Open Access Article
Open Access ArticleProgrammable self-assembly of M13 bacteriophage for micro-color pattern with a tunable colorization†
Thanh Mien Nguyen‡
a,
Won-Geun Kim‡a,
Hyun-Ju Ahnc,
Minjun Kim c,
Young Do Kimd,
Vasanthan Devaraj
c,
Young Do Kimd,
Vasanthan Devaraj b,
Ye-Ji Kima,
Yujin Leea,
Jong-Min Lee
b,
Ye-Ji Kima,
Yujin Leea,
Jong-Min Lee *e,
Eun Jung Choi*b and
Jin-Woo Oh
*e,
Eun Jung Choi*b and
Jin-Woo Oh *ab
*ab
aDepartment of Nano Fusion Technology, BK21 Plus Nano Convergence Division, Pusan National University, Busan 46214, Republic of Korea. E-mail: ojw@pusan.ac.kr
bBio-IT Fusion Technology Research Institute, Pusan National University, Busan, 46241, Republic of Korea. E-mail: eunjung721203@gmail.com
cDepartment of Physics, Chungnam National University, Daejeon, 34134, Republic of Korea
dSamsung Display Co., Ltd., Yongin 17113, Republic of Korea
eSchool of Nano Convergence Technology, Hallym University, Chuncheon, Gangwon-do 24252, Republic of Korea. E-mail: jongminlee1984@gmail.com
First published on 30th September 2021
Abstract
Over the last decade, the M13 bacteriophage has been used widely in various applications, such as sensors, bio-templating, and solar cells. The M13 colorimetric sensor was developed to detect toxic gases to protect the environment, human health, and national security. Recent developments in phage-based colorimetric sensor technologies have focused on improving the sensing characteristics, such as the sensitivity and selectivity on a large scale. On the other hand, few studies have examined precisely controllable micro-patterning techniques in phage-based self-assembly. This paper developed a color patterning technique through self-assembly of the M13 bacteriophages. The phage was self-assembled into a nanostructure through precise temperature control at the meniscus interface. Furthermore, barcode color patterns could be fabricated using self-assembled M13 bacteriophage on micrometer scale areas by manipulating the grooves on the SiO2 surface. The color patterns exhibited color tunability based on the phage nano-bundles reactivity. Overall, the proposed color patterning technique is expected to be useful for preparing new color sensors and security patterns.
1. Introduction
Viruses are a promising natural resource for constructing multi-functional nanoscale materials in many applications.1–3 The M13 bacteriophage has been demonstrated to be a biological building block with unique features for biomimetic self-templating processes. The bacteriophage provides a highly anisotropic shape and monodispersed properties, making it attractive for self-assembly into a highly ordered structure.4,5 In addition, bacteriophages are safe in humans and can generate identical copies of themselves by infecting bacterial host cells.6 These unique characteristics have prompted researchers to evaluate the use of M13 phages as a primary building block in self-assembly processes.7–9 The M13 phage has been used successfully in sensors,10–17 solar cells,18,19 piezoelectric devices,9,20,21 tissue regeneration,4,7,22 and batteries.23–25M13 phage-based colorimetric sensors have attracted considerable attention because of their high sensitivity and selectivity.11,12,17 Previously, the M13 bacteriophage films were applied to color sensors using a vertical pulling system to produce desired colored gaps depending on the pulling rate to change the diameter of the virus bundles.10 Recent approaches have applied self-assembly technologies using a spin coating method for phage-based colorimetric sensors to improve the sensing characteristics, such as sensitivity and selectivity at a large scale.11,12 On the other hand, these methods have been limited in self-assembled M13 bacteriophage at a micro-pattering scale. Hence, the accurate, straightforward, and low-cost fabrication of high crystallinity nanostructures for micro-patterning is still challenging.
Micro-patterns are used commonly in various application fields, such as tissue engineering, optics, and sensors, because of their unique capability properties.26–28 For example, micropatterns can provide precise shape control similar to the extracellular matrix topography in nature for tissue engineering to examine the cell behavior. In sensor applications, sensor micro-patterns can simultaneously overcome low-cost manufacturing, sensitivity, and selectivity.29,30 On the other hand, micro-patterning fabricating requires typically complicated processing steps, resulting in high-cost manufacturing.31
This study developed the programmable color pattern through the self-assembly of M13 bacteriophages. A simple, highly crystallinity method, called meniscus-dragging deposition (MDD), which can be used in the self-assembly of M13 phages with highly ordered nanostructures, was first generated. The phage was self-assembled into nanostructures through programmable temperature conditions at the meniscus interface. Smectic helicoidal nanofilaments were observed in a repeatable color matrix on a SiO2 substrate due to low cost, stability, and high reflectance in white light.8 To extend the applicable area of color pattern, the SiO2 substrate was implemented to modify the reflectance by controlling the deposited thickness. Finally, excellent barcode-like color patterns were fabricated for precise humidity detection on the micrometer scale. This simple, low-cost method can be used for sensor system development to protect human health and the living environment. Moreover, barcode patterning with color tunability and micrometer-sized pattern allowed high-security level information encoding applications.32
2. Results and discussion
2.1. Principle and characteristic of M13 color film fabrication
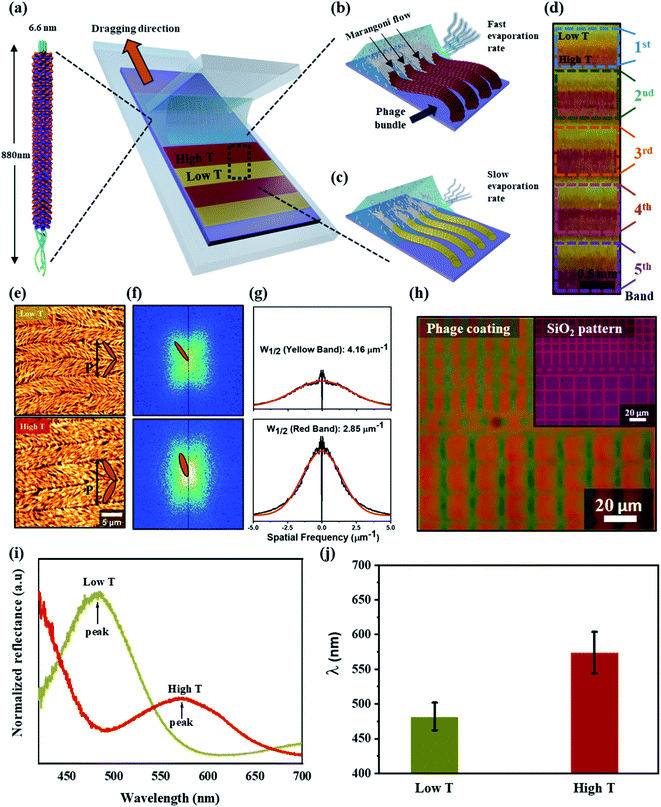
The color matrices were fabricated from M13 phages using a simple method, meniscus-dragging deposition (MDD), as shown in Fig. 1a (Fig. S1 in ESI†). Phage solution consumption and fabrication time were decreased significantly because the MDD is a microliter-scale solution process.33–35 For color matrix preparation, the micro-litter phage solution was injected onto a SiO2 substrate, resulting in a highly ordered M13 phage layer. The M13 phage exists as filamentous nanofibers, 6.6 nm in diameter and 880 nm in length, which is similar to the shape of nanowires. The single-stranded DNA is covered with a cylindrical coat made up of 2700 copies of the major coat protein (pVIII), and there are five copies. The single-stranded DNA is covered with a cylindrical coat made up of 2700 copies of the major coat protein (pVIII), and there are five copies each of the minor proteins (pIII and pVI) and other minor proteins (pVII and pIX) at both ends of the cylinder, respectively. The surface of the M13 phage can be modified genetically and chemically to introduce specific peptides that enable absorption with molecules and organic materials. Therefore, the self-templating of M13 phages with highly anisotropic shapes and monodispersed properties can easily mimic helical macromolecules in nature. From that, color films can be developed from self-assembled M13 phages using the MDD technique for many applications.In self-assembly processing, the M13 phage nanostructures were tunable by varying the kinetics and thermodynamics of assembly, such as phage condition, dragging speed, and temperature in the MDD system. These parameters are essential, affecting the crystal phase transition from an isotropic phage to a crystalline phase at the interfacial meniscus.36,37 The present study developed smectic helicoidal nanofilaments with zigzag morphologies, which have highly ordered, periodic grip-like structures. On the other hand, the self-templated phages were dragged horizontally using a stationary blade in the MDD method and assembled parallel to the dragging direction. During the MDD process, the water evaporation rate and phage density correlated according to the temperature at the interfacial meniscus due to the Marangoni effect.38,39 Consequently, higher temperature (high T) leads to higher density phages at the meniscus surface, enhancing the M13 bundle diameter due to the dense packing of M13 phages, as shown in Fig. 1b. In contrast, the M13 density at the interfacial meniscus at lower temperatures (low T) decreased (Fig. 1c), resulting in an alteration of the bundle diameter of phage nanostructures. The mechanism for the self-assembly of M13 phages was examined by implementing the self-assembled M13 at the same phage concentration and dragging speed. By controlling the temperature condition during the self-assembly process (Fig. S2a in ESI†), a repeatable color gap was fabricated precisely by altering the bundle diameter, as shown in Fig. 1d.
Atomic force microscopy (AFM) was performed for surface characterization of the self-assembled M13 bacteriophage (see the Materials and methods for details). The smectic helicoidal nanofilaments were layered with zigzag morphologies (Fig. 1e). Under the high T condition, the phage bundle size increased dramatically, resulting in a narrower-spaced self-assembled virus. AFM revealed a difference in bundle size in the low T and high T cases (Fig. S2b in ESI†). Significant differences in the periodic pitches (P) of low and high temperature (8.0 ± 0.3 μm and 10.3 ± 0.3 μm, respectively) were observed. Fast Fourier Transform (FFT) analysis from the AFM images was performed to confirm the alternating angle θ in the SHN structures, as shown in Fig. 1f. The alternating angle θ of the high T case was larger than the low T case. The widths of the spatial frequency were compared for more quantitative analysis. The width of the spatial spectrum decreased with increasing temperature because of an increase in phage bundle size (Fig. 1g). The behaviors of the smectic helicoidal nanofilaments in the self-assembly process cause coherent scattering to form the desired color from the zigzag nanofilaments. Therefore, the highly efficient temperature control in the phage self-assembly process can produce a full-color film using the simple MDD method. The self-assembly coating process of M13 phages on a micro-pattern up to the 10 μm scale was produced by mimicking a checkered stripe structure by etching the SiO2-300 substrate to make grooves (Fig. S3 in ESI†). The result was a colored micro-pattern after coating an M13 phage layer on the surface, shown in Fig. 1h.
Reflectance optical microscopy images indicated that the coherent scattering was different at low T and high T at each band. Fig. 1i presents the reflectance spectrum at 470 nm and red-shifted at 580 nm under the low T and high T conditions, respectively. The coherent scattering wavelengths of each band at low T and high T cases were almost unaltered, as shown in Fig. 1j. This suggests that a color film could be fabricated with uniformity and precision over a small area and at each band under temperature control during MDD processing. Similar to the surface characterization of the self-assembled M13 bacteriophage by AFM analysis, these results show that the phage color matrices are strongly dependent on the diameter of the bundled nanostructures. Under the low T condition, the phage bundle with a thin size exhibited a shorter reflectance wavelength. In contrast, a red-shifted wavelength occurs at a thicker bundle size under high T conditions. Therefore, tunable color films can be produced easily by controlling the bundle diameter through temperature control.
2.2. Micro-patterning of color film fabrication
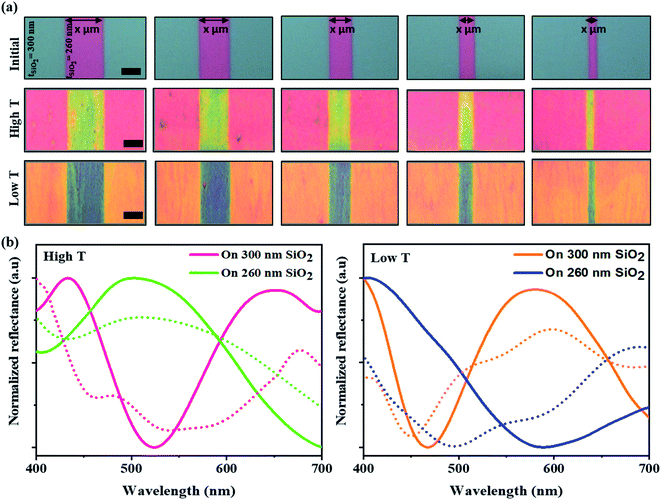
The efficiency and development of color films from the self-assembly of M13 bacteriophages using the MDD system were optimized for various applications by changing the structural color based on the flexibly diversified reflectance substrate properties. Diverse color films with many substrate structures were produced by changing the reflectance properties of the SiO2 substrates through film thickness adjustment. From that, a barcode-like structure was developed with grooves etched with different widths (Fig. S4a, ESI†). Those grooves had a controlled width from 10 to 50 μm, and a depth of 40 μm (as shown in Fig. S4b, ESI†) compared to the initial substrate. Consequently, the initial pattern was coated on 300 nm-thick SiO2 layers on a Si substrate, and the groove areas were coated with 260 nm-thick SiO2 layers. Indeed, the virus colors at the grooves were distinctly altered compared to the virus color on the initial SiO2 substrate, even though the self-assembly condition was the same. A brilliant multi-color virus film and a color change were achieved by varying the temperature conditions during MDD processing. A color barcode-like exhibited four distinct color areas: high T 300 nm, high T 260 nm, low T 300 nm, and low T 260 nm. Specifically, the phage color shows a yellow-shifted color from pink to yellow at the high T area. In contrast, in the low T area, the structural color changed from orange to blue in the grooves in Fig. 2a. The reflectance measurements and simulation data in Fig. 2b helped clarify the optical properties in color barcode-like structure. Therefore, a complex and colorful micro-patterning substrate could be produced by controlling the self-assembly condition, providing great potential for color sensors, anti-counterfeiting barcodes, and high-security level information encoding applications.2.3. M13 phage color sensor for humidity detection
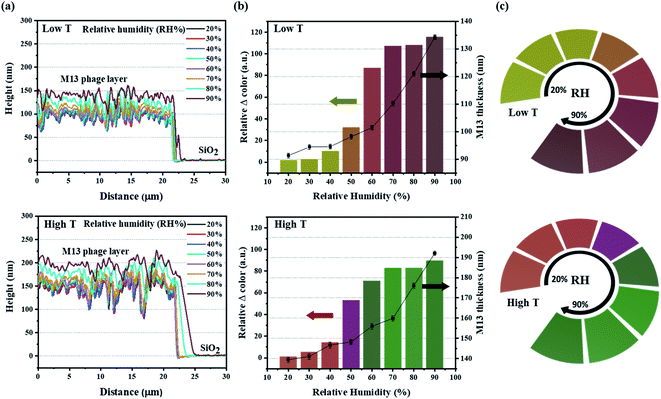
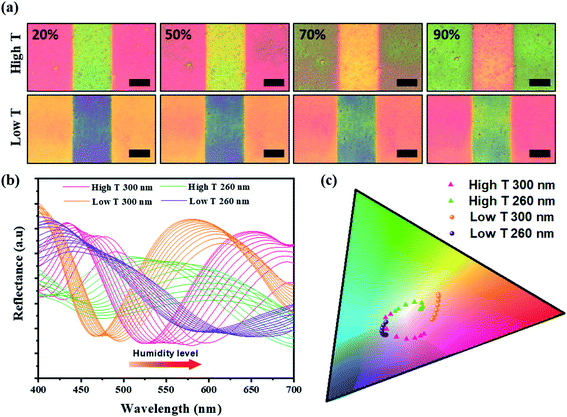
The sensitivity of the color film was evaluated by implementing a humidity-sensing test over a wide range from 20% to 90%. For rigorous testing of the responsive M13 layer, atomic force microscopy (AFM) was used to measure the thickness of the M13 layer for the dynamic changes with water absorption (Fig. 3a). The results revealed an impressive increase in the phage layer with water absorption. For quantitative analysis of the color change intensity, the difference in intensity (Fig. 3b) was calculated based on the ΔRGB (see Fig. S5 in ESI† for details) exhibited under the bar chart corresponding to the real-time change color in Fig. 3c. The scatter chart representing the M13 layer thickness determined by AFM also showed a gradual increase consistent with the water absorption of the M13 layer and a change in color intensity. Specifically, the M13 layer thickness at low T and the high T areas became larger from 90 nm (20%) to 135 nm (90%) and from 140 nm (20%) to 190 nm (90%), respectively. Fig. 3c presents the real-time changed digitized color images at the low T and high T areas. The color of the low T area red-shifted with increasing humidity, whereas the color green-shifted at the high T area. The real-time color image changed significantly from 20 to 40% and increased rapidly at 50%, showing a significant color change consistent with the color change intensity in Fig. 3b. All colors of each band were recorded and analyzed using a charge-coupled device (CCD) video camera controlled by the MATLAB program in real-time.A color change of barcode-like M13 color film was observed from four different color areas: high T 300 nm, high T 260 nm, low T 300 nm, and low T 260 nm. All four color areas changed according to the different humidity conditions, as shown in Fig. 4a and S6 (ESI).† The reflectance spectra in Fig. 4b showed a wide dispersion in the visible light region. The chromatic values of each color area from the reflectance spectra were plotted using the RGB color gamut in CIE 1931 coordinates (Fig. 4c and S7 in ESI†). Each color area showed a distinct change at different humidities from 20% to 90% on the RGB color gamut. The color barcode-like structure showed a dramatic and obvious change at different color areas through further developments of smart color structures from the M13 phage. Comparing with the color films based on M13 bacteriophage by pulling and spin coating methods, the color film using MDD improved a variety in choosing the coating substrate (planar or 3D structure). Besides, the reactivity of color film on SiO2 substrate under humidity conditions showed high sensitivity compare with the color film on the gold (Au) substrate.
3. Conclusion
A programmable color pattern was fabricated on a self-assembled M13 bacteriophage using a simple meniscus-dragging deposition (MDD) method. In the self-assembly process of the M13 phage, the temperature at the interfacial meniscus played a vital role in the color-generation film. A repeatable color film was produced easily by controlling the bundle diameter. A color film for various applications was optimized by first fabricating a precisely controllable micropattern as a barcode-like structure by forming grooves on the initial SiO2 surface. The M13 phages were then coated on the barcode-like structure to generate a distinct array color that responded rapidly to the relative humidity over a wide range (20–90%). The color film fabricating system will be a catalyst for further research on color sensor fabricating, which is commonly used to detect many harmful chemicals to protect human health and the environment. Furthermore, color micro-patterning of barcode-like structures could be useful for high-security level information encoding applications in the future.4. Materials and methods
4.1. Preparation of M13 phage color films and micro-patterned color films
M13 phage color films were fabricated on SiO2 wafers using the meniscus-dragging deposition (MDD) technique.26–28 The SiO2 wafers were first cleaned by rinsing with acetone, ethanol, isopropyl alcohol, and deionized water, followed by an O2 plasma process (50 W, 80 sccm, and 30 s). In micro pattern preparation, the micropattern film was fabricated by lithography using a contact aligner. The MDD method was used with a facile system to control the moving speed of the SiO2 substrates (Kinesis K-Cube Brushless DC Servo Controller with DDSM100/M – Compact 100 mm Travel Direct Drive Stage, Metric, THORLABS, USA). A wild M13 phage dispersed at various concentrations in a Tris-buffered saline (TBS) buffer (12.5 mM of Tris and 37.5 mM of NaCl, pH 7.5) was used to prepare the samples. Briefly, the deposition plate was placed on the moving specimen. 50 μL of the M13 phage solution was injected into the wedge between the stationary blade and SiO2 film at an angle of 50°. Finally, the M13 phage color film was achieved by moving linearly at a constant speed (0.005 mm s−1).4.2. Humidity detection system
Real-time humidity sensing was implemented over a wide range from 20% to 90% using a home-built detection system.10 All colors were recorded and analyzed using a charge-coupled device (CCD) video camera controlled by the MATLAB program in real-time. A MATLAB program was used to control the CCD video camera to capture the image and display the RGB values calculated in real-time. External light interference was prevented by conducting humidity detection in a darkroom to accept simple LED lighting from the CCD camera. For humidity sensing preparation, dry N2 was first passed through the chamber to remove any moisture present. Subsequently, the moisture flow from the humidifier was injected into the chamber controlled by a humidity sensor. The real-time color changes were analyzed under the RGB signals, and color images were captured for 3 s. The phage color changes were displayed and calculated based on the RGB data difference between the initial color and phage color film at each relative humidity level using the Photoshop program (Adobe System Inc.).4.3. Atomic force microscopy (AFM) measurements
An NX10 AFM system (Park Systems, Suwon, Korea) operated by the data acquisition program XEP 3.0.4 (Park Systems, Suwon, Korea) was used to examine the topography of the M13 films. To characterize the M13 surface and measure the bundle of M13 phages from AFM images, the data were analyzed using the XEI 1.8.2 image-processing program (Park Systems, Suwon, Korea). All images were collected in true non-contact mode. A specialized probe for non-contact mode was selected for the measurement (PPP-NCHR, NANOSENSORS, Neuchatel, Switzerland).4.4. Reflectance measurement of M13 phage color film
The virus color film was illuminated by a white light generated using a xenon lamp (X-Cite, Exfo, Mississauga, Canada). The reflected light was obtained using a fiber-optic spectrophotometer (USB2000+, Ocean Optics, Dunedin, FL). A Y-shaped reflection/backscattering optical fiber (QR400-7-VIS-NIR, Ocean Optics, Dunedin, FL) was used for illumination and obtaining reflected light. An optical fiber fixed on a z stage was positioned normal to the virus color film that had been fixed on a hollow rotation stage.4.5. Reflectance simulation of M13 phage color film
Three-dimensional optical simulations were carried out using an ANSYS Lumerical FDTD electromagnetic solver.40,41 Multi-layered films were surrounded by perfectly matched layer boundary conditions in XYZ directions. Broadband plane wave source (Total-Field Scattered-Field (TFSF) source) was used to excite the film with an incident electric field (E0) in the normal direction from the top (+Z direction). Two mesh sizes were used: 5 nm size covering the whole simulated region and a mesh over-ride of 0.5 nm covering the M13 bacteriophage, SiO2, and Si. To extract scattering data, we used a box-shaped power monitor to record the field. The refractive indices of materials used are as follows: SiO2 and Si (Palik database); M13 bacteriophage (n = 1.37).42Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Advanced Production Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (318104-3).References
- J.-S. Moon, W.-G. Kim, C. Kim, G.-T. Park, J. Heo, S. Y. Yoo and J.-W. Oh, Mini-Rev. Org. Chem., 2015, 12, 271–281 CrossRef CAS PubMed.
- R. Selvakumar, N. Seethalakshmi, P. Thavamani, R. Naidu and M. Megharaj, RSC Adv., 2014, 4, 52156–52169 RSC.
- N.-M. D. Courchesne, M. T. Klug, P.-Y. Chen, S. E. Kooi, D. S. Yun, N. Hong, N. X. Fang, A. M. Belcher and P. T. Hammond, Adv. Mater., 2014, 26, 3398–3404 CrossRef CAS PubMed.
- W.-J. Chung, A. Merzlyak and S.-W. Lee, Soft Matter, 2010, 6, 4454–4459 RSC.
- J. Han, V. Devaraj, C. Kim, W.-G. Kim, D.-W. Han, S. W. Hong, Y.-C. Kang and J.-W. Oh, ACS Appl. Nano Mater., 2018, 1, 2851–2857 CrossRef CAS.
- S. H. Yang, W.-J. Chung, S. McFarland and S.-W. Lee, Chem. Rec., 2013, 13, 43–59 CrossRef CAS PubMed.
- A. Merzlyak, S. Indrakanti and S.-W. Lee, Nano Lett., 2009, 9, 846–852 CrossRef CAS PubMed.
- W.-G. Kim, K. Kim, S.-H. Ha, H. Song, H.-W. Yu, C. Kim, J.-M. Kim and J.-W. Oh, Sci. Rep., 2015, 5, 13757 CrossRef PubMed.
- B. Y. Lee, J. Zhang, C. Zueger, W.-J. Chung, S. Y. Yoo, E. Wang, J. Meyer, R. Ramesh and S.-W. Lee, Nat. Nanotechnol., 2012, 7, 351–356 CrossRef CAS PubMed.
- J.-W. Oh, W.-J. Chung, K. Heo, H.-E. Jin, B. Y. Lee, E. Wang, C. Zueger, W. Wong, J. Meyer and C. Kim, Nat. Commun., 2014, 5, 3043 CrossRef PubMed.
- Y. J. Yoo, J. H. Ko, W.-G. Kim, Y. J. Kim, D.-J. Kong, S. Kim, J.-W. Oh and Y. M. Song, ACS Appl. Nano Mater., 2020, 3, 6636–6644 CrossRef CAS.
- Y. J. Yoo, W.-G. Kim, J. H. Ko, Y. J. Kim, Y. Lee, S. G. Stanciu, J.-M. Lee, S. Kim, J.-W. Oh and Y. M. Song, Adv. Sci., 2020, 7, 2000978 CrossRef CAS PubMed.
- K.-H. Kim, T. M. Nguyen, S.-H. Ha, E. J. Choi, Y. Kim, W.-G. Kim, J.-W. Oh and J.-M. Kim, ACS Appl. Mater. Interfaces, 2020, 12, 45590–45601 CrossRef CAS PubMed.
- J.-S. Moon, M. Park, W.-G. Kim, C. Kim, J. Hwang, D. Seol, C.-S. Kim, J.-R. Sohn, H. Chung and J.-W. Oh, Sens. Actuators, B, 2017, 240, 757–762 CrossRef CAS.
- J.-W. Oh, J. Choi, B. T. Luong and N. Kim, Macromol. Res., 2010, 18, 8–13 CrossRef CAS.
- J. H. Lee, B. Fan, T. D. Samdin, D. A. Monteiro, M. S. Desai, O. Scheideler, H.-E. Jin, S. Kim and S.-W. Lee, ACS Nano, 2017, 11, 3632–3641 CrossRef CAS PubMed.
- J.-M. Lee, Y. Lee, V. Devaraj, T. M. Nguyen, Y.-J. Kim, Y. H. Kim, C. Kim, E. J. Choi, D.-W. Han and J.-W. Oh, Biosens. Bioelectron., 2021, 188, 113339 CrossRef CAS PubMed.
- H.-S. Lin, J.-M. Lee, J. Han, C. Lee, S. Seo, S. Tan, H. M. Lee, E. J. Choi, M. S. Strano, Y. Yang, S. Maruyama, I. Jeon, Y. Matsuo and J.-W. Oh, Adv. Sci., 2020, 2000782 CrossRef CAS PubMed.
- H. B. Lee, W.-G. Kim, M. Lee, J.-M. Lee, S. He, N. Kumar, V. Devaraj, E. J. Choi, I. Jeon, M. Song, J.-W. Oh and J.-W. Kang, Adv. Opt. Mater., 2020, 8, 1902080 CrossRef CAS.
- D.-M. Shin, H. J. Han, W.-G. Kim, E. Kim, C. Kim, S. W. Hong, H. K. Kim, J.-W. Oh and Y.-H. Hwang, Energy Environ. Sci., 2015, 8, 3198–3203 RSC.
- Y. Yan, W.-G. Kim, X. Ma, T. Tegafaw, T. M. Nguyen, J.-M. Lee, E.-J. Choi, H. Ahn, S.-H. Ha, K. Kim, J.-M. Kim, H. K. Kim, J.-W. Oh, D.-M. Shin and Y.-H. Hwang, Nano Energy, 2021, 81, 105607 CrossRef CAS.
- E. S. Place, N. D. Evans and M. M. Stevens, Nat. Mater., 2009, 8, 457–470 CrossRef CAS PubMed.
- Y. J. Lee, H. Yi, W. J. Kim, K. Kang, D. S. Yun, M. S. Strano, G. Ceder and A. M. Belcher, Science, 2009, 324, 1051–1055 CAS.
- D. Oh, J. Qi, Y.-C. Lu, Y. Zhang, Y. Shao-Horn and A. M. Belcher, Nat. Commun., 2013, 4, 2756 CrossRef PubMed.
- G. Zhang, S. Wei and A. M. Belcher, ACS Appl. Nano Mater., 2018, 1, 5631–5639 CrossRef CAS.
- T. Xu, L.-P. Xu, X. Zhang and S. Wang, Chem. Soc. Rev., 2019, 48, 3153–3165 RSC.
- M. Ermis, E. Antmen and V. Hasirci, Bioact. Mater., 2018, 3, 355–369 CrossRef PubMed.
- Y. Jung, H. Lee, T.-J. Park, S. Kim and S. Kwon, Sci. Rep., 2015, 5, 15629 CrossRef CAS PubMed.
- L. Liu, Y. Wang, F. Sun, Y. Dai, S. Wang, Y. Bai, L. Li, T. Li, T. Zhang and S. Qin, Microsyst. Nanoeng., 2020, 6, 31 CrossRef CAS PubMed.
- S. H. Lee, H. J. Jin, H. S. Song, S. Hong and T. H. Park, J. Biotechnol., 2012, 157, 467–472 CrossRef CAS PubMed.
- S. H. Lee, W.-Y. Rho, S. J. Park, J. Kim, O. S. Kwon and B.-H. Jun, Sci. Rep., 2018, 8, 16763 CrossRef PubMed.
- H. Liu, D. Xie, H. Shen, F. Li and J. Chen, Adv. Polym. Technol., 2019, 2019, 6519018 Search PubMed.
- Y. Ko, N. H. Kim, N. R. Lee and S. T. Chang, Carbon, 2014, 77, 964–972 CrossRef CAS.
- J. Cho, Y. Ko, K. H. Cheon, H.-J. Yun, H.-K. Lee, S.-K. Kwon, Y.-H. Kim, S. T. Chang and D. S. Chung, J. Mater. Chem. C, 2015, 3, 2817–2822 RSC.
- L. Boinovich and A. Emelyanenko, Adv. Colloid Interface Sci., 2011, 165, 60–69 CrossRef CAS PubMed.
- Z. Dogic and S. Fraden, Curr. Opin. Colloid Interface Sci., 2006, 11, 47–55 CrossRef CAS.
- X. Gu, L. Shaw, K. Gu, M. F. Toney and Z. Bao, Nat. Commun., 2018, 9, 534 CrossRef PubMed.
- Z. Zhang, B. Peng, X. Ji, K. Pei and P. K. L. Chan, Adv. Funct. Mater., 2017, 27, 1703443 CrossRef.
- S. B. Lee, S. Lee, D. G. Kim, S. H. Kim, B. Kang and K. Cho, Adv. Funct. Mater., 2021, 31, 2100196 CrossRef CAS.
- V. Devaraj, J.-M. Lee, S. Adhikari, M. Kim, D. Lee and J.-W. Oh, Nanoscale, 2020, 12, 22452–22461 RSC.
- V. Devaraj, N.-N. Jeong, J.-M. Lee, Y.-H. Hwang, J.-R. Sohn and J.-W. Oh, J. Korean Phys. Soc., 2019, 75, 313–318 CrossRef CAS.
- E. D. Palik, in Handbook of Optical Constants of Solids, ed. E. D. Palik, Academic Press, Boston, 1985, pp. 3–9, DOI:10.1016/B978-0-08-054721-3.50006-X.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04302a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |