Open Access Article
Open Access ArticleThe role of β-hairpin conformation in ester hydrolysis peptide catalysts based on a TrpZip scaffold†
Xinyu Liu a,
Riley Watersb,
Hannah E. Gilbertb,
Gage T. Barroso
a,
Riley Watersb,
Hannah E. Gilbertb,
Gage T. Barroso b,
Kelsey M. Boyleb and
Leah S. Witus
b,
Kelsey M. Boyleb and
Leah S. Witus *b
*b
aDepartment of Chemistry, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA
bDepartment of Chemistry, Macalester College, Saint Paul, Minnesota 55105, USA. E-mail: lwitus@macalester.edu
First published on 6th July 2021
Abstract
To explore the role of peptide conformation on catalytic activity in the context of ester hydrolysis catalysts, pairs of sequences were designed that contained or lacked β-hairpin character. For the hydrolysis of para-nitrophenylacetate in aqueous media, we found small but consistent trends wherein His-containing sequences based on a TrpZip scaffold showed higher catalytic activity without β-hairpin character.
Introduction
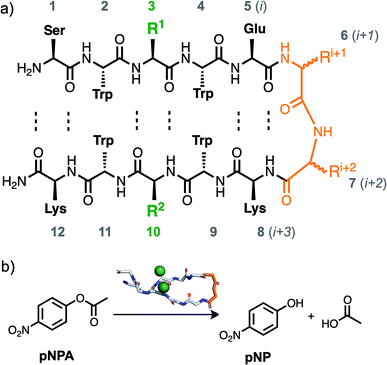
Peptide and peptidomimetic catalysts are important for understanding principles of enzymatic catalysis, and they are also useful catalysts in their own right in a variety of synthetic applications, including in the synthesis of natural products.1–4 Peptide catalysts provide a middle ground between small molecule organocatalysts and full-length enzymes, retaining the synthetic accessibility of the former, and some aspects of the complex multifunctional, three-dimensional, and chiral environment of the latter. While the rate enhancement and stereoselectivity of most current peptide catalysts is not on the scale of enzymatic catalysis, studying catalytic peptides can increase their synthetic utility.3,4 Elements of secondary structure have often been used in the design of peptide catalysts,5–10 which can serve to orient reactive residues near each other in space to mimic the catalytic triads and dyads of enzyme active sites. The study of ester hydrolysis catalysts has been used as a starting point for more challenging hydrolysis reactions,11 and has important practical applications itself,12 such as in the depolymerization of polyester plastics.13,14 Peptide ester hydrolysis catalysts have been reported using a variety of structural scaffolds including β-hairpins,7,15,16 polyproline helices,9,17 α-helices,5 cyclic peptides,18 dendrimers,19 and more.20 Further investigation of the effect of secondary structure is important for determining the role of conformation on catalysis and in guiding the design of future catalysts.β-Hairpins are among the smallest design motifs that form well-folded monomeric structures.21,22 β-Hairpins form two faces, as the side chains alternate above and below the plane of hydrogen bonding, with cross-strand residues oriented on the same face (Scheme 1a). In the case of β-hairpin ester hydrolysis catalysts, catalytic sequences have been reported that were designed de novo,15 and that were identified from combinatorial screening.7,16 Combinatorial screens have been performed using invariant turn-nucleating residues to create a β-hairpin scaffold, and variable positions in the residues oriented on the same face of the β-hairpin (either in the hydrogen bonding or non-hydrogen bonding positions). Sequences that are active ester hydrolysis catalysts can result in rate transformation through a number of effects, including binding to the substrate, altering side chain pKa, and directly playing a role in the catalysis of the transformation as a nucleophile or base. Thus, studies that used different solvents and ester substrates may not result in sequences that can be directly compared. However, it is notable that prior reports in different reaction conditions are consistent in identifying sequences with multiple His residues as the most active catalysts7,16,20 even in work where it was hypothesized that a ‘catalytic triad’ similar to the classical His Ser Asp might emerge from screening. Imidazole functional groups, present in His side chains, are known to catalyze ester hydrolysis,23,24 so we were curious about the role played by the peptide conformation. Therefore, we sought to explore closely the role of β-hairpin conformation for histidine-containing ester hydrolysis catalysts. We hypothesized that peptides with a β-hairpin conformation would be more active catalysts than closely related analogues without β-hairpin conformation, yet we found that this was not the case for the sequences we investigated as catalysts for the hydrolysis of para-nitrophenylacetate (pNPA) in water (Scheme 1b).
Results and discussion
To design sequences that would adopt a β-hairpin conformation, we used a highly stable TrpZip β-hairpin sequence as a scaffold. TrpZip is a sequence that has been reported to form unusually stable β-hairpins, and contains four tryptophan residues in the non-hydrogen bonding positions.25 To turn this scaffold sequence into an ester hydrolysis catalyst, we substituted two histidine residues in cross-strand positions, 3 and 10 (Scheme 1a). Other cross-strand pairings were also tested with His in position 3 and either Lys or Ala in position 10. For each of these cross-strand dyads, two versions were compared that varied in the i + 1 and i + 2 turn residues in order to study the role of β-hairpin conformation. To further bias the TrpZip scaffold toward a β-hairpin conformation, D-Pro–Gly (pG, the lowercase one-letter code refers to a D amino acid) was used in the i + 1 and i + 2 turn residues (residues 6 and 7), since this heterochiral pG combination is known to be strongly β-hairpin nucleating.26–28 For comparison sequences that would not have β-hairpin character, analogue peptides were investigated with Pro–Gly (PG) in residues 6 and 7, as the homochiral combination of Pro and Gly has been found to eliminate β-hairpin formation in peptides.26,27,29 These design principles resulted in pairs of sequences (Table 1) identical in every respect save for a single chiral center (pG vs. PG). Analogues of these sequences with a truncation of the N- and C-termini were also studied (see ESI†).| Peptide | Sequence |
|---|---|
| TrpZip | NH2–SWTWEGNKWTWK–NH2 |
| 3H10H-pG | NH2–SWHWEpGKWHWK–NH2 |
| 3H10H-PG | NH2–SWHWEPGKWHWK–NH2 |
| 3H10K-pG | NH2–SWHWEpGKWKWK–NH2 |
| 3H10K-PG | NH2–SWHWEPGKWKWK–NH2 |
| 3H10A-pG | NH2–SWHWEpGKWAWK–NH2 |
| 3H10A-PG | NH2–SWHWEPGKWAWK–NH2 |
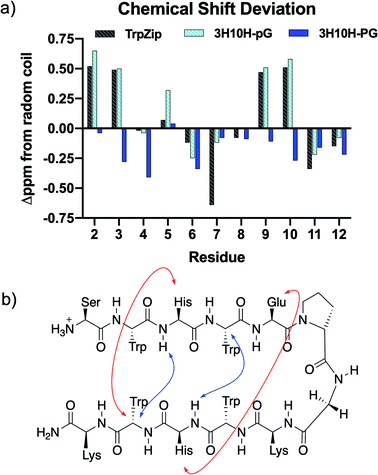
To confirm that the pair of sequences contained one with β-hairpin conformation and one without, the conformational behaviours of TrpZip, 3H10H-pG, and 3H10H-PG in solution were determined using NMR spectroscopy. TOCSY and NOESY/ROESY 2D-NMR experiments were performed for sequential proton resonance assignment (ESI Fig. S5–S13 and Tables S2–S5†). Determination of the secondary structure was accomplished using chemical shift deviation (CSD) analysis,30–32 in which the α-proton chemical shift for each residue was compared to random coil, with positive deviations in the strands indicative of β-sheet conformation. As seen in Fig. 1a, the CSDs of peptide 3H10H-pG were very similar to the TrpZip sequence which has been reported to form a stable β-hairpin,25 indicating β-hairpin character, while the 3H10H-PG sequence did not show any signs of β-hairpin character via positive CSDs. Additionally, cross-strand NOEs were observed for 3H10H-pG, shown in Fig. 1b, further indicating β-hairpin conformation. With this confirmation of the effect of the pG and PG turn residues on conformation (as well as circular dichroism, ESI Fig. S25†), the conformation of other sequences with D-Pro–Gly in the turn positions were assumed to also adopt β-hairpin conformation similarly to 3H10H-pG, and the sequences with Pro–Gly in the turn positions were assumed to not have β-hairpin character similarly to the 3H10H-PG sequence.
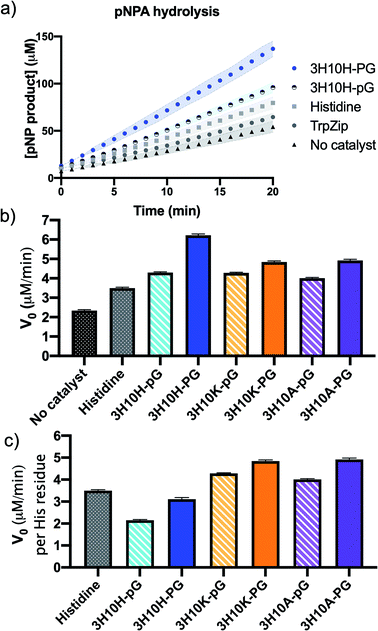
Each peptide sequence was evaluated for its catalytic efficiency in the hydrolysis of pNPA by monitoring the absorbance of the resulting product para-nitrophenol (pNP) to determine the initial velocity of the reaction (V0). The reactions were performed using 5 mM pNPA and 0.1 mM peptide catalyst (2 mol%) in 100 mM HEPES buffer (pH 7.4) with 5% acetonitrile for substrate solubility (Fig. 2). The catalytic performance of the individual amino acid His was used as a comparison. We found that the peptide 3H10H-PG, which lacks a β-hairpin conformation, led to increased rate enhancement of pNPA hydrolysis compared to the identical sequence containing a β-hairpin conformation, 3H10H-pG (Fig. 2a).
Since the closer spatial proximity of the His residues in the β-hairpin catalyst 3H10H-pG did not show increased catalytic activity relative to 3H10H-PG, we wondered how the effect of the His–His cross-strand interaction would compare to the effects of pairings of His in position 3 across from other amino acids. Therefore, we investigated β-hairpin sequences that paired His in position 3 across from Lys or Ala in position 10 (3H10A-pG and 3H10K-pG). When comparing the catalytic performance of these three β-hairpin catalysts (striped bars in Fig. 2b), the V0 values were very similar, but 3H10H-pG contained two His residues per peptide, while 3H10A-pG and 3H10K-pG contained one each. Therefore, we compared the initial velocities per His residue in Fig. 2c, which showed the His–His pairing led to lower rate enhancement than His–Ala or His–Lys in a β-hairpin structure on a per His residue basis (stripped bars). This enforced the finding from the 3H10H-pG and 3H10H-PG comparison that for this combination of catalyst scaffold, substrate, and reaction conditions, the positioning of His residues in close spatial proximity had a detrimental effect on catalytic activity. When compared on a per-His equivalent, the His–His containing peptides 3H10H-pG, and 3H10H-PG were actually less active than the single amino acid.
The 3H10A-pG and 3H10K-pG sequences were also compared to nearly identical sequences without β-hairpin conformation, 3H10A-PG and 3H10K-PG. In all of the cross-strand pairings (His–His, His–Ala, His–Lys) the sequence without β-hairpin character (PG turn) had slightly higher rate enhancement than the analogous hairpin sequence (pG turn), a small effect but perceptible relative to the error tolerance. In a linear peptide, the identity of residue 10 would not be expected to affect the reactivity of residue 3, such that on a per His equivalence basis all of the non-hairpin (PG containing) sequences (solid bars) would be expected to have similar rate enhancement. Since the reactions catalyzed by sequences with a single His residue (3H10A-PG and 3H10K-PG) had higher initial velocities per His residue than the sequence with multiple His residues (3H10H-PG), the apparent influence of position 10 on position 3 indicates some secondary structure may be forming. However, the CSD analysis indicates that it is not a β-hairpin (Fig. 1a). Similar reactivity trends were observed for the truncated sequences (data in ESI†), indicating that the trends, while small, are consistent.
Conclusions
In this work, we applied design principles from previous reports on ester hydrolysis catalysts – namely the use of a β-hairpin scaffold, the positioning of catalytically active residues on one face, and the inclusion of multiple His residues as the reactive groups – and studied the effect of these sequences on the rate of the hydrolysis of pNPA in aqueous media. Through the careful design of analogous sequences for comparison, we observed small yet interesting and consistent trends. A difference in conformation between otherwise identical sequences (caused by the difference in a single chiral center) was observed to lead to differences in catalytic performance. Under the reaction conditions studied here, we found that the sequences without β-hairpin character resulted in increased rate enhancement of pNPA hydrolysis compared to the corresponding β-hairpin sequences. Furthermore, while we found sequences with greater His content had rate enhancements, when comparing V0 per His equivalent, they were less active catalysts. The potential for cooperative activity based on His proximity should be further explored for other substrate, media, and structural scaffolds, but this study indicates that cooperativity should not be assumed – an important consideration in the design of future combinatorial libraries. Future studies will examine His pairings with aromatic groups,33 investigate the role of pH and His accessibility, and explore further the solution conformation of the non-hairpin peptides.Experimental section
Reagents for the para-nitrophenylacetate (pNPA) hydrolysis assay were purchased from Sigma Aldrich unless otherwise noted. Peptides were purchased from Biomatik with >95% HPLC purity, and the quality control data (mass and HPLC) were verified using a Waters Aquity Arc UHPLC with UV/Vis and QDa detector Liquid Chromatography Mass Spectrometer (LCMS), data in the ESI.† NMR experiments were carried out on a Bruker Advance III-HD-600 NMR spectrometer. UV-Vis absorbance measurements (well plate and cuvette) were carried out using a Tecan M Nano Plus multimode plate reader. Water was purified using a Milli-Q Advantage A10 Water Purification System. The molecular rendering in Scheme 1b was made using the PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.The hydrolysis of pNPA was monitored by measuring the absorbance of the resulting para-nitrophenol (pNP) product in a multiwell plate. Each catalyst was evaluated nine times – three separate experiments with each catalyst tested in triplicate in three wells. Each well had contained 0.1 mM catalyst and 5 mM pNPA in aqueous media buffered by 100 mM HEPES (pH 7.4) with 5% acetonitrile for substrate solubility. The concentration of the peptide catalysts was determined using the absorbance at 280 nm and converting to concentration using the extinction coefficient for the four Trp residues. A standard curve containing known concentrations of pNP was used in each plate to convert the absorbance measurements to concentrations. To prepare each multiwell plate experiment, 190 μL of catalyst dissolved in HEPES buffer was added to each well, and 10 μL of pNPA substrate dissolved in acetonitrile was added last to all wells immediately before measuring the absorbance at 405 nm in the plate reader every minute for twenty minutes at 25 °C.
Author contributions
X. L. and L. S. W. designed the sequences and experiments. X. L. performed the NMR experiments, L. S. W., H. E. G., R. W., G. T. B. and K. M. B. performed the pNPA hydrolysis experiments. L. S. W. and X. L. analyzed the data and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Professor Sam Gellman for his support, discussions, and access to instrumentation. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R15GM129703. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Efforts by X. L. were supported in part by NSF grant CHE-1904940. The Bruker Advance III-HD-600 NMR spectrometer was funded by NIH grant S10 OD012245 (2013). L. S. W. is supported by a Cottrell Scholar Award from the Research Corporation for Science Advancement (ID no: 27499). Publication of this article was funded by the Macalester College Dewitt Wallace Library Open Access Fund.Notes and references
- A. Dermenci, P. S. Selig, R. A. Domaoal, K. A. Spasov, K. S. Anderson and S. J. Miller, Quasi-biomimetic ring contraction promoted by a cysteine-based nucleophile: Total synthesis of Sch-642305, some analogs and their putative anti-HIV activities, Chem. Sci., 2011, 2, 1568–1572 RSC.
- Z. C. Girvin, M. K. Andrews, X. Liu and S. H. Gellman, Foldamer-templated catalysis of macrocycle formation, Science, 2019, 366, 1528–1531 CrossRef CAS.
- E. A. C. Davie, S. M. Mennen, Y. Xu and S. J. Miller, Asymmetric Catalysis Mediated by Synthetic Peptides, Chem. Rev., 2007, 107, 5759–5812 CrossRef.
- A. J. Metrano, A. J. Chinn, C. R. Shugrue, E. A. Stone, B. Kim and S. J. Miller, Asymmetric Catalysis Mediated by Synthetic Peptides, Version 2.0: Expansion of Scope and Mechanisms, Chem. Rev., 2020, 120, 11479–11615 CrossRef CAS.
- S. Bezer, M. Matsumoto, M. W. Lodewyk, S. J. Lee, D. J. Tantillo, M. R. Gagné and M. L. Waters, Identification and optimization of short helical peptides with novel reactive functionality as catalysts for acyl transfer by reactive tagging, Org. Biomol. Chem., 2014, 12, 1488–1494 RSC.
- J. D. Revell and H. Wennemers, Investigating Sequence Space: How Important is the Spatial Arrangement of Functional Groups in the Asymmetric Aldol Reaction Catalyst H-Pro-Pro-Asp-NH2?, Adv. Synth. Catal., 2008, 350, 1046–1052 CrossRef CAS.
- M. Matsumoto, S. J. Lee, M. L. Waters and M. R. Gagné, A Catalyst Selection Protocol That Identifies Biomimetic Motifs from β-Hairpin Libraries, J. Am. Chem. Soc., 2014, 136, 15817–15820 CrossRef CAS PubMed.
- A. J. Metrano, N. C. Abascal, B. Q. Mercado, E. K. Paulson and S. J. Miller, Structural studies of β-turn-containing peptide catalysts for atroposelective quinazolinone bromination, Chem. Commun., 2016, 52, 4816–4819 RSC.
- P.-Y. Hung, Y.-H. Chen, K.-Y. Huang, C.-C. Yu and J.-C. Horng, Design of Polyproline-Based Catalysts for Ester Hydrolysis, ACS Omega, 2017, 2, 5574–5581 CrossRef CAS PubMed.
- K. Akagawa and K. Kudo, Development of Selective Peptide Catalysts with Secondary Structural Frameworks, Acc. Chem. Res., 2017, 50, 2429–2439 CrossRef CAS PubMed.
- M. J. MacDonald, L. D. Lavis, D. Hilvert and S. H. Gellman, Evaluation of the Ser-His Dipeptide, a Putative Catalyst of Amide and Ester Hydrolysis, Org. Lett., 2016, 18, 3518–3521 CrossRef CAS PubMed.
- C. A. Martinez, S. Hu, Y. Dumond, J. Tao, P. Kelleher and L. Tully, Development of a Chemoenzymatic Manufacturing Process for Pregabalin, Org. Process Res. Dev., 2008, 12, 392–398 CrossRef CAS.
- V. Tournier, C. M. Topham, A. Gilles, B. David, C. Folgoas, E. Moya-Leclair, E. Kamionka, M.-L. Desrousseaux, H. Texier, S. Gavalda, M. Cot, E. Guémard, M. Dalibey, J. Nomme, G. Cioci, S. Barbe, M. Chateau, I. André, S. Duquesne and A. Marty, An engineered PET depolymerase to break down and recycle plastic bottles, Nature, 2020, 580, 216–219 CrossRef CAS PubMed.
- H. P. Austin, M. D. Allen, B. S. Donohoe, N. A. Rorrer, F. L. Kearns, R. L. Silveira, B. C. Pollard, G. Dominick, R. Duman, K. E. Omari, V. Mykhaylyk, A. Wagner, W. E. Michener, A. Amore, M. S. Skaf, M. F. Crowley, A. W. Thorne, C. W. Johnson, H. L. Woodcock, J. E. McGeehan and G. T. Beckham, Characterization and engineering of a plastic-degrading aromatic polyesterase, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4350–E4357 CrossRef CAS PubMed.
- B. Goyal, K. Patel, K. R. Srivastava and S. Durani, De novo design of stereochemically-bent sixteen-residue β-hairpin as a hydrolase mimic, RSC Adv., 2015, 5, 105400–105408 RSC.
- C. Schmuck, U. Michels and J. Dudaczek, Hydrolytic activity of histidine-containing octapeptides in water identified by quantitative screening of a combinatorial library, Org. Biomol. Chem., 2009, 7, 4362 RSC.
- K.-Y. Huang, C.-C. Yu and J.-C. Horng, Conjugating Catalytic Polyproline Fragments with a Self-Assembling Peptide Produces Efficient Artificial Hydrolases, Biomacromolecules, 2020, 21, 1195–1201 CrossRef CAS PubMed.
- M. Z. Atassi and T. Manshouri, Design of peptide enzymes (pepzymes): surface-simulation synthetic peptides that mimic the chymotrypsin and trypsin active sites exhibit the activity and specificity of the respective enzyme, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8282–8286 CrossRef PubMed.
- A. Clouet, T. Darbre and J.-L. Reymond, Combinatorial synthesis, selection, and properties of esterase peptide dendrimers, Biopolymers, 2006, 84, 114–123 CrossRef CAS PubMed.
- H. B. Albada and R. M. J. Liskamp, TAC-Scaffolded Tripeptides as Artificial Hydrolytic Receptors: A Combinatorial Approach Toward Esterase Mimics, J. Comb. Chem., 2008, 10, 814–824 CrossRef CAS PubMed.
- S. H. Gellman, Minimal model systems for β-sheet secondary structure in proteins, Curr. Opin. Chem. Biol., 1998, 2, 717–725 CrossRef CAS.
- P. Morales and M. A. Jiménez, Design and structural characterisation of monomeric water-soluble α-helix and β-hairpin peptides: State-of-the-art, Arch. Biochem. Biophys., 2019, 661, 149–167 CrossRef CAS PubMed.
- M. L. Bender and B. W. Turnquest, General Basic Catalysis of Ester Hydrolysis and Its Relationship to Enzymatic Hydrolysis1, J. Am. Chem. Soc., 1957, 79, 1656–1662 CrossRef CAS.
- L. Schoonen, K. S. van Esterik, C. Zhang, R. V. Ulijn, R. J. M. Nolte and J. C. M. van Hest, Alternative application of an affinity purification tag: hexahistidines in ester hydrolysis, Sci. Rep., 2017, 7, 14772 CrossRef PubMed.
- A. G. Cochran, N. J. Skelton and M. A. Starovasnik, Tryptophan zippers: Stable, monomeric β-hairpins, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5578–5583 CrossRef CAS.
- T. S. Haque and S. H. Gellman, Insights on β-Hairpin Stability in Aqueous Solution from Peptides with Enforced Type I‘ and Type II‘ β-Turns, J. Am. Chem. Soc., 1997, 119, 2303–2304 CrossRef CAS.
- H. E. Stanger and S. H. Gellman, Rules for Antiparallel β-Sheet Design: D-Pro-Gly Is Superior to L-Asn-Gly for β-Hairpin Nucleation1, J. Am. Chem. Soc., 1998, 120, 4236–4237 CrossRef CAS.
- A. C. Gibbs, T. C. Bjorndahl, R. S. Hodges and D. S. Wishart, Probing the Structural Determinants of Type II‘ β-Turn Formation in Peptides and Proteins, J. Am. Chem. Soc., 2002, 124, 1203–1213 CrossRef CAS PubMed.
- H. E. Stanger, F. A. Syud, J. F. Espinosa, I. Giriat, T. Muir and S. H. Gellman, Length-dependent stability and strand length limits in antiparallel β-sheet secondary structure, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 12015–12020 CrossRef CAS PubMed.
- D. S. Wishart, B. D. Sykes and F. M. Richards, The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy, Biochemistry, 1992, 31, 1647–1651 CrossRef CAS.
- D. S. Wishart, C. G. Bigam, A. Holm, R. S. Hodges and B. D. Sykes, 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects, J. Biomol. NMR, 1995, 5, 67–81 CrossRef CAS PubMed.
- R. M. Fesinmeyer, F. M. Hudson, K. A. Olsen, G. W. N. White, A. Euser and N. H. Andersen, Chemical Shifts Provide Fold Populations and Register of β Hairpins and β Sheets, J. Biomol. NMR, 2005, 33, 213–231 CrossRef CAS PubMed.
- M. Matsumoto, S. J. Lee, M. R. Gagné and M. L. Waters, Cross-strand histidine–aromatic interactions enhance acyl-transfer rates in beta-hairpin peptide catalysts, Org. Biomol. Chem., 2014, 12, 8711–8718 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04288b |
| This journal is © The Royal Society of Chemistry 2021 |