Open Access Article
Open Access ArticleTwo putative parallel pathways for naringenin biosynthesis in Epimedium wushanense†
Yating Liu‡
,
Linrui Wu‡,
Zixin Deng and
Yi Yu *
*
Key Laboratory of Combinatory Biosynthesis and Drug Discovery (Ministry of Education), School of Pharmaceutical Sciences, Wuhan University, Wuhan 430071, China. E-mail: yu_yi@whu.edu.cn
First published on 13th April 2021
Abstract
Flavonoids that exhibit various biological activities such as antioxidant, antitumor, antiviral, antibacterial and anti-inflammatory properties are found in a wide range of medicinal plants. Among the flavonoid-producing plants identified so far, the genus Epimedium is recognised as a group of prolific prenyl-flavonoid glycoside producers with high economic value in the global dietary supplement market. To date, the biosynthetic genes for prenyl-flavonoid glycosides still remain elusive in Epimedium. Here, we identified five genes in Epimedium wushanense responsible for the biosynthesis of naringenin, the common precursor for flavonoid natural products. We successfully set up the biosynthetic pathway of naringenin using L-tyrosine as the precursor through enzymatic assays of these genes' encoding products, including phenylalanine ammonia-lyase (EwPAL), 4-coumarate-CoA ligase (Ew4CL1), chalcone synthase (EwCHS1), chalcone isomerase (EwCHI1) and CHI-like protein (EwCHIL3). Intriguingly, in vitro characterisation of the above catalytic enzymes' substrate specificity indicated a route parallel to naringenin biosynthesis, which starts from L-phenylalanine and ends in pinocembrin. The fact that there is no pinocembrin or pinocembrin-derived flavonoid accumulated in E. wushanense prompted us to propose that pinocembrin is likely converted into naringenin in vivo, constituting two parallel biosynthetic pathways for naringenin. Therefore, our study provides a basis for the full elucidation of the biosynthetic logic of prenyl-flavonoid glycoside in Epimedium, paving the way for future metabolite engineering and molecular breeding of E. wushanense to acquire a higher titre of desired, bioactive flavonoid compounds.
Introduction
Among many plant-derived secondary metabolites, flavonoids represent a particular group of compounds with over 9000 members identified so far.1 Terrestrial plants have evolved the biosynthesis ability of flavonoids to persist their habitant sustainability on dry land, utilising them in UV protection, plant architecture, pigment generation, sexual reproduction, defence response and other applications.2 On the other hand, the growing interest in the research of the biosynthesis of flavonoid compounds can be attributed to their vast pharmaceutical applicable activities, such as anti-oxidant, anti-inflammatory, antibacterial, antifungal and other therapeutic properties.3The biosynthesis of flavonoids is widely distributed in higher plants, among which Epimedium has been recognised as a prominent flavonoid-producing genus since the first identification of icariin from E. grandiflorum.4 The prenylated flavonoid glycoside has been recognised as an indicative chemotaxonomic marker of Epimedium,5 after which over 140 other flavonoids, 31 lignins, 12 ionones, nine phenol glycosides, six phenylethanoid glycosides, five sesquiterpenes and some other active compounds were characterised.6 The secondary metabolite profiling of Epimedium revealed their superior capability for generating distinctive flavonoids with prominent pharmacological values in anti-osteoporosis, antioxidation, anti-tumour and immunoregulation.6 These findings also corroborate the extended ethnopharmacological use of Epimedium, known as “horny goat weed” in traditional Chinese medicine. Of the over 50 species of Epimedium, E. sagittatum Maxim., E. koreanum Nakai, E. pubescens Maxim., E. brevicornum Maxim. and E. wushanense are the five most commonly found and thoroughly examined Epimedium species.6,7
The pre-clinical studies of Epimedium-derived flavonoids were based upon the detailed understanding of their chemical constituents. Prenyl-flavonoids iteratively modified with sugar moieties, such as glucose, rhamnose and xylose (e.g., icariin, ikarisoside, epimedin, epimedoside), have been recognised as the main active components in Epimedium crude extract (Scheme 1).5 Owing to the vast occurrences of flavonoids in land plant species, a wealth of knowledge regarding the biosynthesis of flavonoids has been accumulated, laying the groundwork for a deeper understanding of plant-derived secondary metabolites. Comprising the flavonol backbone as the core structure, flavonoid glycosides found in Epimedium are proposed to be assembled from the common pathway for flavonoid biosynthesis, starting from phenylalanine.8 Phenylalanine would go through deamination, hydroxylation and coenzyme A acetylation to give 4-coumaroyl-CoA, which then enters the specialised pathway for flavonoid biosynthesis under the catalysis of chalcone synthase (CHS), a typical type III PKS.9–11 The resulting naringenin chalcone forms the flavonol backbone through cyclisation under the catalysis of chalcone isomerase (CHI) and gives naringenin.12 Furthermore, from the characterised flavonoid biosynthetic pathways, it has been established that naringenin serves as a vital branching point and general precursor. When taken up by specialised enzymes, such as flavone synthase (FNS), isoflavone synthase (IFS) and flavanone 3-hydroxylase (F3H), it can be transformed to the corresponding intermediates in their designated path to generate different types of secondary metabolites (Scheme 1).1 The flux of naringenin therefore profoundly affects the titre of the downstream flavonoid production, and becomes a research hotspot for its metabolic role.13 Furthermore, the standalone pharmacological activities of naringenin, including inversing cardiovascular risk and reducing endothelial dysfunction, have also been well-studied, indicating its promising clinical use.14 Studies demonstrated its outstanding activity against health-threatening viruses, such as Zika virus, dengue virus and SARS-CoV-1.15–17 It prompted naringenin to be a promising candidate against COVID-19 under the pandemic, benefitting also from its anti-inflammatory activity.18 However, despite the extensive studies surrounding flavonoid compounds from major flavonoid-producing plant families, how prenyl-flavonoid glycosides are assembled in Epimedium remains largely unknown.
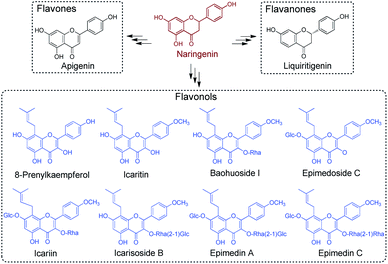 | ||
| Scheme 1 Representative flavonoids derived from naringenin. Representative prenyl-flavonoids identified from Epimedium are indicated in blue. | ||
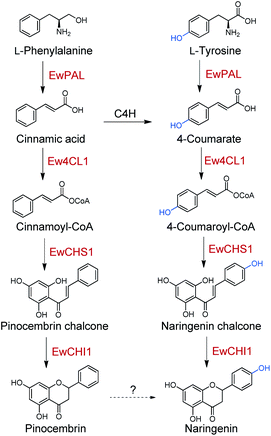
In this study, we identified five enzymes responsible for the biosynthesis of naringenin in E. wushanense. Biochemical characterisation of these enzymes established the early stage of prenyl-flavonoid glycoside biosynthesis. Based on the substrate specificity test, we further revealed that these enzymes could also transform L-phenylalanine into pinocembrin, another direct precursor of many types of flavonoid. Furthermore, our results suggest two parallel pathways for naringenin biosynthesis in E. wushanense.
Experimental
Chemicals and reagents
Standards used in this study, including naringenin, naringenin chalcone and pinocembrin chalcone were obtained from Chengdu DeSiTe Biological Technology. L-Phenylalanine, L-tyrosine, acetyl-CoA, malonyl-CoA, ATP and isopropyl β-D-1-thiogalactopyranoside, kanamycin and other reagents were purchased from Sigma-Aldrich. TRIzol reagent was purchased from Thermo Scientific.Plant material, sampling, RNA extraction and cDNA synthesis
For RNA isolation, fresh leaves were collected from E. wushanense grown in a greenhouse, flash frozen in liquid nitrogen and stored at −80 °C until use. Total RNA was extracted separately from E. wushanense leaf tissue using the TRIzol reagent. The first-strand cDNA was amplified by reverse transcription PCR using total RNA samples as templates. Genomic DNA was removed using the TransScript II One-Step, and cDNA was synthesised by SuperMix with the oligo(dT) primer (TransGen Biotech).RNA sequencing and data processing
The integrity of the extracted total RNA was assessed by Agilent 2100 Bioanalyzer. An ABI StepOnePlus Real-Time PCR System was used in the quantification and quality control of the sample library. For the RNA-seq experiment, the libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina), and sequenced on a HiSeq2000 sequencer (Illumina) in paired-end mode (PE100) by Shanghai Majorbio Bio-pharm Technology Co., Ltd. Sequence reads (FASTQ files) from three leaf samples were assessed with FastQC.19 The reads were then trimmed to exclude sequencing adaptors and low quality reads using Trimmomatic with default parameters.20 The trimmed reads were used to assemble de novo merged transcriptome using Trinity,21 resulting in 112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 545 transcript sequences. The completeness of the combined transcriptome was evaluated using BUSCO,22 from which we identified 73
545 transcript sequences. The completeness of the combined transcriptome was evaluated using BUSCO,22 from which we identified 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 328 unique transcripts and 67
328 unique transcripts and 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191 putative open reading frames (ORFs) longer than 100 amino acids using Transdecoder (http://transdecoder.github.io). The annotation of these ORFs was performed with Trinotate pipeline (http://trinotate.github.io), and transcriptome mining was performed on a local BLAST server.23
191 putative open reading frames (ORFs) longer than 100 amino acids using Transdecoder (http://transdecoder.github.io). The annotation of these ORFs was performed with Trinotate pipeline (http://trinotate.github.io), and transcriptome mining was performed on a local BLAST server.23
Sequence alignment and phylogenetic analysis
The protein multiple sequence alignments were generated using ClustalX2.24 ESPript 3.0 was used to display the alignment results.25 The phylogeny was inferred using the maximum-likelihood method in MEGA7 with default parameters.26 Gene bank accession numbers for sequences used in constructing the PAL phylogenetic tree: SbPTAL, XP_021319560.1; LrPAL, AWW24969.1; CmPAL, P45726.1; SlPAL, P35511.1; GmPAL1, P27991.1; NtPAL, P35513.2; TaPAL, Q43210.1; BdPTAL1, XP_003575396.1; PvPTAL, AFY17067.1; SiPTAL, XP_004973667.1; ZmPTAL, XP_008678828.3; ZmPTAL6, NP_001105334.2; ZmPAL1, NP_001241797.1; BoPTAL4, ADB97626.1; SiPAL6, XP_004976241.1; ZmPAL10, XP_008645952.1; OsPAL7, A2X7F7.1; BoPAL3, ACN62413.1; SiPAL7, XP_004953154.1; PcPAL2, P45728.1; PcPAL1, P24481.1; PcPAL3, P45729.1; SiPAL1, XP_004953153.1; SiPAL4, XP_004976238.1; PpPAL, AAP85250.1; Fungi PTAL, P11544.2; Fungi_PAL, P10248.2; Bacteria TAL, Q1LRV9.1; Bacteria_PAL, Q3M5Z3.1. Gene bank accession numbers for sequences used in constructing the 4CL phylogenetic tree: St4CL1, AAA33842.1; At4CL1, AAA82888.1; Pta4CL1, AAA92668.1; Pta4CL2, AAA92669.1; Nt4CL1, AAB18637.1; Nt4CL2, AAB18638.1; Ptr4CL1, AAC24503.1; Ptr4CL2, AAC24504.1; Pd4CL1, AAC39365.1; Pd4CL2, AAC39366.1; Gm4CL3, AAC97599.1; Gm4CL2, AAC97600.1; St4CL1a, SAAD40664.1; At4CL2, AAD47193.1; At4CL3, AAD47195.1; Lp4CL1, AAF37732.1; Lp4CL2, AAF37733.1; Lp4CL3, AAF37734.1; Ri4CL3, AAF91308.1; Ri4CL2, AAF91309.1; Ri4CL1, AAF91310.1; Gm4CL1, AAL98709.1; Ac4CL, AAS48417.1; Ar4CL, AAT02218.1; Cs4CL, ABA40922.1; Pv4CL, ACD02135.1; Pr4CL, ACF35279.1; Sa4CL3, ADE96997.1; Ot4CL, ADO16242.1; Pa4CL1, ADZ54779.1; Pv4CL2, ADZ96250.1; Ob4CL, AGP02119.1; As4CL, AMP18194.1; Le4CL1, BAA08365.1; Le4CL2, BAA08366.2; Sb4CL, BAD90937.1; Pc4CL1, CAA31696.1; Pc4CL2, CAA31697.1; Gm4CL4, CAC36095.1; Zm4CL, AAS67644.1; Os4CL, CAA36850.1; Vp4CL, O24540.1. Gene bank accession numbers for sequences used in constructing the CHI phylogenetic tree: ThEFP, BAJ10401.1; PhEFP, BAJ10400.1; InEFP, BAO58581.1; AtCHIL, NP_568154.1; AtFAP1, NP_567140.1; AtFAP2, NP_001324880.1; AtFAP3, NP_175757.1; AtCHI, NP_191072.1; PtCHI, XP_002315258.1; GhCHI, ABM64798.1; PlCHI, Q43056.1; LjCHI, CAD69022.1; LjCHI2, Q8H0G1.1; MsCHI, P28012.1; AcCHI, AAU11843.1; PcCHI, A5HBK6.1; PhCHIB, CAA32730.1; PhCHIA, AAF60296.1; PvCHI, P14298.2; GmCHI1A, AAT94358.1; GmCHI1B1, AAT94359.1; GmCHI2, AAT94360.1; GmCHI3, AAT94361.1; GmCHI4, AAT94362.1; FaCHI, Q4AE11.1; CsCHI, BAA36552.1; PsCHI, ADK55061.1; VvCHI, P51117.1; VvCHIL, XP_002280158.1; ZmCHI, Q08704.1; ZmCHIL, NP_001151452.1.Molecular cloning and recombinant protein overproduction
The coding sequences (CDS) of the candidate genes were amplified from cDNA by PCR using gene-specific primers (Table S3†). In-Fusion Cloning was used to ligate PCR amplicons into the vector pET28a(+). E. coli containing appropriate constructs were grown in Terrific Broth at 37 °C, 220 rpm until the OD600 reached 0.7–0.8, and then induced with 0.1 mM isopropyl-β-D-thiogalactoside (IPTG). The culture was then allowed to grow for an additional 20 h at 18 °C, 220 rpm. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl, 25 mM imidazole), and lysed with a French press. The crude protein lysate was clarified by centrifugation and filtration prior to GE nickel–nitrilotriacetic acid (Ni–NTA) gravity flow chromatographic purification. After loading the lysate, His6-tagged recombinant protein-bound Ni–NTA resin was washed with 10 column volume (CV) of lysis buffer, and eluted with 2 CV of elution buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl and 250 mM imidazole). The desired protein fractions were combined, dialysed, concentrated by Amicon Ultra-15 Centrifugal Filters (Millipore) and stored in storage buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl, and 5% glycerol).Enzyme assays
The 4-coumarate-CoA ligase assays using Ew4CL1 were carried out in 100 μL of reaction buffer (200 mM Tris, pH 8.0) in the presence of 0.5 mM 4-coumarate or cinnamic acid, 5 mM ATP, 0.5 mM acetyl-CoA, 5 mM MgCl2 and 2 μM recombinant enzyme. The reactions were incubated at 30 °C for 30 min, and quenched with an equal volume of ice-cold methanol. The reaction mixture was then centrifuged, and the supernatant was collected for HPLC and LC-MS analysis using water with 0.1% formic acid as solvent A and methanol with 0.1% formic acid as solvent B. Reverse-phase separation was performed on a C18 column (250 × 4.6 mm, Phenomenex) with a ramp gradient of solvent A and solvent B: 8% solvent B for 3 min, 8–95% solvent B over 17 min, 95% solvent B for 3 min, followed by a final equilibration of 8% solvent B for 7 min with a flow rate at 0.8 mL min−1. Chromatograms were obtained by monitoring the absorbance at 300 nm for cinnamoyl-CoA and 330 nm for 4-coumaroyl-CoA.Activity assays of chalcone synthase were performed by adding 2 μM EwCHS1 (and 2 μM EwCHIL3) and 1 mM malonyl-CoA in the diluted Ew4CL1 reaction mixture after incubation, as described above. Reactions were initiated by the addition of the recombinant enzyme, conducted at 30 °C for 30 min, and finally quenched with 200 μL of methanol. The reaction mixture was then centrifuged, and the supernatant was collected for HPLC and LC-MS analysis using water with 0.1% formic acid as solvent A and methanol with 0.1% formic acid as solvent B. The HPLC analysis for the detection of pinocembrin and naringenin was performed with the below condition: 15% solvent B for 3 min, 15–90% solvent B over 17 min, 90% solvent B for 3 min, followed by a final equilibration of 15% solvent B for 7 min with a flow rate at 0.8 mL min−1. Chromatograms were obtained by monitoring the absorbance at 290 nm, and compared with analytical standards.
Enzyme assays for chalcone isomerase were performed in 50 mM Tris–HCl buffer (pH 8.0) containing 1 μM EwCHI1 and 0.5 mM initial substrate (naringenin chalcone or pinocembrin chalcone). The mixture was extracted by an equal volume of ethyl acetate for HPLC analysis after incubation for 2 min. Compounds were separated by reversed-phase chromatography with a ramp gradient of solvent A (0.1% formic acid in H2O) and solvent B (0.1% formic acid in acetonitrile): 40% solvent B for 3 min, 40–80% solvent B over 17 min, 80% solvent B for 3 min, followed by a final equilibration of 40% solvent B for 7 min with a flow rate at 0.8 mL min−1. For the determination of the stereochemistry of CHI-generated products, the corresponding reaction mixture was extracted by ethyl acetate. The resulting organic phase was evaporated and residues were re-dissolved in isopropanol. The sample was subjected to HPLC analysis using a CHIRALPAK IA column (250 × 4.6 mm) and developed by mobile solvent 80% n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20% isopropanol (v/v) at a flow rate of 0.8 mL min−1. The chromatograms were obtained by monitoring the absorbance at 280 nm.
20% isopropanol (v/v) at a flow rate of 0.8 mL min−1. The chromatograms were obtained by monitoring the absorbance at 280 nm.
HPLC and LC-MS analysis
HPLC analysis was carried out on a Shimadzu (Kyoto, Japan) HPLC instrument equipped with a degasser (DGU-20A3), an autosampler (SIL-20A), a column oven (CTO-20A) and two pumps (LC-20AT). The separation was performed using a Phenomenex C18 column (250 mm × 4.6 mm). LC-MS analysis was carried out in positive ion mode using a Thermo Scientific LTQ XL Orbitrap mass spectrometer equipped with a Thermo Scientific Accela 600 pump (Thermo Fisher Scientific Inc.). The LC conditions for each product were as described above. The MS analysis parameters were as follows: 45 V capillary voltage, 45 °C capillary temperature, auxiliary gas flow rate 10 arbitrary units, sheath gas flow rate 40 arbitrary units, 3.5 kV spray voltage, and 50–1000 amu mass range (maximum resolution 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000).
000).
Results and discussion
Transcriptome analysis of E. wushanense led to the characterisation of EwPAL
The transcriptome data of E. wushanense were first acquired by subjecting RNA samples extracted from fresh leaves for RNA-sequencing, which yielded about 45 million paired-end reads per sample (Table S1†). A total of 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 328 unique transcripts were assembled from three biological repeat samples. The assembled transcriptome was evaluated as 81% complete by the metric of Benchmarking Universal Single-Copy Orthologs (BUSCO).27 Coding sequences (CDS) were then predicted and further annotated by BLAST search against the UniProtKB, Swiss-Prot and Pfam database.
328 unique transcripts were assembled from three biological repeat samples. The assembled transcriptome was evaluated as 81% complete by the metric of Benchmarking Universal Single-Copy Orthologs (BUSCO).27 Coding sequences (CDS) were then predicted and further annotated by BLAST search against the UniProtKB, Swiss-Prot and Pfam database.
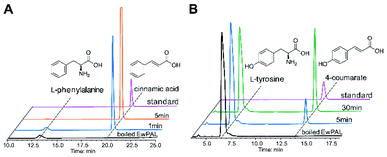
With the transcriptome data at hand, we set out to identify putative proteins that might be involved in naringenin biosynthesis. As almost all flavonoid compounds are derived from the primary metabolite L-phenylalanine, a BLAST search was conducted using the previously reported Sorghum bicolor phenylalanine ammonia-lyase (SbPAL, NCBI accession number XP_021319560.1) as query against the assembled E. wushanense transcriptome.28 The search identified a full-length candidate with 76% amino acid sequence identity to the query, and the putative protein was hence denoted as EwPAL. The corresponding open reading frame fragment was then amplified from E. wushanense cDNA, and cloned into an E. coli expression vector for His6-tagged fusion protein overproduction. The recombinant protein was purified to near homogeneity (Fig. S1A†) and subjected to enzyme assay. Compared to the negative control using boil-inactivated enzyme, incubating the purified EwPAL with L-phenylalanine gave rise to the generation of cinnamic acid, as confirmed by HPLC and MS analysis (Fig. 1A and S2A†).
After establishing the bioactivity of EwPAL, a phylogenetic tree was built for evaluating its phylogenetic relationship with PAL homologously characterised from various sources (Fig. S3†). EwPAL clusters with other dicot plant-derived PALs, suggesting its substrate preference for phenylalanine.29,30 Multiple sequence alignment of EwPAL with representative PALs revealed that it comprises a Phe residue at the established substrate selectivity switch site, instead of the His residue found in tyrosine ammonia-lyases (TALs) or bifunctional phenylalanine/tyrosine ammonia-lyase (PTALs) (Fig. S4†).31 Nonetheless, as demonstrated in previous studies, several PALs identified from dicot plants such as Arabidopsis thaliana can also convert L-tyrosine and produce the hydroxylated 4-coumarate accordingly.32 When tested, as shown in Fig. 1B and S2B,† EwPAL showed decent transformational activity towards L-tyrosine. Unfortunately, although many studies have indicated that the phenylalanine derived pathway can be converged into the tyrosine derived pathway by the catalysis of cinnamate 4-hydroxylase (C4H),33 we failed to identify any active C4H from E. wushanense after various attempts. However, it is still reasonable to propose that, similar to many established naringenin biosynthetic pathways, E. wushanense also incorporates a functional C4H so that phenylalanine contributes the major influx for naringenin biosynthesis for its preferred selectivity. As the biological roles of TAL and PTAL, which exhibits greater affinity towards tyrosine than that of PAL, are yet to be fully elucidated, it remains to be a mystery why dicot plants preserved the PAL activity when the tyrosine pathway can bypass the hydroxylation step of C4H and achieve higher energetic efficiency. Maeda et al. proposed that the phenylalanine pathway-specific cinnamic acid is critical for downstream metabolite synthesis, such as benzenoid volatiles and the plant hormone salicylic acid.34 Also, a study into a bifunctional PTAL in the model grass species Brachypodium distachyon demonstrated that nearly half of the lignin monomers generated were provided by the tyrosine it employed, suggesting the critical involvement of PTAL in lignin biosynthesis, and a complex regulatory mechanism is at play to modulate the influx of different branches.35 Since PAL is more prevalent in flavonoid biosynthetic pathways, we speculate that many plants, as represented by dicots, retained the energy-costly phenylalanine branch to preserve a more specific flavonoid precursor synthesis and avoid excessive alternative carbon-flux hijacking.
Identification and biochemical characterisation of 4-coumarate-CoA ligase (4CL)
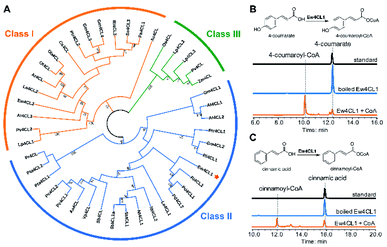
To probe the next step for naringenin biosynthesis, we focused on 4-coumarate, where the tyrosine and phenylalanine pathways putatively converge. A BLAST search using Nicotiana tabacum origin 4-coumarate-coenzyme A ligase (Nt4CL2, NCBI accession number AAB18638.1) as the query identified two 4CL homologs in E. wushanense, denoted as Ew4CL1 and Ew4CL2, respectively.36 In the plant, 4CL plays a significant role in the phenylpropanoid metabolic biosynthesis, and catalyses the conversion of differently substituted cinnamic acids.37,38 4CLs for branched pathways have also diverged from each other phylogenetically.39 To identify the specific 4CL-like enzyme for naringenin biosynthesis, a phylogenetic tree was built with the above two 4CL candidates, along with representative 4CLs identified from higher plant species (Fig. 2A). As shown, monocot 4CLs form their own group (Class III, coloured in green), while 4CLs from dicot plants can be grouped into two major clades: Class I (coloured in orange) are mainly constituted of 4CLs characterised to be responsible for lignin biosynthesis, and Class II (coloured in blue), where Ew4CL1 resides, comprises enzymes that participate in flavonoid biosynthesis.Based on this, Ew4CL1 was deemed as the target 4CL involved in naringenin biosynthesis and further cloned from E. wushanense cDNA for overexpression in E. coli. The recombinant protein was purified to near homogeneity (Fig. S1B†), and assayed against 4-coumarate with co-enzyme A. As a result, 4-coumarate was acetylated to give 4-coumaroyl-CoA as confirmed by HPLC and LC-MS analysis (Fig. 2B and S5A†). Interestingly, Ew4CL1 also showed undiscriminating activity towards cinnamic acid and the corresponding cinnamoyl-CoA was transformed (Fig. 2C and S5B†). Such substrate flexibility of 4CL has been examined in other plants, finally leading to the production of (2S)-pinocembrin, the 4-deoxy analogue of naringenin.40,41 To this point, we postulated that the pathway originated from phenylalanine parallels with the one originated from tyrosine, leading to the biosynthesis of pinocembrin instead of naringenin. From this point, both products of Ew4CL1 were examined for further characterisation.
Identification of chalcone synthase (CHS) for naringenin chalcone synthesis
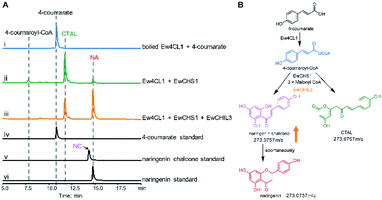
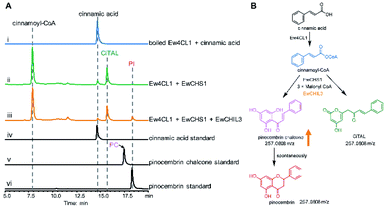
Chalcone synthase (CHS)-mediated iterative decarboxylative condensation is extensively established to be the first committed step for flavonoid biosynthesis.42 The tetraketide intermediate after condensing with malonyl-CoA molecules will then go through Claisen condensation to yield the chalcone backbone.43 At this point, three putative CHS encoding transcripts were identified from the annotated E. wushanense transcriptome using the characterised CHS from Arabidopsis thaliana as a query. The candidates share 78% to 86% amino acid sequence identity to the query (Table S2†).44 Of the three candidates, we managed to amplify the encoding sequence for EwCHS1 and EwCHS3 from E. wushanense cDNA. We then overexpressed and purified recombinant EwCHS1 and EwCHS3, respectively, from E. coli and tested in enzyme assay (Fig. S1C†). Using malonyl-CoA as the co-substrate and 4-coumaroyl-CoA as the starter unit generated by Ew4CL1, EwCHS3 did not show any transformation activity as compared to the negative control (data not shown), while incubation with EwCHS1 generated two new compounds, one of which kept the same retention time on HPLC as the naringenin standard (Fig. 3A). As both compounds showed the same [M + H]+ ion at m/z 273.0757, MS2 spectra were employed to analyse the identity of the two compounds, which were confirmed to be the expected naringenin and a shunt product, p-coumaroyltriacetic acid lactone (CTAL), respectively (Fig. 3B and S6†). The failure of detecting the direct product of EwCHS1, naringenin chalcone, was attributed to its rapid spontaneous cyclisation. In parallel, testing with cinnamoyl-CoA demonstrated a similar catalysis derailment pattern. The cyclised pinocembrin from pinocembrin chalcone and shunt product cinnamoyltriacetic acid lactone (CiTAL) could also be observed from HPLC and the MS2 spectrum (Fig. 4 and S7†). Similar reactions dominantly producing shunt products have also been observed in other plant-derived CHSs, such as kava, snapdragon and soybean.45,46Stereo-specific cyclisation catalysed by chalcone isomerase (CHI)
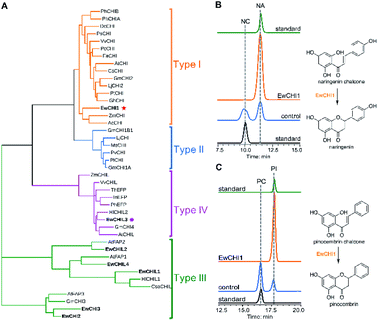
Naringenin biosynthesis is branched from the general phenylpropanoid pathway, whose completion relies on the catalysis of chalcone isomerase (CHI) to transform naringenin chalcone to the final product.47 From the transcriptome data of E. wushanense, seven putative CHI and CHI-like (CHIL) protein-encoding sequences were identified according to the Pfam annotation (PF02431 for CHI, PF16035 and PF16036 for CHIL), showing 23% to 85% amino acid sequence identity to the three known CHI and CHILs (Table S2†).44,48 The diversified CHIs and CHILs phylogenetically build up four major types,49 with type I and II groups comprising characterised bona fide CHIs that show intramolecular and stereo-specific cyclisation activity towards chalcones to yield flavanones.50 To identify the CHI specifically responsible for naringenin synthesis in E. wushanense, we performed a phylogenetic analysis using the above seven CHI candidates, along with representative sequences of four canonical types of CHIs. As shown in Fig. 5A, only EwCHI1 clustered with type I CHIs, while the other candidates grouped closer to non-catalytic type III and IV CHILs. Multiple sequence alignment further revealed that EwCHI1 contains two residues Thr190 and Met191 conserved in CHIs of type I and type II that have been verified to be responsible for contacting (2S)-naringenin (Fig. S8†).12 We therefore hypothesised that EwCHI1 is responsible for the final cyclisation of naringenin chalcone. The full-length cDNA of EwCHI1 was then cloned, heterologously overexpressed in E. coli, and subjected to biochemical characterisation (Fig. S1D†). When incubated with the naringenin chalcone (NA) and pinocembrin chalcone (PC) standards separately, EwCHI1 exhibited catalytic activity towards both substrates and converted them into the corresponding flavanone, naringenin (NA) and pinocembrin (PI) (Fig. 5B and C), whose identities were confirmed by comparing with the retention time of the standards and LC-MS analysis (Fig. S9†).As demonstrated above, the cyclisation of naringenin chalcone and pinocembrin chalcone can occur spontaneously in vitro, rendering the catalysis of CHI seemingly redundant. However, the spontaneous isomerisation has been proven to produce both 2S- and 2R-isomers as a racemic mixture, yet only the (2S)-stereoisomer can be accepted in the downstream flavonoid biosynthesis.47 We then employed chiral HPLC analysis to examine the stereo-chemistry of EwCHI1-generated products according to the established elution order of the two isomers.47 As shown in Fig. S10,† the (2S)-stereoisomer dominated both enzymatic products, while a near-equal ratio of the 2S and 2R stereoisomer presented in the spontaneous reaction mixture. The stereoselectivity of EwCHI1 ensured the conformational specificity of the resulting (2S)-pinocembrin and (2S)-naringenin, suggesting that plants have evolved a special catalyst to maintain an efficient assembly line by eliminating the unwanted product formation. The stereo purity of the flavonoids is also vital for their preclinical activity in various aspects of pharmaceutical use.14,51
CHILs have shown their vital role as auxiliary proteins by interacting with CHS or CHI to enhance protein–protein interactions and stabilise chalconoids, therefore maintaining the substrate specificity of CHS.48 Among the two types of CHILs, type III CHILs exhibit binding activity towards fatty acids, and hence play a role in fatty acid metabolism.52 Of the rest of the CHI and CHIL candidates from E. wushanense, only EwCHIL3 was phylogenetically grouped with type IV CHILs, which have been linked to flavonoid biosynthesis (Fig. 5A).48 We therefore expressed and purified recombinant EwCHIL3 from E. coli (Fig. S1C†) and tested its catalytic activity against naringenin chalcone. As expected, racemic naringenin was observed from the chiral HPLC spectrum due to the spontaneous reaction (data not shown). However, when supplementing EwCHIL3 to the Ew4CL1/EwCHS1 reaction mixture, the formation of the shunt product CTAL was repressed, while the production of naringenin increased significantly (Fig. 3A). The same phenomenon was also observed when cinnamoyl-CoA was used as the substrate (Fig. 4A). These results demonstrated that though EwCHIL3 is not directly involved in the naringenin biosynthesis, it may deviate evolutionarily from canonical CHIs and develop the function as a rectifier to maintain an efficient and economical production of flavonoids in vivo.
Naringenin, the ubiquitous flavanone found in many families of plants, has been recognised as a primary C15 intermediate whose biosynthesis can profoundly affect plant development, as well as total flavanone biosynthesis.53 In light of this, exploring the biosynthetic and regulatory logic of naringenin in flavonoid-rich plants has been a research hotspot, upon which further efforts on overcoming limited flavonoid production and flavonoid semi-synthesis from promising species can build. In this work, a total of five enzymes involved in naringenin biosynthesis were identified from E. wushanense, a renowned prenyl-flavonoid glycoside producer (Scheme 2). Starting from L-phenylalanine and L-tyrosine, albeit with differences in the substrate affinity, EwPAL can efficiently transform them into cinnamic acid and 4-coumarate, respectively. The two products then go through acetylation in parallel under the catalysis of Ew4CL1 to form the corresponding CoA thioesters. As the gatekeeper for flavonoid biosynthesis, CHS is considered to be the representative of the Type III plant PKSs (polyketide synthases) family.54 There has been over 20 functionally characterised CHS since its first identification in parsley.55–57 The production of CTAL/CiTAL by EwCHS1 as observed in biochemical assays may not honestly reflect how the pipeline performs in vivo, as a much more stringent regulation may control this branching step. The promiscuity nature of CHSs is believed to be intentionally preserved, serving as a basis for the derivatisation of other plant-specific PKSs, such as stilbene synthase (STS) and p-coumaroyltriacetic acid synthase (CTAS).58,59 However, it also presents a problem for flavonoid-producing plants, and hence forces them to develop a separate mechanism to enhance the efficiency of the reaction. In such sense, E. wushanense may have employed EwCHIL3 to interact with EwCHS1 as a rectifier. A similar observation has been made in HlCHIL1 from Humulus lupulus L., which plays a significant role in DMX biosynthesis by stabilising the ring-opening conformation of the substrate and enhancing the catalytic efficiency of CHS through protein–protein interaction.48 The interactions between CHSs and CHILs demonstrate conserved species-specificity, implying that EwCHIL3 is likely derived from the gene duplication of CHI during a recent speciation event.46 To date, much effort has been made for the heterologous reconstruction of naringenin, and by extension, flavonoid biosynthetic pathways. The efficiency-enhancing CHILs were indicated to work in a species-specific manner.46 Thus, the identification of EwCHIL3 provides a basis for pathway optimisation in Epimedium, so as to achieve a higher titre of the desired flavonoids. The isomerisation reaction, on the other hand, is catalysed by EwCHI1, the homolog of EwCHIL3. Although such a thermodynamically favoured reaction can occur spontaneously to produce a racemic mixture, nature has employed bona fide CHIs as an asymmetric catalyst to take the responsibility of specifically and efficiently generating stereochemically pure (2S)-pinocembrin and (2S)-naringenin.52 Such strategy has ensured a high-functioning assembly line for flavonoids. The findings provide a basis for future exploration in the backbone modification on naringenin, and the ultimate synthesis of prenyl-flavonoid glycosides. The asymmetric nature of the reaction under the catalysis of EwCHI1 implied its potential application as a chemoenzymatic catalyst in organic synthesis, in replacement of expansive inorganic catalysts, which might require demanding reaction conditions. Also, by rationally incorporating upstream backbone constructing enzymes and modifying downstream modification enzymes through synthetic biology and enzyme engineering, we can achieve the precise production of structurally diverse non-natural flavonoids with desired biological activity.
 | ||
| Scheme 2 The putative parallel pathways for naringenin biosynthesis in E. wushanense. Red font indicates the enzymes characterised in this study. | ||
Taken together, the identified enzyme rectifier EwCHIL3 and four catalytic enzymes all exhibited a certain extent of substrate flexibility, generating naringenin and pinocembrin in parallel. Nevertheless, as a contradiction, no pinocembrin or 4-deoxy-flavonoid has been identified in the genus Epimedium. We also failed to detect pinocembrin production in E. wushanense. A common and direct explanation for such discrepancy is that in vivo, an underlying C4H would timely hydroxylate cinnamic acid to give 4-coumarate, thus preventing the upstream pinocembrin flux. However, there are examples (such as Cephalocereus senilis) harbouring a similar naringenin/pinocembrin biosynthetic enzyme set that is also able to produce a considerable amount of pinocembrin in vivo.41,60 Furthermore, a comparative study regarding the relationship between the expression level of C4H and pinocembrin-derived flavonoid biosynthesis in Datisca glomerata and Medicago spp. concluded that, C4H presents as a gatekeeper, controlling the relative flux of the two-branched pathways, rather than arbitrarily diminishing the pinocembrin route.61 In light of this, it seems that C4H is not the culprit to be blamed for the silenced pinocembrin synthesis in E. wushanense. We therefore propose that there might be an undiscovered hydroxylation mechanism for the conversion of pinocembrin to naringenin, which converges the phenylalanine and tyrosine pathways, channelising two parallel routes for naringenin biosynthesis (Scheme 2). Blount et al. proved that in plants, PAL is feedback downregulated through the production of cinnamic acid.62 Cinnamic acid has also shown to be able to down-regulate the transcription level of CHS.63 A parallel pathway would ease the stringent regulation and flexibly modulate the influx of the two starting primary metabolites, which could profoundly affect the downstream lignin content and other phenylpropanoid titres. Future validation and characterisation could deepen our understanding of the global regulation of phenylpropanoid biosynthesis for further molecular breeding and metabolite engineering reference.
Conclusions
In this study, five enzymes involved in naringenin biosynthesis were identified from E. wushanense. Biochemical assays revealed their relatively flexible substrate specificity, which enabled them to transform phenylalanine and tyrosine into (2S)-pinocembrin and (2S)-naringenin, respectively. Moreover, EwCHIL3 showed rectifying activity by interacting with EwCHS1 to diminish shunt product formation, and ensured an efficient biosynthetic route for the flavonoid precursor synthesis. The lack of pinocembrin or pinocembrin-derived flavonoid in Epimedium implied the existence of a hydroxylation mechanism that may transform the produced pinocembrin to naringenin, hence constituting a parallel pathway for naringenin biosynthesis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Key Research and Development Program of China (2018YFA0900400) for funding this study.Notes and references
- K. Yonekura-Sakakibara, Y. Higashi and R. Nakabayashi, Front. Recent Dev. Plant Sci., 2019, 10, 943 CrossRef PubMed
.
- R. E. Koes, F. Quattrocchio and J. N. Mol, BioEssays, 1994, 16, 123–132 CrossRef CAS
.
- M. M. Jucá, F. M. S. Cysne Filho, J. C. de Almeida, D. d. S. Mesquita, J. R. d. M. Barriga, K. C. F. Dias, T. M. Barbosa, L. C. Vasconcelos, L. K. A. M. Leal and J. E. Ribeiro, Nat. Prod. Res., 2020, 34, 692–705 CrossRef PubMed
.
- S. Akai, J. Pharm. Soc. Jpn., 1935, 55, 537–599 CrossRef CAS
.
- H. Wu, E. J. Lien and L. L. Lien, Prog. Drug Res., 2003, 60, 1–57 CAS
.
- H. Ma, X. He, Y. Yang, M. Li, D. Hao and Z. Jia, J. Ethnopharmacol., 2011, 134, 519–541 CrossRef CAS PubMed
.
- W. T. Stearn, J. Shaw, P. S. Green, B. Mathew and W. T. Stearn, Genus Epimedium and other herbaceous Berberidaceae, Timber Press, 2002 Search PubMed
.
- W. Huang, S. Zeng, G. Xiao, G. Wei, S. Liao, J. Chen, W. Sun, H. Lv and Y. Wang, Front. Recent Dev. Plant Sci., 2015, 6, 689 Search PubMed
.
- M. B. Austin and J. P. Noel, Nat. Prod. Rep., 2003, 20, 79–110 RSC
.
- B. Winkel-Shirley, Plant Physiol., 2001, 126, 485–493 CrossRef CAS
.
- I. Abe and H. Morita, Nat. Prod. Rep., 2010, 27, 809–838 RSC
.
- J. M. Jez, M. E. Bowman, R. A. Dixon and J. P. Noel, Nat. Struct. Biol., 2000, 7, 786–791 CrossRef CAS PubMed
.
- Q. Wang, J. Yang, X.-m. Zhang, L. Zhou, X.-L. Liao and B. Yang, J. Chem. Res., 2015, 39, 455–457 CrossRef CAS
.
- B. Salehi, P. V. T. Fokou, M. Sharifi-Rad, P. Zucca, R. Pezzani, N. Martins and J. Sharifi-Rad, Pharmaceuticals, 2019, 12, 11 CrossRef CAS
.
- A. H. D. Cataneo, D. Kuczera, A. C. Koishi, C. Zanluca, G. F. Silveira, T. B. de Arruda, A. A. Suzukawa, L. O. Bortot, M. Dias-Baruffi and W. A. Verri, Sci. Rep., 2019, 9, 1–15 CAS
.
- S. Frabasile, A. C. Koishi, D. Kuczera, G. F. Silveira, W. A. Verri, C. N. D. Dos Santos and J. Bordignon, Sci. Rep., 2017, 7, 1–11 CrossRef PubMed
.
- N. Clementi, C. Scagnolari, A. D’Amore, F. Palombi, E. Criscuolo, F. Frasca, A. Pierangeli, N. Mancini, G. Antonelli and M. Clementi, Pharmacol. Res., 2020, 163, 105255 CrossRef PubMed
.
- R. W. Alberca, F. M. E. Teixeira, D. R. Beserra, E. A. de Oliveira, M. M. de Souza Andrade, A. J. Pietrobon and M. N. Sato, Front. Immunol., 2020, 11, 570919 CrossRef CAS PubMed
.
- S. W. Wingett and S. Andrews, F1000Research, 2018, 7, 1338 Search PubMed
.
- A. M. Bolger, M. Lohse and B. Usadel, Bioinformatics, 2014, 30, 2114–2120 CrossRef CAS PubMed
.
- M. G. Grabherr, B. J. Haas, M. Yassour, J. Z. Levin, D. A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury and Q. Zeng, Nat. Biotechnol., 2011, 29, 644 CrossRef CAS PubMed
.
- M. Seppey, M. Manni and E. M. Zdobnov, in Gene Prediction, Springer, 2019, pp. 227–245 Search PubMed
.
- C. Camacho, G. Coulouris, V. Avagyan, N. Ma, J. Papadopoulos, K. Bealer and T. L. Madden, BMC Bioinf., 2009, 10, 421 CrossRef PubMed
.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm and R. Lopez, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS
.
- X. Robert and P. Gouet, Nucleic Acids Res., 2014, 42, W320–W324 CrossRef CAS
.
- S. Kumar, G. Stecher and K. Tamura, Mol. Biol. Evol., 2016, 33, 1870–1874 CrossRef CAS PubMed
.
- F. A. Simão, R. M. Waterhouse, P. Ioannidis, E. V. Kriventseva and E. M. Zdobnov, Bioinformatics, 2015, 31, 3210–3212 CrossRef PubMed
.
- S.-Y. Jun, S. A. Sattler, G. S. Cortez, W. Vermerris, S. E. Sattler and C. Kang, Plant Physiol., 2018, 176, 1452–1468 CrossRef CAS PubMed
.
- L. A. Wanner, G. Li, D. Ware, I. E. Somssich and K. R. Davis, Plant Mol. Biol., 1995, 27, 327–338 CrossRef CAS PubMed
.
- M. J. MacDonald and G. B. D'Cunha, Biochem. Cell Biol., 2007, 85, 273–282 CrossRef CAS PubMed
.
- K. T. Watts, B. N. Mijts, P. C. Lee, A. J. Manning and C. Schmidt-Dannert, Chem. Biol., 2006, 13, 1317–1326 CrossRef CAS PubMed
.
- F. C. Cochrane, L. B. Davin and N. G. Lewis, Phytochemistry, 2004, 60, 1–57 Search PubMed
.
- M. Mizutani, D. Ohta and R. Sato, Plant Physiol., 1997, 113, 755–763 CrossRef CAS PubMed
.
- H. A. Maeda, Nat. Plants, 2016, 2, 1–2 Search PubMed
.
- J. Barros, J. C. Serrani-Yarce, F. Chen, D. Baxter, B. J. Venables and R. A. Dixon, Nat. Plants, 2016, 2, 1–9 Search PubMed
.
- D. Lee and C. J. Douglas, Plant Physiol., 1996, 112, 193–205 CrossRef CAS PubMed
.
- C. J. Douglas, Trends Plant Sci., 1996, 1, 171–178 CrossRef
.
- J. Ehlting, D. Büttner, Q. Wang, C. J. Douglas, I. E. Somssich and E. Kombrink, Plant J., 1999, 19, 9–20 CrossRef CAS PubMed
.
- X.-Y. Liu, P.-P. Wang, Y.-F. Wu, A.-X. Cheng and H.-X. Lou, Molecules, 2018, 23, 595 CrossRef PubMed
.
- L. Guo, X. Chen, L.-N. Li, W. Tang, Y.-T. Pan and J.-Q. Kong, Microb. Cell Fact., 2016, 15, 1–19 CrossRef CAS
.
- Q. Liu, M. S. Bonness, M. Liu, E. Seradge, R. A. Dixon and T. J. Mabry, Arch. Biochem. Biophys., 1995, 321, 397–404 CrossRef CAS PubMed
.
- M. L. Durbin, B. McCaig and M. T. Clegg, Plant Mol. Biol., 2000, 79–92 CrossRef CAS
.
- D. M. Macoy, W.-Y. Kim, S. Y. Lee and M. G. Kim, Plant Biotechnol. Rep., 2015, 9, 269–278 CrossRef
.
- B. W. Shirley, W. L. Kubasek, G. Storz, E. Bruggemann, M. Koornneef, F. M. Ausubel and H. M. Goodman, Plant J., 1995, 8, 659–671 CrossRef CAS PubMed
.
- T. Pluskal, M. P. Torrens-Spence, T. R. Fallon, A. De Abreu, C. H. Shi and J.-K. Weng, Nat. Plants, 2019, 5, 867–878 CrossRef PubMed
.
- T. Waki, R. Mameda, T. Nakano, S. Yamada, M. Terashita, K. Ito, N. Tenma, Y. Li, N. Fujino and K. Uno, Nat. Commun., 2020, 11, 1–14 Search PubMed
.
- A. X. Cheng, X. Zhang, X. J. Han, Y. Y. Zhang, S. Gao, C. J. Liu and H. X. Lou, New Phytol., 2018, 217, 909–924 CrossRef CAS PubMed
.
- Z. Ban, H. Qin, A. J. Mitchell, B. Liu, F. Zhang, J.-K. Weng, R. A. Dixon and G. Wang, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E5223–E5232 CrossRef CAS PubMed
.
- Y.-c. Yin, X.-d. Zhang, Z.-q. Gao, T. Hu and Y. Liu, Mol. Biotechnol., 2019, 61, 32–52 CrossRef CAS PubMed
.
- L. Ralston, S. Subramanian, M. Matsuno and O. Yu, Plant Physiol., 2005, 137, 1375–1388 CrossRef CAS PubMed
.
- Y.-J. Sun, J.-M. He and J.-Q. Kong, BMC Plant Biol., 2019, 19, 195 CrossRef PubMed
.
- M. N. Ngaki, G. V. Louie, R. N. Philippe, G. Manning, F. Pojer, M. E. Bowman, L. Li, E. Larsen, E. S. Wurtele and J. P. Noel, Nature, 2012, 485, 530–533 CrossRef CAS PubMed
.
- G. d. S. Bido, M. d. L. L. Ferrarese, R. Marchiosi and O. Ferrarese-Filho, Braz. Arch. Biol. Technol., 2010, 53, 533–542 CrossRef CAS
.
- S. A. Pandith, S. Ramazan, M. I. Khan, Z. A. Reshi and M. A. Shah, Planta, 2020, 251, 15 CrossRef CAS PubMed
.
- H.-N. Yu, L. Wang, B. Sun, S. Gao, A.-X. Cheng and H.-X. Lou, Plant Cell Rep., 2015, 34, 233–245 CrossRef CAS PubMed
.
- Y. Wang, Y. Dou, R. Wang, X. Guan, Z. Hu and J. Zheng, Gene, 2017, 635, 16–23 CrossRef CAS PubMed
.
- J. Li, C. Tian, Y. Xia, I. Mutanda, K. Wang and Y. Wang, Metab. Eng., 2019, 52, 124–133 CrossRef CAS PubMed
.
- S. Tropf, T. Lanz, S. Rensing, J. Schröder and G. Schröder, J. Mol. Evol., 1994, 38, 610–618 CrossRef CAS PubMed
.
- T. Akiyama, M. Shibuya, H. M. Liu and Y. Ebizuka, Eur. J. Biochem., 1999, 263, 834–839 CrossRef CAS PubMed
.
- L. Qin, K. R. Markham, P. W. Paré, R. A. Dixon and T. J. Mabry, Phytochemistry, 1993, 32, 925–928 CrossRef
.
- I. Gifford, K. Battenberg, A. Vaniya, A. Wilson, L. Tian, O. Fiehn and A. M. Berry, Front. Recent Dev. Plant Sci., 2018, 9, 1463 CrossRef PubMed
.
- J. W. Blount, K. L. Korth, S. A. Masoud, S. Rasmussen, C. Lamb and R. A. Dixon, Plant Physiol., 2000, 122, 107–116 CrossRef CAS PubMed
.
- G. J. Loake, A. D. Choudhary, M. J. Harrison, M. Mavandad, C. J. Lamb and R. A. Dixon, Plant Cell, 1991, 3, 829–840 CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00866h |
| ‡ Yating Liu and Linrui Wu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |