.53–5.49 (m, 1H), 4.99 (dd, J = 9.2, 2.2 Hz, 1H), 4.25–4.16 (m, 2H), 3.98–3.95 (m, 1H), 3.31 (dt, J = 11.5, 5.7 Hz, 1H), 3.02–2.98 (m, 2H), 1.73 (s, 3H), 1.49 (s, 9H), 1.53–1.43 (m, 13H), 1.27 (t, J = 7.1 Hz, 3H), 0.91 (d, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). ESIMS calcd for C29H42N5O6 (M + H)+: 556.3. Found: 556.2.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-[5-(3-tert-butoxycarbonylaminophenyl)-1H-1,2,3-triazol-1-yl]cyclohex-1-ene-1-carboxylate (6j). Using the general procedure for RuAAC, compound 6j was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and tert-butyl (3-ethynylphenyl)carbamate (130 mg, 0.6 mmol, 1.5 eq.) as a white foam (73 mg, 33%). 1H NMR (500 MHz, CDCl3) δ 7.64 (s, 1H), 7.58 (s, 1H), 7.37 (d, J = 4.9 Hz, 2H), 7.02 (d, J = 4.1 Hz, 2H), 6.83 (s, 1H), 6.39 (d, J = 3.7 Hz, 1H), 5.36 (td, J = 11.0, 5.8 Hz, 1H), 4.83 (d, J = 8.1 Hz, 1H), 4.18–4.15 (m, 3H), 3.31 (t, J = 5.6 Hz, 1H), 2.88 (td, J = 17.5, 8.1 Hz, 2H), 1.77 (s, 3H), 1.49 (s, 9H), 1.49–1.44 (m, 4H), 1.26–1.23 (m, 3H), 0.89 (d, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C29H42N5O6 (M + H)+: 556.3130. Found: 556.3140.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-[5-(2-aminophenyl)-1H-1,2,3-triazol-1-yl]cyclohex-1-ene-1-carboxylate (6k). Using the general procedure for RuAAC, compound 6k was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 2-ethynylaniline (71 mg, 0.6 mmol, 1.5 eq.) as a white foam (84 mg, 46%). 1H NMR (500 MHz, CDCl3) δ 7.77 (s, 1H), 7.33 (t, J = 8.5 Hz, 1H), 7.06–6.96 (m, 3H), 6.82 (s, 1H), 5.21 (dd, J = 15.3, 10.0 Hz, 1H), 4.85 (d, J = 6.4 Hz, 1H), 4.19 (q, J = 7.1 Hz, 2H), 4.07 (s, 1H), 3.32–3.27 (m, 1H), 2.98–2.82 (m, 2H), 1.79 (s, 3H), 1.52–1.45 (m, 4H), 1.27 (t, J = 7.1 Hz, 3H), 0.90 (d, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C24H34N5O4 (M + H)+: 456.2611. Found: 456.2609.
Ethyl (3R,4R,5S)-4-acetamido-5-[5-(4-dimethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (6l). Using the general procedure for RuAAC, compound 6l was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynyl-N,N-dimethylaniline (87 mg, 0.6 mmol, 1.5 eq.) as a white foam (114 mg, 59%). 1H NMR (500 MHz, CDCl3) δ 7.61 (s, 1H), 7.24 (d, J = 8.8 Hz, 2H), 6.84–6.80 (m, 3H), 5.73 (d, J = 6.4 Hz, 1H), 5.58–5.52 (m, 1H), 5.06 (d, J = 8.9 Hz, 1H), 4.15–4.25 (m, 2H), 4.02–3.97 (m, 1H), 3.32–3.30 (m, 1H), 3.01 (s, 4H), 2.83–2.80 (m, 2H), 1.74 (s, 3H), 1.53–1.45 (m, 4H), 1.27 (t, J = 7.1 Hz, 3H), 0.91 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). ESIMS calcd for C26H38N5O4 (M + H)+: 484.3. Found: 484.3.
Ethyl (3R,4R,5S)-4-acetamido-5-[5-(4-diethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylate (6m). Using the general procedure for RuAAC, compound 6m was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and 4-ethynyl-N,N-diethylaniline (134 mg, 0.6 mmol, 1.5 eq.) as a white foam (135 mg, 66%). 1H NMR (500 MHz, CDCl3) δ 7.57 (s, 1H), 7.22 (d, J = 8.4 Hz, 2H), 6.85 (s, 1H), 6.72 (d, J = 8.4 Hz, 2H), 5.81 (d, J = 6.6 Hz, 1H), 5.58–5.54 (m, 1H), 5.10 (d, J = 8.7 Hz, 1H), 4.23–4.13 (m, 4H), 4.03 (ddd, J = 11.6, 9.2, 6.7 Hz, 1H), 3.47–3.29 (m, 5H), 2.86–2.70 (m, 2H), 1.75 (s, 3H), 1.56–1.45 (m, 4H), 1.26 (t, J = 7.1 Hz, 3H), 1.19 (t, J = 7.0 Hz, 6H), 0.93–0.87 (m, 6H). ESIMS calcd for C28H42N5O4 (M + H)+: 512.3. Found: 512.2.
Ethyl (3R,4R,5S)-4-acetamido-3-(pentan-3-yloxy)-5-{5-{[4-((S)-2-tert-butoxycarbonylamino-3-methoxy-3-oxopropyl)phenoxy]methyl}-1H-1,2,3-triazol-1-yl}cyclohex-1-ene-1-carboxylate (6n). Using the general procedure for RuAAC, compound 6n was afforded from 3 (135 mg, 0.4 mmol, 1.0 eq.) and methyl(S)-2-[(tert-butoxycarbonyl)amino]-3-[4-(prop-2-yn-1-yloxy)phenyl]propanoate (200 mg, 0.6 mmol, 1.5 eq.) as a white foam (99 mg, 37%). 1H NMR (500 MHz, CDCl3) δ 7.71 (s, 1H), 7.00 (d, J = 8.0 Hz, 2H), 6.86 (d, J = 8.1 Hz, 3H), 6.53 (d, J = 7.3 Hz, 1H), 5.36 (td, J = 11.1, 6.3 Hz, 1H), 5.14–5.07 (m, 2H), 5.01 (d, J = 8.1 Hz, 1H), 4.67 (d, J = 8.3 Hz, 1H), 4.50 (d, J = 6.9 Hz, 1H), 4.19 (q, J = 7.1 Hz, 2H), 3.98 (dd, J = 18.5, 8.8 Hz, 1H), 3.67 (s, 3H), 3.34 (q, J = 5.5 Hz, 1H), 3.05–2.93 (m, 4H), 1.67 (s, 3H), 1.52–1.44 (m, 4H), 1.39 (s, 9H), 1.26 (t, J = 7.1 Hz, 3H), 0.89 (t, J = 7.4 Hz, 3H), 0.84 (t, J = 7.4 Hz, 3H). HRESIMS calcd for C34H50N5O9 (M + H)+: 672.3603. Found: 672.3602.
(3R,4R,5S)-4-Acetamido-5-[4-(1,1′-biphenyl-4-yl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1a). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5a (165 mg, 0.32 mmol) gave 1a as a white foam (123 mg, 79%). IR (neat) νmax 3338.84, 2925.77, 2851.90, 1654.08, 1555.73, 1431.87, 1344.86, 1078.97 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.45 (s, 1H), 7.92–7.67 (m, 6H), 7.47 (t, J = 7.7 Hz, 2H), 7.39–7.36 (m, 1H), 6.89 (s, 1H), 5.10–5.07 (m, 1H), 4.45–4.36 (m, 2H), 3.49–3.44 (m, 1H), 3.11–3.08 (m, 2H), 1.75 (s, 3H), 1.60–1.53 (m, 4H), 0.98 (t, J = 7.4 Hz, 3H), 0.90 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.65, 142.48, 139.88, 129.20, 128.66, 127.62, 127.52, 126.68, 125.35, 119.13, 82.13, 71.18, 56.48, 55.54, 31.60, 25.85, 25.23, 21.36, 8.52, 8.09. HRESIMS calcd for. C28H32N4NaO4 (M + Na)+: 511.2316. Found: 511.2318.
(3R,4R,5S)-4-Acetamido-3-(pentan-3-yloxy)-5-[4-(4-phenoxyphenyl)-1H-1,2,3-triazol-1-yl]cyclohex-1-ene-1-carboxylic acid (1b). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5b (139 mg, 0.26 mmol) gave 1b as a white foam (99 mg, 75%). IR (neat) νmax 3318.93, 2939.20, 1628.36, 1420.34, 1167.99, 1036.75 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.24 (s, 1H), 7.70–6.92 (m, 9H), 6.77 (s, 1H), 4.98–4.92 (m, 1H), 4.33–4.23 (m, 2H), 3.35–3.31 (m, 1H), 3.00–2.90 (m, 2H), 1.62 (s, 3H), 1.48–1.39 (m, 4H), 0.84 (t, J = 7.4 Hz, 3H), 0.77 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.00, 172.34, 158.83, 154.88, 138.11, 129.45, 128.03, 122.44, 118.47, 81.93, 70.75, 52.76, 52.32, 35.02, 29.42, 28.98, 25.25, 9.46, 8.91. HRESIMS calcd for C28H31N4O5 (M + H)+: 503.2300. Found: 503.2296.
(3R,4R,5S)-4-Acetamido-5-[4-(2-naphthyloxy)methyl-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1c). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5c (183 mg, 0.35 mmol) gave 1c as a white foam (107 mg, 62%). IR (neat) νmax 3295.60, 2965.95, 2877.58, 1655.98, 1576.99, 1413.19, 1345.39, 1078.65 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.14 (s, 1H), 7.81–7.73 (m, 3H), 7.42 (t, J = 7.5 Hz, 1H), 7.37 (d, J = 2.3 Hz, 1H), 7.32 (t, J = 7.1 Hz, 1H), 7.17 (dd, J = 8.9, 2.5 Hz, 1H), 6.63 (s, 1H), 5.28 (s, 2H), 4.97 (dd, J = 18.0, 9.1 Hz, 1H), 4.30 (dd, J = 18.4, 7.3 Hz, 2H), 3.40 (dd, J = 11.2, 5.6 Hz, 1H), 3.01 (d, J = 7.3 Hz, 2H), 1.57 (s, 3H), 1.55–1.46 (m, 4H), 0.93 (t, J = 7.4 Hz, 3H), 0.84 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.23, 157.60, 144.68, 136.05, 133.45, 130.68, 130.47, 128.56, 128.01, 127.37, 124.80, 124.51, 119.86, 108.25, 83.41, 77.34, 62.32, 60.95, 56.45, 34.03, 27.31, 26.62, 22.48, 9.93, 9.52. HRESIMS calcd for C27H32N4NaO5 (M + Na)+: 515.2265. Found: 515.2268.
(3R,4R,5S)-4-Acetamido-5-[4-(4-methylphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1d). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5d (169 mg, 0.37 mmol) gave 1d as a white foam (122 mg, 77%). IR (neat) νmax 3393.75, 2946.75, 2835.71, 1645.75, 1597.51, 1416.94, 1026.59 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.34 (s, 1H), 7.68 (d, J = 8.1 Hz, 2H), 7.24 (d, J = 8.0 Hz, 2H), 6.70 (s, 1H), 5.03–4.95 (m, 1H), 4.37 (s, 2H), 3.46–3.40 (m, 1H), 3.06–3.01 (m, 2H), 2.36 (s, 3H), 1.70 (s, 3H), 1.59–1.49 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.30, 148.67, 139.34, 134.49, 130.52, 128.95, 126.64, 120.87, 83.53, 77.32, 60.89, 56.34, 33.77, 27.32, 26.66, 22.59, 21.27, 9.94, 9.54. HRESIMS calcd for C23H31N4O4 (M + H)+: 427.2340. Found: 427.2340.
(3R,4R,5S)-4-Acetamido-3-(pentan-3-yloxy)-5-(4-phenyl-1H-1,2,3-triazol-1-yl)cyclohex-1-ene-1-carboxylic acid (1e). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5e (173 mg, 0.39 mmol) gave 1e as a white foam (130 mg, 81%). IR (neat) νmax 3396.19, 2947.34, 2836.81, 1646.87, 1417.34, 1025.08 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.39 (s, 1H), 7.81 (d, J = 7.4 Hz, 2H), 7.43 (t, J = 7.6 Hz, 2H), 7.34 (t, J = 7.4 Hz, 1H), 6.92 (s, 1H), 5.08 (dd, J = 17.4, 10.5 Hz, 1H), 4.44 (d, J = 7.5 Hz, 1H), 4.38–4.31 (m, 1H), 3.47–3.40 (m, 1H), 3.11–2.99 (m, 2H), 1.71 (s, 3H), 1.60–1.47 (m, 4H), 0.96 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.43, 168.80, 148.68, 139.47, 131.70, 129.97, 129.74, 129.38, 126.69, 121.34, 83.84, 76.67, 60.22, 56.16, 32.44, 27.27, 26.71, 22.59, 9.94, 9.52. HRESIMS calcd for C22H29N4O4 (M + H)+: 413.2183. Found: 413.2179.
(3R,4R,5S)-4-Acetamido-5-[4-(4-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1f). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5f (171 mg, 0.34 mmol) gave 1f as a white foam (112 mg, 72%). IR (neat) νmax 3361.77, 2963.24, 2929.43, 2875.61, 1658.95, 1550.11, 1409.77, 1287.15, 1074.26 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.45 (s, 1H), 8.02 (d, J = 8.2 Hz, 2H), 7.82 (d, J = 8.2 Hz, 2H), 6.67 (s, 1H), 5.01 (dd, J = 18.5, 8.6 Hz, 1H), 4.35 (dd, J = 18.7, 7.9 Hz, 2H), 3.43 (dt, J = 11.1, 5.4 Hz, 1H), 3.06 (d, J = 8.1 Hz, 2H), 1.71 (s, 3H), 1.61–1.45 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 174.74, 174.17, 173.31, 148.19, 138.52, 135.51, 133.63, 130.98, 125.98, 121.80, 83.48, 77.34, 61.00, 56.55, 33.92, 27.34, 26.67, 22.60, 9.94, 9.55. HRESIMS calcd for C23H29N4O6 (M + H)+: 457.2082. Found: 457.2088. C23H28N4NaO6 (M + Na)+: 479.1901. Found: 479.1904.
(3R,4R,5S)-4-Acetamido-5-[4-(3-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1g). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5g (157 mg, 0.32 mmol) gave 1g as a white foam (99 mg, 69%). IR (neat) νmax 3387.01, 1654.02, 1548.54, 1389.71, 1017.78 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.37 (d, J = 3.6 Hz, 2H), 7.90 (dd, J = 17.6, 7.7 Hz, 2H), 7.43 (t, J = 7.7 Hz, 1H), 6.66 (s, 1H), 5.00 (dd, J = 17.4, 10.1 Hz, 1H), 4.45–4.29 (m, 2H), 3.47–3.41 (m, 1H), 3.11–3.03 (m, 2H), 1.71 (s, 3H), 1.60–1.47 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 174.92, 174.28, 173.24, 148.47, 140.04, 135.75, 133.44, 131.36, 130.06, 129.35, 128.27, 127.69, 121.59, 83.48, 77.40, 60.96, 56.61, 33.98, 27.33, 26.65, 22.63, 9.95, 9.54. HRESIMS calcd for C23H29N4O6 (M + H)+: 457.2082. Found: 457.2092.
(3R,4R,5S)-4-Acetamido-5-[4-(2-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1h). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5h (163 mg, 0.3 mmol) gave 1h as a white foam (104 mg, 70%). IR (neat) νmax 3295.18, 2964.19, 2933.58, 1649.54, 1397.34, 1076.56 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.16 (s, 1H), 7.80 (dd, J = 7.2, 0.8 Hz, 1H), 7.63–7.58 (m, 2H), 7.50–7.46 (m, 1H), 6.74 (s, 1H), 5.00–5.06 (m, 1H), 4.45–4.25 (m, 2H), 3.45–3.41 (m, 1H), 3.11–3.03 (m, 2H), 1.77 (s, 3H), 1.58–1.48 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 172.03, 171.10, 168.67, 145.58, 134.04, 131.16, 130.77, 130.19, 129.91, 129.34, 128.06, 122.04, 82.16, 75.80, 59.30, 54.90, 32.32, 25.89, 25.26, 21.34, 8.54, 8.12. HRESIMS calcd for C23H29N4O6 (M + H)+: 457.2082. Found: 457.2083.
(3R,4R,5S)-4-Acetamido-5-[4-(4-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1i). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5i (178 mg, 0.39 mmol) gave 1i as a white foam (92 mg, 55%). IR (neat) νmax 3374.09, 2947.67, 2835.94, 1656.64, 14449.41, 1410.50, 1025.83 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.05 (s, 1H), 7.43 (d, J = 8.4 Hz, 1H), 6.70 (s, 1H), 6.66 (d, J = 8.4 Hz, 1H), 4.92–4.87 (m, 1H), 4.31–4.18 (m, 2H), 3.35–3.30 (m, 1H), 2.99–2.87 (m, 2H), 1.62 (s, 3H), 1.46–1.38 (m, 4H), 0.84 (t, J = 7.4 Hz, 3H), 0.77 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.88, 171.13, 147.65, 146.67, 129.45, 126.39, 119.67, 118.26, 114.75, 82.15, 75.59, 59.01, 54.85, 32.47, 26.75, 25.87, 21.19, 8.55, 8.14. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2292. Found: 428.2292.
(3R,4R,5S)-4-Acetamido-5-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1j). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5j (124 mg, 0.27 mmol) gave 1j as a white foam (67 mg, 58%). IR (neat) νmax 3278.09, 2965.28, 2877.64, 1658.25, 1556.55, 1374.81, 1079.39 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.27 (s, 1H), 7.16 (s, 1H), 7.14 (d, J = 7.7 Hz, 1H), 7.11 (s, 1H), 6.70 (d, J = 7.6 Hz, 1H), 6.66 (s, 1H), 5.01–4.93 (m, 1H), 4.40–4.32 (m, 2H), 3.43 (p, J = 5.5 Hz, 1H), 3.06–2.98 (m, 2H), 1.71 (s, 3H), 1.57–1.47 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 174.26, 173.29, 149.40, 148.98, 135.69, 133.47, 132.33, 130.62, 121.12, 116.51, 116.45, 113.40, 83.51, 77.45, 60.96, 56.39, 34.06, 27.31, 26.64, 22.62, 9.93, 9.55. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2292. Found: 428.2295.
(3R,4R,5S)-4-Acetamido-5-[4-(2-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1k). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5k (101 mg, 0.22 mmol) gave 1k as a white foam (58 mg, 61%). IR (neat) νmax 3397.63, 2948.65, 2937.92, 1678.89, 1449.47, 1203.89, 1020.50 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.29 (s, 1H), 7.42 (d, J = 7.7, 1.3 Hz, 1H), 7.10–7.07 (m, 1H), 6.91 (s, 1H), 6.82 (d, J = 8.0 Hz, 1H), 6.71 (t, J = 7.3 Hz, 1H), 5.09–5.03 (m, 1H), 4.43–4.36 (m, 2H), 3.43 (p, J = 5.6 Hz, 1H), 3.07–3.04 (m, 2H), 1.71 (s, 3H), 1.58–1.48 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 172.07, 167.48, 147.19, 144.95, 138.02, 128.67, 128.46, 127.87, 120.26, 117.41, 116.53, 114.45, 82.45, 75.35, 58.79, 54.76, 31.12, 25.88, 25.33, 21.25, 8.54, 8.14. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2298. Found: 428.2291.
(3R,4R,5S)-4-Acetamido-5-[4-(4-dimethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1l). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5l (150 mg, 0.31 mmol) gave 1l as a white foam (95 mg, 67%). IR (neat) νmax 3361.70, 2963.53, 2841.42, 1645.74, 1423.30, 1018.09 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.16 (s, 1H), 7.63 (d, J = 8.6 Hz, 1H), 6.84 (s, 1H), 6.81 (d, J = 8.6 Hz, 1H), 4.43–4.30 (m, 2H), 3.44–4.42 (m, 1H), 3.04–2.97 (m, 8H), 1.72 (s, 3H), 1.60–1.49 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.87 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.88, 150.92, 147.87, 129.45, 126.15, 118.27, 112.45, 109.92, 82.38, 72.32, 58.90, 55.96, 39.05, 31.62, 29.40, 28.88, 21.16, 8.53, 8.10. HRESIMS calcd for C22H30N5O4 (M + H)+: 456.2611. Found: 456.2608.
(3R,4R,5S)-4-Acetamido-5-[4-(4-diethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1m). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 5m (113 mg, 0.22 mmol) gave 1m as a white foam (67 mg, 63%). IR (neat) νmax 3339.48, 2968.29, 2875.82, 1657.91, 1613.80, 1505.16, 1269.25, 1077.65 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.05 (s, 1H), 7.50 (d, J = 8.9 Hz, 1H), 6.73 (s, 1H), 6.64 (d, J = 8.9 Hz, 1H), 4.94–4.88 (m, 1H), 4.31–4.22 (m, 2H), 3.35–3.29 (m, 5H), 2.99–2.88 (m, 2H), 1.62 (s, 3H), 1.47–1.40 (m, 4H), 1.07 (t, J = 7.0 Hz, 6H), 0.85 (t, J = 7.4 Hz, 3H), 0.77 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 172.06, 150.73, 147.80, 129.40, 126.20, 118.40, 118.21, 112.38, 82.37, 75.50, 55.56, 54.93, 39.30, 29.44, 25.88, 25.30, 21.23, 11.63, 8.52, 8.13. HRESIMS calcd for C22H30N5O4 (M + H)+: 484.2924. Found: 484.2922.
(3R,4R,5S)-4-Acetamido-5-{4-{[4-((S)-2-amino-2-carboxyethyl)phenoxy]methyl}-1H-1,2,3-triazol-1-yl}-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (1n). Using the general procedure B for the synthesis of 1 or 2, deprotection of compound 5n (168 mg, 0.25 mmol) gave 1n as a white foam (104 mg, 79%). IR (neat) νmax 3287.59, 2927.55, 1659.25, 1512.02, 1435.81, 1202.45 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.08 (s, 1H), 7.22 (d, J = 8.6 Hz, 2H), 7.00 (d, J = 8.6 Hz, 2H), 6.90 (d, J = 1.5 Hz, 1H), 5.17 (d, J = 0.7 Hz, 2H), 5.05–5.00 (m, 1H), 4.38 (d, J = 7.3 Hz, 1H), 4.35–4.26 (m, 1H), 4.19 (dd, J = 7.5, 5.4 Hz, 1H), 3.45–3.39 (m, 1H), 3.25 (dd, J = 14.6, 5.3 Hz, 1H), 3.11 (dd, J = 14.6, 7.6 Hz, 1H), 3.02 (d, J = 8.3 Hz, 2H), 1.66 (s, 3H), 1.57–1.46 (m, 4H), 0.94 (t, J = 7.4 Hz, 3H), 0.85 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.32, 171.46, 168.86, 159.27, 144.66, 139.51, 131.70, 129.64, 128.10, 124.81, 116.56, 83.89, 76.79, 62.26, 60.18, 55.97, 55.36, 36.49, 32.27, 27.20, 26.66, 22.58, 9.91, 9.49. HRESIMS calcd for C26H36N5O7 (M + H)+: 530.2609. Found: 530.2607.
(3R,4R,5S)-4-Acetamido-3-(pentan-3-yloxy)-5-(5-phenyl-1H-1,2,3-triazol-1-yl)cyclohex-1-ene-1-carboxylic acid (2e). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6e (157 mg, 0.36 mmol) gave 2e as a white foam (115 mg, 78%). IR (neat) νmax 3424.69, 2842.65, 1643.99, 1427.59, 1256.33, 1017.10 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.75 (s, 1H), 7.56–7.44 (m, 5H), 6.86 (d, J = 1.3 Hz, 1H), 5.16–5.14 (m, 1H), 5.54–5.52 (m, 1H), 4.22–4.18 (m, 1H), 3.35–3.33 (m, 1H), 3.07–3.04 (m, 2H), 1.68 (s, 3H), 1.52–1.44 (m, 4H), 0.91 (t, J = 7.4 Hz, 3H), 0.84 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.01, 168.81, 140.91, 139.57, 133.26, 130.86, 130.41, 130.15, 129.42, 127.88, 83.47, 75.15, 62.65, 57.76, 32.85, 27.22, 26.61, 22.74, 9.94, 9.48. HRESIMS calcd for C22H29N4O4 (M + H)+: 413.2183. Found: 413.2180.
(3R,4R,5S)-4-Acetamido-5-[5-(4-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2f). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6f (106 mg, 0.21 mmol) gave 2f as a white foam (64 mg, 66%). IR (neat) νmax 3383.81, 2950.17, 2839.14, 1649.36, 1393.10, 1020.19 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.45 (s, 1H), 8.02 (d, J = 8.3 Hz, 2H), 7.83 (d, J = 8.3 Hz, 2H), 6.67 (s, 1H), 5.04–4.97 (m, 1H), 4.41–4.33 (m, 2H), 3.44–3.41 (m, 1H), 3.07–3.04 (m, 2H), 1.71 (s, 3H), 1.57–1.47 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 174.01, 173.31, 148.17, 135.44, 133.72, 132.35, 130.98, 129.87, 126.00, 121.82, 83.47, 77.33, 60.99, 56.55, 33.90, 27.34, 26.67, 22.60, 9.94, 9.55. HRESIMS calcd for C23H28N4NaO6 (M + Na)+: 479.1901. Found: 479.1904.
(3R,4R,5S)-4-Acetamido-5-[5-(3-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2g). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6g (76 mg, 0.15 mmol) gave 2g as a white foam (40 mg, 58%). IR (neat) νmax 3377.18, 2966.02, 2935.50, 1657.54, 1656.63, 1397.48, 1077.71 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.12 (d, J = 7.1 Hz, 1H), 7.99 (s, 1H), 7.78 (s, 1H), 7.62–7.54 (m, 2H), 6.77 (s, 1H), 4.98–4.93 (m, 1H), 4.35–4.33 (m, 2H), 3.36–3.33 (m, 1H), 3.24–3.15 (m, 1H), 3.11 (dd, J = 17.6, 5.7 Hz, 1H), 1.69 (s, 3H), 1.52–1.43 (m, 4H), 0.91 (t, J = 7.4 Hz, 3H), 0.82 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 172.79, 172.23, 140.61, 137.50, 132.87, 131.70, 130.97, 130.28, 127.86, 111.40, 83.55, 79.61, 64.41, 57.73, 33.25, 27.24, 26.63, 22.74, 9.87, 9.52. HRESIMS calcd for C23H29N4O6 (M + H)+: 457.2082. Found: 457.2090.
(3R,4R,5S)-4-Acetamido-5-[5-(2-carboxyphenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2h). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6h (63 mg, 0.12 mmol) gave 2h as a white foam (32 mg, 56%). IR (neat) νmax 3334.80, 2836.90, 2525.99, 1678.81, 1449.44, 1203.80, 1143.36, 1024.27 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.09 (s, 1H), 7.58 (d, J = 7.4 Hz, 1H), 7.36 (d, J = 7.0 Hz, 1H), 7.13 (d, J = 8.4 Hz, 1H), 6.97 (t, J = 7.2 Hz, 1H), 6.73 (s, 1H), 5.06 (d, J = 6.7 Hz, 1H), 4.37–4.24 (m, 2H), 3.41–3.37 (m, 1H), 3.05–2.97 (m, 2H), 1.63 (s, 3H), 1.53–1.47 (m, 4H), 0.91 (t, J = 7.4 Hz, 3H), 0.83 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 172.77, 171.63, 169.65, 139.28, 138.67, 135.84, 132.79, 130.82, 130.74, 128.49, 127.51, 121.34, 82.76, 74.47, 60.08, 55.97, 30.24, 24.14, 23.77, 20.31, 7.08, 6.99. HRESIMS calcd for C23H28N4NaO6 (M + Na)+: 479.1901. Found: 479.1905.
(3R,4R,5S)-4-Acetamido-5-[5-(4-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2i). Using the general procedure B for the synthesis of 1 or 2, deprotection of compound 6i (83 mg, 0.15 mmol) gave 2i as a white foam (46 mg, 72%). IR (neat) νmax 3359.53, 2967.00, 2937.43, 2878.38, 1660.22, 1557.67, 1499.89, 1200.77 1077.20 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.63 (s, 1H), 7.20 (d, J = 8.5 Hz, 1H), 6.89 (s, 1H), 6.86 (d, J = 8.5 Hz, 1H), 5.17–5.11 (m, 1H), 4.56–4.54 (m, 1H), 4.25–4.21 (m, 1H), 3.39–3.35 (m, 1H), 3.10–2.98 (m, 2H), 1.70 (s, 3H), 1.55–1.48 (m, 4H), 0.94 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.62, 167.56, 148.25, 140.03, 138.16, 132.39, 131.28, 129.72, 128.15, 115.56, 82.16, 74.03, 56.14, 55.47, 31.45, 25.83, 25.24, 21.34, 8.53, 8.09. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2292. Found: 428.2293.
(3R,4R,5S)-4-Acetamido-5-[5-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2j). Using the general procedure B for the synthesis of 1 or 2, deprotection of compound 6j (74 mg, 0.13 mmol) gave 2j as a white foam (41 mg, 73%). IR (neat) νmax 3392.69, 2970.90, 2749.20, 1678.74, 1435.55, 1204.21, 1143.03, 1019.03 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.84 (s, 1H), 7.70 (t, J = 7.9 Hz, 1H), 7.58–7.53 (m, 3H), 6.87 (s, 1H), 5.19–5.18 (m, 1H), 4.55–4.54 (m, 1H), 4.16 (t, J = 9.6 Hz, 1H), 3.35–3.31 (m, 1H), 3.10–3.01 (m, 2H), 1.71 (s, 3H), 1.51–1.43 (m, 4H), 0.90 (t, J = 7.4 Hz, 3H), 0.83 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.37, 168.89, 139.62, 132.22, 129.97, 129.41, 125.00, 124.24, 117.14, 114.87, 83.61, 75.12, 57.86, 54.45, 32.84, 27.20, 26.59, 22.78, 9.91, 9.46. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2292. Found: 428.2293.
(3R,4R,5S)-4-Acetamido-5-[5-(2-aminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2k). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6k (108 mg, 0.24 mmol) gave 2k as a white foam (70 mg, 69%). IR (neat) νmax 3391.31, 2950.09, 2838.83, 1645.74, 1417.23, 1203.74, 1021.06 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.69 (s, 1H), 7.24–7.21 (m, 1H), 7.03 (d, J = 7.5 Hz, 1H), 6.85 (d, J = 8.0 Hz, 1H), 6.78 (t, J = 7.4 Hz, 1H), 6.61 (s, 1H), 4.65–4.64 (m, 1H), 4.38–4.30 (m, 2H), 3.45–3.38 (m, 1H), 3.17–3.03 (m, 2H), 1.73 (s, 3H), 1.51–1.42 (m, 4H), 0.91 (t, J = 7.4 Hz, 3H), 0.82 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.34, 165.61, 146.57, 136.58, 132.89, 130.77, 129.48, 118.06, 117.46, 115.80, 112.58, 110.66, 82.05, 75.35, 57.07, 55.81, 32.28, 25.88, 25.21, 21.56, 8.48, 8.15. HRESIMS calcd for C22H30N5O4 (M + H)+: 428.2298. Found: 428.2297.
(3R,4R,5S)-4-Acetamido-5-[5-(4-dimethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2l). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6l (98 mg, 0.20 mmol) gave 2l as a white foam (62 mg, 67%). IR (neat) νmax 3271.17, 2963.35, 2933.31, 1657.42, 1506.99, 1362.62, 1227.19, 1021.24 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.53 (s, 1H), 7.18 (d, J = 8.6 Hz, 1H), 6.78 (d, J = 8.6 Hz, 1H), 6.72 (s, 1H), 5.11–4.92 (m, 1H), 4.45–4.36 (m, 1H), 4.23–4.11 (m, 1H), 3.27 (t, J = 5.5 Hz, 1H), 3.00–2.89 (m, 8H), 1.60 (s, 3H), 1.44–1.35 (m, 4H), 0.83 (t, J = 7.4 Hz, 3H), 0.75 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.66, 151.38, 140.19, 137.57, 131.32, 129.60, 129.43, 113.18, 112.25, 82.12, 73.96, 56.32, 55.37, 39.01, 31.57, 25.85, 25.24, 21.33, 8.52, 8.09. HRESIMS calcd for C22H30N5O4 (M + H)+: 456.2611. Found: 456.2612.
(3R,4R,5S)-4-Acetamido-5-[5-(4-diethylaminophenyl)-1H-1,2,3-triazol-1-yl]-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2m). Using the general procedure A for the synthesis of 1 or 2, deprotection of compound 6m (113 mg, 0.22 mmol) gave 2m as a white foam (64 mg, 60%). IR (neat) νmax 3362.32, 2966.94, 2932.70, 1657.04, 1613.50, 1504.52, 1268.94, 1076.93 cm−1. 1H NMR (500 MHz, CD3OD) δ 7.62 (s, 1H), 7.25 (d, J = 8.9 Hz, 1H), 6.85–6.82 (m, 3H), 5.14–5.09 (m, 1H), 4.56–4.51 (m, 1H), 4.30–4.24 (m, 1H), 3.46 (q, J = 7.1 Hz, 4H), 3.37–3.35 (m, 1H), 3.11–3.01 (m, 2H), 1.70 (s, 3H), 1.54–1.46 (m, 4H), 1.20 (t, J = 7.0 Hz, 6H), 0.93 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 171.54, 168.72, 148.53, 140.24, 137.49, 131.03, 129.68, 129.07, 112.04, 111.61, 81.20, 74.36, 56.05, 55.66, 43.97, 31.69, 25.85, 25.25, 21.36, 11.44, 8.53, 8.13. HRESIMS calcd for C22H30N5O4 (M + H)+: 484.2924. Found: 484.2923.
(3R,4R,5S)-4-Acetamido-5-{5-{[4-((S)-2-amino-2-carboxyethyl)phenoxy]methyl}-1H-1,2,3-triazol-1-yl}-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid (2n). Using the general procedure B for the synthesis of 1 or 2, deprotection of compound 6n (99 mg, 0.15 mmol) gave 2n as a white foam (61 mg, 78%). IR (neat) νmax 3433.92, 3013.45, 2929.50, 1686.89, 1512.86, 1436.01, 1296.51, 1205.47, 1137.42, 1025.52 cm−1. 1H NMR (500 MHz, CD3OD) δ 8.08 (s, 1H), 7.22 (d, J = 8.6 Hz, 2H), 7.01 (d, J = 8.6 Hz, 2H), 6.90 (s, 1H), 5.17 (d, J = 1.8 Hz, 2H), 5.05–4.99 (m, 1H), 4.38–4.19 (m, 3H), 3.44–3.39 (m, 1H), 3.26 (dd, J = 14.7, 5.2 Hz, 1H), 3.13–3.07 (m, 1H), 3.04–2.95 (m, 2H), 1.65 (s, 3H), 1.56–1.48 (m, 4H), 0.94 (t, J = 7.4 Hz, 3H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (125 MHz, CD3OD) δ 173.38, 171.27, 168.80, 159.35, 144.81, 139.47, 131.68, 129.66, 127.95, 124.53, 116.65, 83.87, 76.77, 62.27, 60.22, 55.97, 55.21, 36.48, 32.38, 27.22, 26.67, 22.55, 9.92, 9.49. HRESIMS calcd for C26H36N5O7 (M + H)+: 530.2609. Found: 530.2606.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This work was financially supported by the National Science & Technology Major Project of China (No. 2018ZX09711002-001-007), National Natural Science Foundation of China (Grant No. 31770862 and 91953116).
Notes and references
- M. P. Girard, J. S. Tam, O. M. Assossou and M. P. Kieny, Vaccine, 2010, 28, 4895–4902 CrossRef PubMed.
- T. M. Uyeki and N. J. Cox, N. Engl. J. Med., 2013, 368, 1862–1864 CrossRef CAS PubMed.
- J. Stevens, O. Blixt, L. M. Chen, R. O. Donis, J. C. Paulson and I. A. Wilson, J. Mol. Biol., 2008, 381, 1382–1394 CrossRef CAS PubMed.
- C. Liu, M. C. Eichelberger, R. W. Compans and G. M. Air, J. Virol., 1995, 69, 1099–1106 CrossRef CAS PubMed.
- K. McClellan and C. M. Perry, Drugs, 2001, 61, 263–283 CrossRef CAS PubMed.
- M. von Itzstein, W. Y. Wu, G. B. Kok, M. S. Pegg, J. C. Dyason, B. Jin, T. Van Phan, M. L. Smythe, H. F. White, S. W. Oliver, P. M. Colman, J. N. Varghese, D. M. Ryan, J. M. Woods, R. C. Bethell, V. J. Hotham, J. M. Cameron and C. R. Penn, Nature, 1993, 363, 418–423 CrossRef CAS PubMed.
- C. J. Dunn and K. L. Goa, Drugs, 1999, 58, 761–784 CrossRef CAS PubMed.
- S. Kubo, T. Tomozawa, M. Kakuta, A. Tokumitsu and M. Yamashita, Antimicrob. Agents Chemother., 2010, 54, 1256–1264 CrossRef CAS PubMed.
- P. Chand, P. L. Kotian, A. Dehghani, Y. El-Kattan, T. H. Lin, T. L. Hutchison, Y. S. Babu, S. Bantia, A. J. Elliott and J. A. Montgomery, J. Med. Chem., 2001, 44, 4379–4392 CrossRef CAS PubMed.
- M. Kiso, K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya and Y. Kawaoka, Lancet, 2004, 364, 759–765 CrossRef CAS.
- Q. M. Le, M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. L. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki and Y. Kawaoka, Nature, 2005, 437, 1108 CrossRef CAS PubMed.
- Y. Hu, S. Lu, Z. Song, W. Wang, P. Hao, J. Li, X. Zhang, H. L. Yen, B. Shi, T. Li, W. Guan, L. Xu, Y. Liu, S. Wang, X. Zhang, D. Tian, Z. Zhu, J. He, K. Huang, H. Chen, L. Zheng, X. Li, J. Ping, B. Kang, X. Xi, L. Zha, Y. Li, Z. Zhang, M. Peiris and Z. Yuan, Lancet, 2013, 381, 2273–2279 CrossRef.
- J. D. Bloom, L. I. Gong and D. Baltimore, Science, 2010, 328, 1272–1275 CrossRef CAS PubMed.
- J. D. Thompson, D. G. Higgins and T. J. Gibson, Comput. Appl. Biosci., 1994, 10, 19–29 CrossRef CAS PubMed.
- R. J. Russell, L. F. Haire, D. J. Stevens, P. J. Collins, Y. P. Lin, G. M. Blackburn, A. J. Hay, S. J. Gamblin and J. J. Skehel, Nature, 2006, 443, 45–49 CrossRef CAS PubMed.
- X. Xu, X. Zhu, R. A. Dwek, J. Stevens and I. A. Wilson, J. Virol., 2008, 82, 10493–10501 CrossRef CAS PubMed.
- G. M. Air, Influenza Other Respir. Viruses, 2012, 6, 245–256 CrossRef CAS PubMed.
- Y. Wu, G. Qin, F. Gao, Y. Liu, C. J. Vavricka, J. Qi, H. Jiang, K. Yu and G. F. Gao, Sci. Rep., 2013, 3, 1551 CrossRef PubMed.
- R. E. Amaro, R. V. Swift, L. Votapka, W. W. Li, R. C. Walker and R. M. Bush, Nat. Commun., 2011, 2, 388 CrossRef PubMed.
- R. E. Amaro, D. D. L. Minh, L. S. Cheng, W. M. Lindstrom, A. J. Olson, J. H. Lin, W. W. Li and J. A. McCammon, J. Am. Chem. Soc., 2007, 129, 7764–7765 CrossRef CAS PubMed.
- R. E. Amaro, X. Cheng, I. Ivanov, D. Xu and J. A. McCammon, J. Am. Chem. Soc., 2009, 131, 4702–4709 CrossRef CAS PubMed.
- N. Han and Y. Mu, PLoS One, 2013, 8, e60995 CrossRef CAS PubMed.
- S. Mohan and B. M. Pinto, Can. J. Chem., 2018, 96, 91–101 CrossRef CAS.
- P. S. Kerry, S. Mohan, R. J. M. Russell, N. Bance, M. Niikura and B. M. Pinto, Sci. Rep., 2013, 3, 2871 CrossRef PubMed.
- V. Zima, C. B. Albiñana, K. Rojíková, J. Pokorná, P. Pachl, P. Řezáčová, J. Hudlicky, V. Navrátil, P. Majer, J. Kinvalinka, M. Kožíšek and A. Machara, Bioorgan. Med. Chem., 2019, 27, 2935–2947 CrossRef CAS PubMed.
- X. Zhu, R. McBride, C. M. Nycholat, W. Yu, J. C. Paulson and I. A. Wilson, J. Virol., 2012, 86, 13371–13383 CrossRef CAS PubMed.
- P. C. Wang, D. C. Chiu, J. T. Jan, W. I. Huang, Y. C. Tseng, T. T. Li, T. J. Cheng, K. C. Tsai and J. M. Fang, Eur. J. Med. Chem., 2018, 145, 224–234 CrossRef CAS PubMed.
- P. H. Hsu, D. C. Chiu, K. L. Wu, P. S. Lee, J. T. Jan, Y. S. E. Cheng, K. C. Tsai, T. J. Cheng and J. M. Fang, Eur. J. Med. Chem., 2018, 154, 314–323 CrossRef CAS PubMed.
- J. Zhang, V. Poongavanam, D. Kang, C. Bertagnin, H. Lu, X. Kong, H. Ju, X. Lu, P. Gao, Y. Tian, H. Jia, S. Desta, X. Ding, L. Sun, Z. Fang, B. Huang, X. Liang, R. Jia, X. Ma, W. Xu, N. A. Murugan, A. Loregian, B. Huang, P. Zhan and X. Liu, J. Med. Chem., 2018, 61, 6379–6397 CrossRef CAS PubMed.
- J. Zhang, N. A. Murugan, Y. Tian, C. Bertagnin, Z. Fang, D. Kang, X. Kong, H. Jia, Z. Sun, R. Jia, P. Gao, V. Poongavanam, A. Loregian, W. Xu, X. Ma, X. Ding, B. Huang, P. Zhan and X. Liu, J. Med. Chem., 2018, 61, 9976–9999 CrossRef CAS PubMed.
- H. Ju, J. Zhang, Z. Sun, Z. Huang, W. Qi, B. Huang, P. Zhan and X. Liu, Eur. J. Med. Chem., 2018, 146, 220–231 CrossRef CAS PubMed.
- R. Jia, J. Zhang, W. Ai, X. Ding, S. Desta, L. Sun, Z. Sun, X. Ma, Z. Li, D. Wang, B. Huang, P. Zhan and X. Liu, Eur. J. Med. Chem., 2019, 178, 64–80 CrossRef CAS PubMed.
- A. S. Chintakrindi, D. J. Gohil, A. S. Chowdhary and M. A. Kanyalkar, Bioorgan. Med. Chem., 2020, 28, 115191 CrossRef CAS PubMed.
- J. Ye, X. Yang, M. Xu, P. K. Chan and C. Ma, Eur. J. Med. Chem., 2019, 182, 111635 CrossRef CAS PubMed.
- W. Ai, J. Zhang, W. A. Zalloum, R. Jia, S. Cherukupalli, X. Ding, Z. Sun, L. Sun, X. Jiang, X. Ma, Z. Li, D. Wang, B. Huang, P. Zhan and X. Liu, Eur. J. Med. Chem., 2020, 191, 112147 CrossRef CAS PubMed.
- A. Mahal, M. Duan, D. S. Zinad, R. K. Mohapatra, A. J. Obaidullah, X. Wei, M. K. Pradhan, D. Das, V. Kandi, H. S. Zinad and Q. Zhu, RSC Adv., 2021, 11, 1804–1840 RSC.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- W. Lew, X. Chen and C. U. Kim, Curr. Med. Chem., 2000, 7, 663–672 CrossRef CAS PubMed.
- L. Zhang, X. Chen, P. Xue, H. H. Y. Sun, I. D. Williams, K. B. Sharpless, V. V. Fokin and G. Jia, J. Am. Chem. Soc., 2005, 127, 15998–15999 CrossRef CAS PubMed.
- M. Potier, L. Mameli, M. Belisle, L. Dallaire and S. B. Melancon, Anal. Biochem., 1979, 94, 287–296 CrossRef CAS PubMed.
- M. Richard, O. Ferraris, A. Erny, M. Barthélémy, A. Traversier, M. Sabatier, A. Hay, Y. P. Lin, R. J. Russell and B. Lina, Antimicrob. Agents Chemother., 2011, 55, 2942–2952 CrossRef CAS PubMed.
- L. Zheng, J. Wei, X. Lv, Y. Bi, P. Wu, Z. Zhang, P. Wang, R. Liu, J. Jiang, H. Cong, J. Liang, W. Chen, H. Cao, W. Liu, G. F. Gao, Y. Du, X. Jiang and X. Li, Biosens. Bioelectron., 2017, 91, 46–52 CrossRef CAS PubMed.
- G. Jones, P. Willett, R. C. Glen, A. R. Leach and R. Taylor, J. Mol. Biol., 1997, 267, 727–748 CrossRef CAS PubMed.
- W. L. DeLano, Proteins: Struct., Funct., Bioinf., 2002, 30, 442–454 Search PubMed.
- M. Wang, J. Qi, Y. Liu, C. J. Vavricka, Y. Wu, Q. Li and G. F. Gao, J. Virol., 2011, 85, 8431–8435 CrossRef CAS PubMed.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307–326 CAS.
- R. J. Read, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2001, 57, 1373–1382 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- G. N. Murshudov, A. A. Vagin and E. J. Dodson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1997, 53, 240–255 CrossRef CAS PubMed.
- P. D. Adams, P. V. Afonine, G. Bunkóczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger and P. H. Zwart, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 213–221 CrossRef CAS PubMed.
- R. A. Laskowski, M. W. MacArthur, D. S. Moss and J. M. Thornton, J. Appl. Crystallogr., 1993, 26, 283–291 CrossRef CAS.
- K. E. Hevener, W. Zhao, D. M. Ball, K. Babaoglu, J. Qi, S. W. White and R. E. Lee, J. Chem. Inf. Model., 2009, 49, 444–460 CrossRef CAS.
- C. Elam, M. Lape and S. Paula, Biophys. Chem., 2010, 150, 88–97 CrossRef PubMed.
- H. Hirota, K. Satou and K. Onodera, J. Chem. Inf. Model., 2007, 47, 1609–1618 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1, copies of NMR and Ms spectra. See DOI: 10.1039/d1ra00472g |
| ‡ These authors contributed equally to this manuscript. |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ac,
Chenning Liab,
Lifeng Fuad,
Shanshan Zhanga,
Jianxun Qiab,
Xun Lv*a and
Xuebing Li
ac,
Chenning Liab,
Lifeng Fuad,
Shanshan Zhanga,
Jianxun Qiab,
Xun Lv*a and
Xuebing Li *abd
*abd

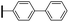
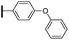
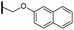

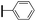
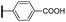
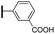
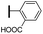
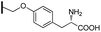
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added freshly prepared 1 M solution of CuSO4·5H2O (24 μL, 0.06 equiv.) in water, followed by the addition of sodium ascorbate (8 mg, 0.04 mmol, 0.1 eq.). The heterogeneous reaction mixture was stirred at room temperature for 6–12 h and monitored by TLC. After the completion, the reaction was evaporated to dryness. The crude product was purified by column chromatography, eluted with 25–75% EtOAc in petroleum ether, yielded the 4′-substituted-1,2,3-triazolylated OSC derivatives 5.
1) was added freshly prepared 1 M solution of CuSO4·5H2O (24 μL, 0.06 equiv.) in water, followed by the addition of sodium ascorbate (8 mg, 0.04 mmol, 0.1 eq.). The heterogeneous reaction mixture was stirred at room temperature for 6–12 h and monitored by TLC. After the completion, the reaction was evaporated to dryness. The crude product was purified by column chromatography, eluted with 25–75% EtOAc in petroleum ether, yielded the 4′-substituted-1,2,3-triazolylated OSC derivatives 5.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The mixture was stirred at rt for 3–4 h and concentrated. The resulting residue was dissolved in deionized water (0.5 mL) and filtered (Millipore Express 0.22 μM filter). Purification with Sephadex G-10 (eluent: 0.1% TFA in H2O) yielded 1 or 2 as a white foam.
1, v/v). The mixture was stirred at rt for 3–4 h and concentrated. The resulting residue was dissolved in deionized water (0.5 mL) and filtered (Millipore Express 0.22 μM filter). Purification with Sephadex G-10 (eluent: 0.1% TFA in H2O) yielded 1 or 2 as a white foam.