Open Access Article
Open Access ArticleSynthesis of covalent bonding MWCNT-oligoethylene linezolid conjugates and their antibacterial activity against bacterial strains†
José A. Alatorre-Barajasa,
Eleazar Alcántar-Zavalaa,
M. Graciela Gil-Rivasa,
Edgar Estrada-Zavalab,
Adrián Ochoa-Terán *a,
Y. Gochi-Ponce*a,
Julio Montes-Ávilab,
Alberto Cabreraa,
Balter Trujillo-Navarrete
*a,
Y. Gochi-Ponce*a,
Julio Montes-Ávilab,
Alberto Cabreraa,
Balter Trujillo-Navarrete a,
Yazmin Yorely Rivera-Lugoa,
Gabriel Alonso-Núñezc,
Edgar A. Reynoso-Sotoa and
J. L. Medina-Francod
a,
Yazmin Yorely Rivera-Lugoa,
Gabriel Alonso-Núñezc,
Edgar A. Reynoso-Sotoa and
J. L. Medina-Francod
aCentro de Graduados e Investigación en Química, Tecnológico Nacional de México/ IT de Tijuana, Tijuana, B. C., Mexico. E-mail: adrian.ochoa@tectijuana.edu.mx; yadira.gochi@tectijuana.edu.mx
bFacultad de Ciencias Químico Biológicas, Universidad Autónoma de Sinaloa, Culiacán, Sin, Mexico
cCentro de Nanociencias y Nanotecnología, Universidad Nacional Autónoma de México, Ensenada, B. C, Mexico
dDepartamento de Farmacia, Facultad de Química, Universidad Nacional Autónoma de México, Ciudad de México, Mexico
First published on 31st August 2021
Abstract
Nowadays, infectious diseases caused by drug-resistant bacteria have become especially important. Linezolid is an antibacterial drug active against clinically important Gram positive strains; however, resistance showed by these bacteria has been reported. Nanotechnology has improved a broad area of science, such as medicine, developing new drug delivery and transport systems. In this work, several covalently bounded conjugated nanomaterials were synthesized from multiwalled carbon nanotubes (MWCNTs), a different length oligoethylene chain (Sn), and two linezolid precursors (4 and 7), and they were evaluated in antibacterial assays. Interestingly, due to the intrinsic antibacterial activity of the amino-oligoethylene linezolid analogues, these conjugated nanomaterials showed significant antibacterial activity against various tested bacterial strains in a radial diffusion assay and microdilution method, including Gram negative strains as Escherichia coli (11 mm, 6.25 μg mL−1) and Salmonella typhi (14 mm, ≤0.78 μg mL−1), which are not inhibited by linezolid. The results show a significant effect of the oligoethylene chain length over the antibacterial activity. Molecular docking of amino-oligoethylene linezolid analogs shows a more favorable interaction of the S2-7 analog in the PTC of E. coli.
1. Introduction
Hospital-acquired infections (HAIs) are a globally significant public health problem and steadily increase.1,2 HAIs are defined as those that were neither present in the patient nor in the incubation period and were acquired during the hospital stay at least 48 hours after hospitalization.3 A common characteristic in most of these pathogens is resistance to commonly used antibiotics, mainly generated by the abuse and inappropriate use of antibiotics. Furthermore, these infections are a significant cause of mortality globally; in the United States, approximately 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths are reported per year.4
000 deaths are reported per year.4
Linezolid was approved by the FDA in 2000 and showed activity against resistant strains as Enterococcus faecalis and E. faecium, vancomycin-resistant Staphylococcus aureus (SAVR), methicillin-resistant Staphylococcus aureus (MRSA), among others.5,6 This drug is characterized by having an oxazolidinone ring with the necessary stereochemistry and an acetamide group responsible for the interaction with the site of action.7 However, linezolid resistant strains have been found, despite their excellent antibacterial characteristics. Then, it is necessary to develop new antibacterial drugs taking advantage of linezolid to improve its qualities.8–11
On the other hand, Multiple-walled carbon nanotubes (MWCNT) are carbon allotrope with excellent thermal, electronic, and mechanical properties. In recent years, MWCNT's have been studied in different areas such as engineering, science, and medicine, being used to develop biological sensors, tissue engineering, drug delivery systems, and composites with antibacterial activity.12,13 Functionalized MWCNTs (f-MWCNTs) with a variety of groups (epoxides, carboxylic acids), organic molecules, and polymers have shown increased activity against bacteria in addition to an improvement in their biocompatibility properties, making them promising candidates for use in these applications.14,15
The binding of drugs on MWCNT has been extensively studied in recent years, showing that this type of formulation is one of the most promising alternatives to treat HAI.16–18 However, it is well known that MWCNTs possess intrinsic toxicity under certain conditions and characteristics, which are reduced by functionalization, considerably improving their solubility in various media, increasing their bioavailability and biocompatibility.19–21 MWCNTs have been covalently bounded with different conventional antibiotics and organic compounds such as ionic liquids for antibacterial purposes.22–26 On the other hand, loading of linezolid into a silica xerogel,27 cadmium selenide/silica nanoparticles,28 silver nanoparticles,29 selenium nanoparticles30 and biopolymer nanoparticles31–36 by non-covalent interactions has shown an improved activity against multidrug-resistant bacteria, including methicillin-resistant Staphylococcus aureus and Mycobacterium tuberculosis. Also linezolid has been adsorbed on a carbon nanomaterial as graphene oxide (GO), where the bactericidal effect increased by the combination with GO.37 To the best of our knowledge, there are not reports regarding to the covalent bonding of linezolid into MWCNT and antibacterial activity studies. Therefore, this work seeks to study the covalent bonding of linezolid-like molecules 4 and 7 on MWCNT functionalized with different variable oligoethylene spacers (C2, C4, and C6) and evaluate the antibacterial activity of these conjugated nanomaterials against different reference and drug-resistant isolated clinical strains.
2. Results y discussion
2.1 Synthesis and characterization of linezolid-type compounds
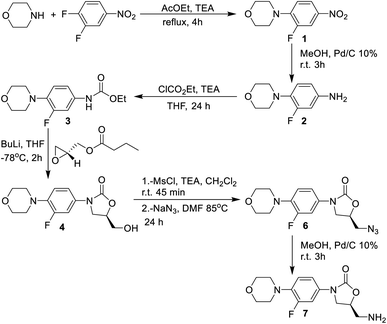
The synthesis of the linezolid type compounds was performed using the methodology reported by Brickner et al. with some variations (Scheme 1).5 For the synthesis of linezolid analogs, 3,4-difluoronitrobenzene was chosen as starting material. The SNAr reaction of 3,4-difluoronitrobenzene with morpholine leads to nitro compound 1 with a 90% yield in trimethylamine, which was then transformed into the arylamine 2 by hydrogenation of the nitro group with H2 and 10% Pd/C with 68% yield. The reaction of 2 with ethyl chloroformate provides the carbamate 3 in a 45% yield. Compound 3 was treated with n-BuLi and (R)-glycidyl butyrate to obtain the oxazolidinone 4 (a hydroxymethyl linezolid analog) in a 50% yield. Mesylation of 4 led to obtaining compound 5 in 98% yield, followed by SN2 displacement of the mesylate by the azide anion, leading to azido derivative 6 in 50% yield. The amino-linezolid 7 was obtained by molecular hydrogenation of the 6 with a 28% yield. The yields are similar to those reported by Bricker despite the changes employed in the reactions. The characterization by FTIR, NMR, and EMIE confirmed the structure of each intermediate compound (Fig. S1 to S25†).2.2 Synthesis and functionalization of MWCNTs
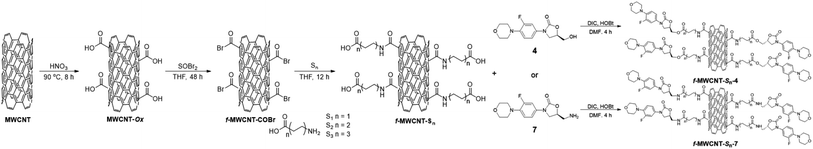
MWCNTs were prepared by the spray pyrolysis technique using toluene as a carbon source and ferrocene as a catalyst. Two different concentrations of ferrocene in toluene were used in the reactor for the synthesis, 0.1 M (MWCNT-1) and 0.2 M (MWCNT-2), respectively, to study the concentration effect over the MWCNTs structural characteristics and biological properties. Then, both MWCNTs were oxidized following the best functionalization route reported by Moreno–Valle,38 using HNO3 at 90 °C for 8 h to obtained MWCNT-Ox-1 and MWCNT-Ox-2, respectively (Scheme 1).After the variable-length oligoethylene amino acids were incorporated, six-functionalized MWCNTs were obtained (f-MWCNT-Sn). MWCNT-Ox-1 and MWCNT-Ox-2 were reacted with SOBr2 in dry THF for 48 h under Ar atmosphere to generate the acyl bromine functionality on the MWCNTs surface (f-MWCNT-COBr). The acylated MWCNTs were manipulated under anhydrous conditions and an inert atmosphere using dry THF. Three oligoethylene amino acids were reacted with the acylated MWCNT-COBr-1 and MWCNT-COBr-2, respectively, introducing different oligoethylene chains (Sn) to the MWCNTs. Finally, f-MWCNT-Sn nanomaterials were subjected to coupling reactions with the amino or hydroxyl linezolid analog (4 or 7) using DIC and HOBt to obtain the MWCNT-oligoethylene linezolid conjugates (Scheme 2). Moreover, the incorporation of amino-linezolid 7 on the MWCNT-Ox-# nanotubes was carried out to study the effect of the oligoethylene chain length on the antibacterial activity. The synthesis was carried out using the methodology employed for the functionalization with the alkylamino acids (Scheme S1†).
2.3 Characterization by Raman spectroscopy
Raman spectroscopy characterization of MWCNT-1 and MWCNT-2 nanotubes showed the characteristic bands of these nanomaterials at 1318 cm−1, 1585 cm−1 and 2621 cm−1, corresponding to D, G and G′ bands, respectively. The first band is related to the amount of disorder or defects and the presence of sp3-hybridized carbon atoms, while the second band is related to the degree of graphitization or structural order of the carbon slides on the material and the presence of carbon atoms with sp2 hybridization. Finally, the last band is an overtone or a second-order vibration of the G band related to order and crystallinity. The ratio ID/G is a value that relates the disorder and order directly associated with the defect density or flaws in MWCNT, while the ratio IG′/G is related to the degree of crystallinity of the nanomaterial. Thus, MWCNT-1 and MWCNT-2 showed a similar degree of defects despite the difference in concentration of ferrocene in toluene used in the reactor for the synthesis, but the crystallinity of MWCNT-1 was higher than MWCNT-2 (Fig. S26†).Considering the ID/G and IG′/G data of the nanomaterials and the synthesis, it can be seen that oxidation and functionalization processes have different effects over the two types of MWCNTs (Table 1). MWCNT-Ox showed considerable changes in the intensity of their characteristic bands, with a decrease in the G and G′ band and an increase in the D band. The latter is explained by the oxidative treatment with nitric acid, which generates carboxyl, hydroxyl, and epoxide groups on the surface of the nanotubes by breaking graphene layers, which leads to an increase in the disorder or higher number of sp3 carbons. The f-MWCNT-Sn showed considerable changes in the intensity of their characteristic bands, with an increase in the D, G, and G′ bands, but a decrease in the ID/G ratio compared to their MWCNT-Ox (Fig. S27 and 28†). This is due to the functionalization that incorporates the oligoethylene chain in the reactive groups, reducing the surface defects of the nanotubes, which leads to an increase in order and a higher crystallinity. Finally, the MWCNTs functionalized with the different oligoethylene amino acids and linezolid precursors 4 and 7 (f-MWCNT-Sn-4, f-MWCNT-Sn-7) did not present considerable changes in the intensity of their characteristic bands, observing that the ID/G and IG′/G values are like those obtained with their precursors regardless the alkyl chain (Fig. S29 and S30†). These results show that the coupling of 4 and 7 occurs on the external carboxylic group or the nanotube surface.
| Nanomaterial | ID/G | IG′/G | Nanomaterial | ID/G | IG′/G |
|---|---|---|---|---|---|
| MWCNT-1 | 0.82 | 1.35 | MWCNT-2 | 0.83 | 1.20 |
| MWCNT-Ox-1 | 0.89 | 1.25 | MWCNT-Ox-2 | 0.87 | 1.37 |
| f-MWCNT-S1-1 | 0.77 | 1.15 | f-MWCNT-S1-2 | 0.93 | 1.07 |
| f-MWCNT-S2-1 | 0.77 | 1.30 | f-MWCNT-S2-2 | 0.89 | 1.28 |
| f-MWCNT-S3-1 | 0.73 | 1.34 | f-MWCNT-S3-2 | 0.84 | 1.18 |
| f-MWCNT-S1-4-1 | 0.69 | 1.06 | f-MWCNT-S1-4-2 | 0.91 | 1.05 |
| f-MWCNT-S2-4-1 | 0.79 | 1.10 | f-MWCNT-S2-4-2 | 0.80 | 1.14 |
| f-MWCNT-S3-4-1 | 0.82 | 1.20 | f-MWCNT-S3-4-2 | 0.81 | 1.12 |
| f-MWCNT-S1-7-1 | 0.67 | 1.08 | f-MWCNT-S1-7-2 | 0.94 | 1.01 |
| f-MWCNT-S2-7-1 | 0.75 | 1.10 | f-MWCNT-S2-7-2 | 0.83 | 1.18 |
| f-MWCNT-S3-7-1 | 0.84 | 1.18 | f-MWCNT-S3-7-2 | 0.76 | 1.09 |
| f-MWCNT-7-1 | 0.71 | 1.27 | f-MWCNT-7-2 | 0.70 | 1.25 |
2.4 TGA analysis
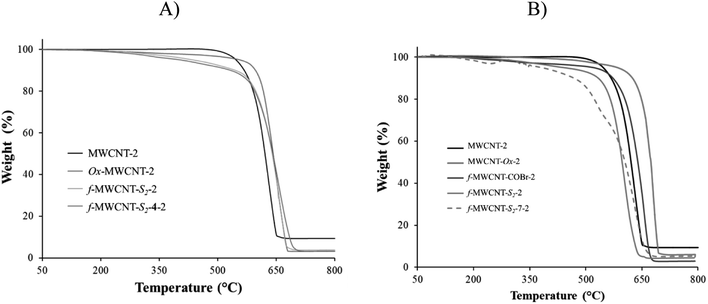
Thermogravimetric analysis of the nanotubes before oxidation showed the typical thermic degradation starting at 650 °C for MWCNT-1, while it was at 620 °C for MWCNT-2. The FeO metal residues were 11.37 and 9.40%, respectively (Fig. S31†). The higher thermic stability of MWCNT-1 may be attributed to its higher crystallinity found by the Raman spectroscopy. On the other hand, after performing the oxidation of both nanotubes, two characteristic thermal decomposition stages are observed in the thermograms; the first stage begins around 300 °C, and it is attributed to the thermal decomposition of the oxygen functional groups such as carboxylic, hydroxyl, among others. The second stage of thermal degradation starts around 500 °C and corresponds to the typical thermal oxidation of carbon nanotube (Fig. S32†). A refractory residue (FeO) derived from the metallic catalyst is observed at the final stage. The metal residue after acid treatment was 5.0 and 4.4% for MWCNT-Ox-1 and MWCNT-Ox-2, respectively. With this data in hand, an approximate functionalization degree of functionalization of 0.70 μmol mg−1 was calculated for both nanotubes.The f-MWCNT-COBr thermogram shows two characteristic thermal decomposition stages; the first stage is around 350 °C, which is 50 °C more than MWCNT-Ox-1 due to the higher bond energy of COBr and CBr functional groups. The second stage of thermal degradation starts around 650 °C, corresponding to the characteristic degradation of nanotubes. The metallic content in f-MWCNT-COBr-1 is 4.0%.
The thermogravimetric analysis of f-MWCNT-Sn shows that weight loss stages are more remarkable as the oligoethylene chain length increases following the trend f-MWCNT-S3 > f-MWCNT-S2 > f-MWCNT-S1 > MWCNT-Ox > MWCNT (Fig. S33 and S34†). All MWCNTs show significant weight losses from 150 °C to 600 °C attributed to the loss of surface defects, carboxylic and amino acid groups on MWCNT. The notorious weight loss indicates that functionalization was efficiently carried out on the nanomaterial surface.
Finally, thermograms of f-MWCNT-Sn-4 and f-MWCNT-Sn-7 indicate a gradual thermic degradation of these nanomaterials from 150 °C to 600 °C due to surface defects, carboxylic groups, amino acids, and linezolid ester or amide on MWCNT. The difference in the degradation pattern is attributed to the presence of Sn-4 and Sn-7. In general, the changes in thermic degradation in each step indicate different structural-chemical characteristics on the carbon nanotube surface. The weight-loss trend was MWCNT < MWCNT-Ox < f-MWCNT-Sn < f-MWCNT-Sn-4 or f-MWCNT-Sn-7. Based on the TGA analysis of f-MWCNT-Sn-7, considering the oligoethylene chains and the oxazolidinone, the percentages of organic material incorporated to the MWCNT surface are approximately 13.1, 15.6, 10.1, 16.7, 14.2 and 15.6% for f-MWCNT-S1-7-1, f-MWCNT-S2-7-1, f-MWCNT-S3-7-1, f-MWCNT-S1-7-2, f-MWCNT-S2-7-2, and f-MWCNT-S3-7-2, respectively (Fig. 1). The f-MWCNT-7-1 and f-MWCNT-7-2 thermograms show percentages of organic material incorporated around 14.1% and 15.4%, indicating the successful functionalization of the MWCNTs with these molecules (Fig. S35 and S36†).
2.5 Characterization by SEM and TEM
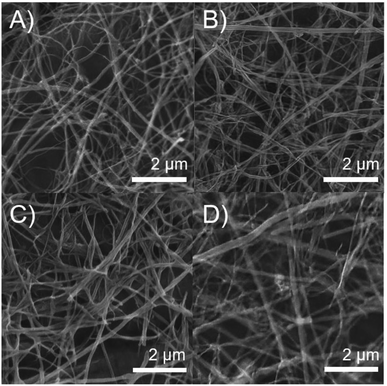
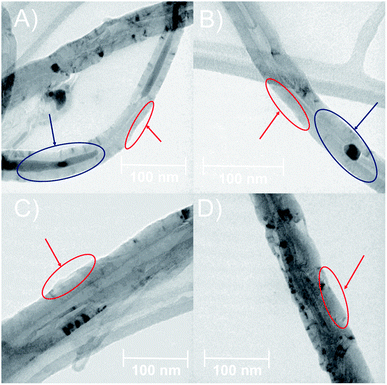
The morphology of MWCNTs obtained in each synthesis stage was analyzed through SEM and TEM. Fig. 2 shows differences in the surface along the chemical modifications sequence of MWCNT-2 by SEM. Analysis of the SEM images in MWCNT-2 showed a smooth surface, while MWCNT-Ox-2 showed a rougher surface due to nitric acid treatment, leading to the cleavage and generation of different functional groups such as carboxylic acids, alcohols, and epoxides on the surface of MWCNTs (Fig. 2A and B). Images of f-MWCNT-S2-2 show the formation of thin layers on the carbon nanotube surface attributed to the functionalization and incorporation of the oligoethylene chain (Fig. 2C). Similar features were observed with the other alkyl amino acids (Fig. S37 to S41†). Finally, images of f-MWCNT-S2-7-2 show the formation of an organic layer on the surface after coupling the amino-linezolid analog 7 with the carboxylic acid groups (Fig. 2D). This behavior was also observed with 4 (Fig. S42†). These images show the incorporation of organic material on the surface of the nanotubes.Fig. 3 shows the TEM images, wherein some differences in the surface and morphologic characteristics at the synthesis stages of MWCNT-2 are observed. The pristine MWCNT-2 have a smooth surface (red) in addition to observing a considerable inside amount of Fe (blue), while MWCNT-Ox-2 shows a rough surface and a notorious decrease of Fe (Fig. 3A and B) with 42.45 ± 5.52 nm and 35.29 ± 3.83 nm diameter, respectively. Because of the oxidative treatment, the MWCNT surface layers are deformed and remove inside and outside Fe in the MWCNT. The images of f-MWCNT-S2-2 and f-MWCNT-S2-7-2 present a homogeneous organic layer on the nanotube surface because organic material is incorporated (Fig. 3C and D), showing 76.31 ± 16.06 nm and 62.41 ± 6.84 nm in diameter, respectively. The increase in the diameter of MWCNTs compared to MWCNT-Ox-2 is attributed to the incorporation of organic molecules on the MWCNT surface, while its decrease after the incorporation of compound 7 compared to f-MWCNT-S2-2 is probably due to that some amount of the oligoethylene amino acid adsorbed on the nanomaterial surface is removed during the reaction and purification.
The results obtained from characterization techniques as TEM, SEM, Raman and TGA agree that the modifications of MWCNTs at different stages were successful.
2.6 Biological assay
The antibacterial activity of the prepared nanomaterials was evaluated against nine Gram-positive and three Gram-negative strains, including four ATCC strains, eight clinically isolated strains, and five methicillin-resistant strains, using the radial diffusion assay and broth microdilution method.MWCNTs were dispersed in distilled water for 1.0 h by sonication before the radial diffusion assay (RDA). An aliquot equivalent to 20 μg of MWCNTs was taken and added directly into Petri dishes with Mueller Hinton agar to achieve greater diffusion of the MWCNTs. linezolid (20 μg) was used as a positive control.
MWCNT-#, MWCNT-Ox-#, f-MWCNT-Sn-#, f-MWCNT-Sn-4-#, 4, and 7 were evaluated against all strains of these materials showing non-inhibition of bacterial growth (inhibition halo) in radial diffusion assays (Fig. S43†).
This result demonstrates that MWCNTs, which are not functionalized with molecules containing essential functional groups, do not have antibacterial activity. For example, MWCNTs bound to compound 4 do not possess an amide functional group which is essential to promote intermolecular hydrogen bond interaction in the site of action, which has vital importance to inhibit bacterial protein synthesis. To prove that acetamide group is indeed essential, MWCNTs were prepared bonding directly compound 7 (f-MWCNT-7), but no activity was observed too because when 7 is released from the nanomaterial, it does not have an amide group in its structure. In addition, the same results were obtained in the evaluation of free 4 and 7 compounds.
Assays with f-MWCNT-Sn-7-1 and f-MWCNT-Sn-7-2 nanomaterials showed that the conjugated nanomaterials possessing the S1 and S2 oligoethylene chains have activity against several of the strains tested. However, the S3-conjugated nanotubes did not exhibit activity against these strains.
Table 2 shows the diameters of the inhibition halos obtained in the radial diffusion test of the f-MWCNT-Sn-7-#, where compound 7 and linezolid were used as control. Noteworthy, 7 does not show activity against the strains tested. In contrast, linezolid showed activity against all Gram-positive strains evaluated, but it was inactive against Gram-negative Escherichia coli and Salmonella typhi strains because of as it is well known, this drug is selective towards Gram-positive bacteria. The most prominent nanomaterial was f-MWCNT-S2-7-2, active against many ATCC strains, MRSA, and Gram-negative strains such as Escherichia coli and Salmonella typhi. Moreover, the f-MWCNT-S1-7-1, f-MWCNT-S2-7-1, and f-MWCNT-S1-7-2 only showed localized inhibition against these same strains, indicating lower antibacterial activity.
| Nanomaterial or compound | Inhibition (mm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 9 | 13 | 29212 | 25922 | R1 | R2 | R3 | R4 | 43300 | 25923 | |
| a 0 = No activity, L = localized inhibition. (1) = A group Streptococcus clinical isolate, (5) = Staphylococcus aureus clinical isolate, (9) = Escherichia coli clinical isolate, (13) = Salmonella typhi clinical isolate, (29212) = Enterococcus faecalis ATCC, (25922) = Escherichia coli ATCC, (R1) = MRSA-01, (R2) = MRSA-02, (R3) = MRSA-03, (R4) = MRSA-04, (43300) = MRSA ATCC, (25923) = Staphylococcus. aureus ATCC. | ||||||||||||
| Linezolid | 17 | 21 | 0 | 0 | 19 | 0 | 21 | 23 | 21 | 21 | 22 | 26 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| f-MWCNT-S1-7-1 | 0 | L | L | L | L | L | 0 | 0 | 0 | L | L | L |
| f-MWCNT-S2-7-1 | 0 | L | L | L | L | L | L | L | L | L | L | L |
| f-MWCNT-S3-7-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| f-MWCNT-S1-7-2 | 0 | 0 | 0 | 0 | 0 | 0 | L | L | L | L | L | L |
| f-MWCNT-S2-7-2 | 7 | 11 | 11 | 14 | L | L | L | 11 | 12 | L | L | L |
| f-MWCNT-S3-7-2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Table 3 shows the minimum inhibitory concentration values (MIC) obtained by the broth microdilution test with the f-MWCNT that showed activity using the RDA using Linezolid and 7 as controls. As previously noted, the f-MWCNTs with S2 spacer stood out among the others because they showed activity against a higher number of strains with MIC values of between 25 and <0.78 μg mL−1. Moreover, the f-MWCNT-S1-7-2 showed a MIC between 25 and 25.6 μg mL−1, whereas the f-MWCNT-S1-7-1 does not show activity against these same strains. In addition, as noted above, compound 7 does not shows activity, and linezolid shows activity only against Gram-positive bacteria.
| Nanomaterial or compound | MIC (μg mL−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 9 | 13 | 29212 | 25922 | R1 | R2 | R3 | R4 | 43300 | 25923 | |
| a 0 = No activity, L = localized inhibition. (1) = A group Streptococcus clinical isolate, (5) = Staphylococcus aureus clinical isolate, (9) = Escherichia coli clinical isolate, (13) = Salmonella typhi clinical isolate, (29212) = Enterococcus faecalis ATCC, (25922) = Escherichia coli ATCC, (R1) = MRSA-01, (R2) = MRSA-02, (R3) = MRSA-03, (R4) = MRSA-04, (43300) = MRSA ATCC, (25923) = Staphylococcus. aureus ATCC. | ||||||||||||
| Linezolid | 0.5 | 4 | >16 | >16 | 4 | >16 | 0.5 | 4 | 8 | 8 | 4 | 4 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| f-MWCNT-S1-7-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| f-MWCNT-S2-7-1 | 0 | 12.5 | 12.5 | 6.25 | 0 | 25 | 25 | 25 | 25 | 25 | 6.25 | 25 |
| f-MWCNT-S1-7-2 | 0 | 0 | 0 | 6.25 | 0 | 12.5 | 12.5 | 0 | 0 | 0 | 6.25 | 25 |
| f-MWCNT-S2-7-2 | 6.25 | 6.25 | 6.25 | <0.78 | 0 | 0 | 50 | 6.25 | 6.25 | 6.25 | 6.25 | 12.5 |
Fig. 4 and 5 show the images obtained in the experiments evaluating the antibacterial activity of f-MWCNT-S2-7-2 against MRSA (R4) and Salmonella typhi clinically isolated strains, respectively.
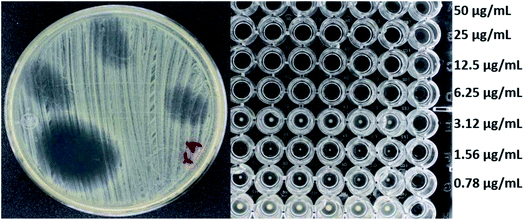 | ||
| Fig. 4 Antibacterial activity of f-MWCNT-S2-7-2 against R4 by radial diffusion (12 mm) and broth microdilution (MIC less than 6.25 μg mL−1). | ||
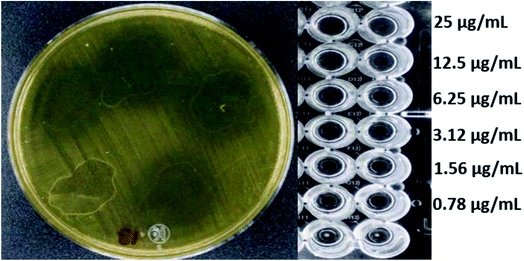 | ||
| Fig. 5 Antibacterial activity of f-MWCNT-S2-7-2 against Salmonella typhi clinically isolated by radial diffusion (14 mm) and by broth microdilution (MIC less than 0.78 μg mL−1). | ||
Noteworthy, based on the results obtained with Linezolid as control, the inhibition halos and MICs obtained with these nanomaterials are quite promising, considering the amount of organic material incorporated in the f-MWCNT-Sn-7-# is around 15% of the total weight. Specifically, f-MWCNT-S2-7-2 has 17.2% weight of organic material, which is approximately the fifth of linezolid amount, and even so, this material showed similar or higher activity against a more significant number of bacteria, indicating a high antimicrobial potential of these nanomaterials and highlighting the importance of the presence of bioactive molecules to display the antibacterial activity. In addition, the above results show that the length of the oligoethylene chain is decisive. The f-MWCNT-7 nanomaterial do not exhibit activity, and those with the shortest do not have significant antibacterial activity. On the other hand, nanomaterials with the longest oligoethylene chain also resulted inactive. Thus, it indicates that there is an optimal length that favors the antibacterial activity. An outstanding result is the activity observed against Gram-negative bacteria such Escherichia coli and Salmonella typhi, even higher than linezolid, which point out the incorporation of an amino-oligoethylene chain to linezolid structure facilitates its internalization through the double cell membrane characteristic of these bacteria, probably due to a higher lipophilic character and the quaternization of the terminal amino group developing a positive charge (Table S1†).
Both radial diffusion assay and broth microdilution test results suggest that the amino-oligoethylene linezolid analogs are released from the nanomaterial surface. A possible mechanism is the participation of exfoliate toxins or ETs, commonly excreted by Gram-positive bacteria such as S. aureus, which possess the catalytic triad of serine, histidine, and aspartic acid capable of breaking the amide bonds of several molecules, and cause the scalded skin syndrome or may be other types of proteases in bacteria (Scheme S2†).39–42
2.7 Molecular docking studies
In order to explain the differences in antibacterial activity of the evaluated nanomaterials at the molecular level, an automated molecular docking study was performed in the Phosphoryl Transferase Center (PTC) of E. coli, which is commonly used to study drug-receptor interactions that inhibit protein synthesis in bacteria.43,44 The structure of amino-linezolid 7, amino-oligoethylene linezolid analogs (Sn-7), and their protonated forms (Sn-7-H+) were modeled in the PTC.In the molecular modeling of 7, the six poses that maintain similarity with the linezolid pose at the PTC and have the lower score values (Table 4). Pose number six of 7 presented the amino group in the same orientation as the amide group of linezolid and an H⋯O hydrogen bond with G2505 and a second σ⋯π bond with G2061 (Fig. 6). However, the morpholine and oxazolidinone rings are spatially displaced compared to linezolid. Therefore, most poses with better score values did not show the same interactions or position as linezolid, directly affecting their interaction, conformation, and score (Fig. S44 to S49†). This behavior explains why the amino-linezolid 7, despite having a similar structure to linezolid, does not show activity in the antibacterial assays.
| Compounds (poses) | Score (kcal mol−1) | Conformational energy (kcal mol−1) | Placement enegy (kcal mol−1) |
|---|---|---|---|
| Linezolid | −7.21 | −3.11 | −70.35 |
| 7 (1) | −6.94 | 65.61 | −54.28 |
| 7 (2) | −6.67 | 66.10 | −50.64 |
| 7 (3) | −6.28 | 63.37 | −61.88 |
| 7 (4) | −6.00 | 63.86 | −43.98 |
| 7 (5) | −5.89 | 63.95 | −53.66 |
| 7 (6) | −5.86 | 62.84 | −44.71 |
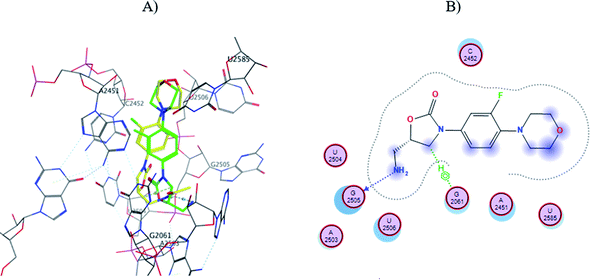 | ||
| Fig. 6 (A) Molecular docking of 7 (pose six, yellow) and linezolid (green) in the PTC of E. coli rRNA and (B) interactions map of 7. | ||
Table 5 shows the score and conformational and placement energies of the linezolid, Sn-7, Sn-7-H+, and amino-linezolid 7 (pose six), where lower or more negative score values indicate higher affinity or binding the site of action. Most of the modeling structures of Sn-7 and Sn-7-H+ (at physiological pH, the free amines can be partially protonated) presented a lower score than linezolid, indicating that they could present bacterial activity if only this parameter is taken into account (Table 5). However, there are significant differences in the conformation and placement energies, S1-7 and S2-7 in the structures having more positive values than linezolid. The structure of protonated analogs S1-7-H+ and S2-7-H+ present more negative values, even the placement energy of S2-7-H+ is more negative or favorable than that for linezolid. As can be seen, the S2-7-H+ structure has a more favorable score and placement energy than linezolid, which justifies the antimicrobial activity observed in the radial diffusion and broth microdilution assays. Interestingly, the S3-7 and S3-7-H+ structures also show thermodynamically favorable scores and placement energies but do not show activity in biological assays. This can be attributed to various factors, such as different intermolecular interactions involved in supramolecular complexes or the steric effect of the spacer chain length with higher units of oligoethylenes.
| Compounds | Score (kcal mol−1) | Conformational energy (kcal mol−1) | Placement enegy (kcal mol−1) |
|---|---|---|---|
| Linezolid | −7.21 | −3.11 | −70.35 |
| 7 | −5.86 | 62.84 | −44.71 |
| S1-7 | −7.52 | 8.24 | −36.95 |
| S1-7-H+ | −7.71 | 11.60 | −65.68 |
| S2-7 | −6.90 | 17.78 | −64.94 |
| S2-7-H+ | −8.55 | 21.58 | −83.14 |
| S3-7 | −7.92 | 24.84 | −73.47 |
| S3-7-H+ | −7.03 | 19.83 | −90.48 |
The S1-7-H+ and S2-7-H+ structures (yellow) presented the typical hydrogen bond interaction (Fig. 7 and 8) and a similar location to linezolid (green) in the PTC; an almost perfect overlap with the linezolid structure is observed, which is directly related to their favorable score values, while the S3-7-H+ structure does not present hydrogen bond interaction (Fig. 9) and the structure is shifted respect to the linezolid structure. The same is observed in the modeling of Sn-7 structures (Fig. S50 to S52†).
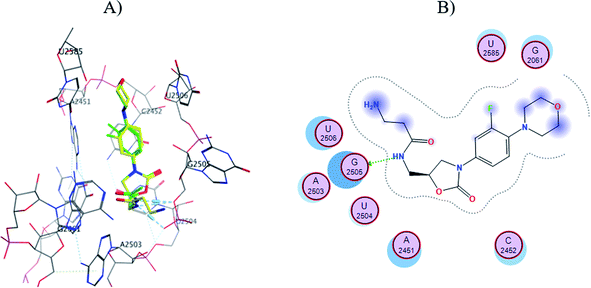 | ||
| Fig. 7 (A) Molecular docking of S1-7-H+ (yellow) and linezolid (green) in the PTC of E. coli rRNA and (B) interactions map of S1-7-H+. | ||
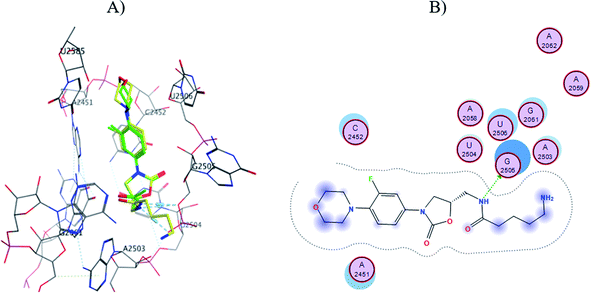 | ||
| Fig. 8 (A) Molecular docking of S2-7-H+ (yellow) and linezolid (green) in the PTC of E. coli rRNA and (B) interactions map of S2-7-H+. | ||
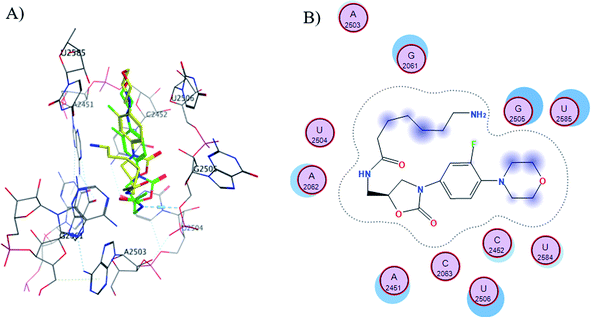 | ||
| Fig. 9 (A) Molecular docking of S3-7-H+ (yellow) and linezolid (green) in the PTC of E. coli rRNA and (B) interactions map of S3-7-H+. | ||
As described above, these results agree with the results obtained in the antibacterial activity studies against various bacterial strains. The higher the number of intermolecular interactions and similarity with the linezolid pose, the higher is the activity. In contrast, compounds with few or no interactions or with displaced poses respect linezolid does not present antibacterial activity. In summary, lower scores linezolid such as S3-7-H+ do not necessarily indicate favorable or good coupling since the placement of the molecule in the PTC directly influences the antibacterial activity.
3. Conclusions
In conclusion, different functionalized carbon nanotubes were synthesized, characterized, and evaluated against different bacterial strains. Some of these nanomaterials show excellent antibacterial activity because of they are covalently bounded to a bioactive linezolid-type molecule with a structure, conformation, and functional groups necessary to establish favorable interactions in the PTC. Antibacterial activity of f-MWCNT-Sn-7-# correlates with the molecular modeling predictions, where the docking of free Sn-7 and Sn-7-H+ structures suggest that antibacterial activity may be related to the oligoethylene (Sn) chain's length, a similar pose than linezolid, and the intermolecular interactions in PTC. These nanomaterials show a significant potency despite they only have a content of about 15% of organic material on the nanomaterial, and their activity is similar or even higher than the observed for linezolid in some of the strains tested. The most significant result is the outstanding activity of f-MWCNT-S2-7-2 against two Gram negative strains, Salmonella typhi and Escherichia coli. The proposed mechanism for releasing the bioactive molecules may be due to the cleavage of the amide bond of f-MWCNT-Sn-7-# by a protease. Moreover, the amino-oligoethylene chain in the structure of these linezolid analogues increases the activity against Gram-negative bacterial strains. These results are relevant considering that both bacterial strains may cause health problems to humans when they are present in food and water. Also give us the opportunity to explore the functionalization of new carbon nanomaterials. Currently, we are working in the functionalization of carbon dots, which have better cytocompatibility properties and dispersity in aqueous media.4. Experimental section
4.1 Materials
Reagents used in this research were obtained from commercial suppliers. The solvents used for the different reactions and studies are HPLC grade and anhydrous. The manipulation of compounds sensitive to moisture or oxygen were performed in the Ar atmosphere. The mixtures used for reactions at −78 °C were prepared into acetone and dry ice bath, while the corresponding at 0 °C was prepared into an ice bath. The organic fractions obtained from aqueous extractions were dried with anhydrous Na2SO4 in all cases. The solvents used in chromatography and extractions were commercially acquired with a high degree of purity. Purifications by column chromatography were performed using silica gel 60 (230–240 mesh) as stationary phase. Thin-layer chromatography was performed using silica gel plates 60 with F254 indicator on aluminum support, being revealed by exposure to UV light and into an iodine chamber.4.2 Characterization
The characterization of all organic compounds was performed by NMR spectroscopy (Bruker 400 MHz spectrometer). Chemical shift values (δ) are expressed in parts per million (ppm). Coupling constants (J) are reported in hertz (Hz) in all cases. Chloroform (CDCl3) or dimethyl sulfoxide-d6 (DMSO-d6) were used as solvent and tetramethylsilane (TMS) as internal reference. Fourier transform infrared spectroscopy (FTIR) spectra were obtained with an infrared spectrophotometer (PerkinElmer FT-IR Spectrum 400). Electron impact (EI) mass spectra were obtained with a mass spectrometer (Agilent, 5975C). The values are expressed in the form m/z (rel intensity).Functional group analysis on the surface of the MWCNT and derivates was performed by FTIR (PerkinElmer FT-IR Spectrum 400) evaluated in the range of 4000 to 700 cm−1 using KBr pellets. Raman spectra were evaluated with a 780 nm laser. Thermogravimetry analysis (TA Instruments Thermogravimetric Scale, model Q500) was carried out with a heating rate of 20 °C min−1 under air or N2 atmosphere. The morphology of MWCNTs and derivates was analyzed by Scanning Electron Microscopy (Tescan Vega 3) operating at 20 kV and transmission electron microscopy (TEM JEOL JEM-2010). The SEM and TEM samples were prepared to suspend MWCNTs in ethanol and depositing few drops on an aluminum strip and a carbon-coated copper grid on a sample holder, respectively.
4.3. Methods
4.3.1.1. Synthesis of 3-fluoro-4-morpholinylnitrobenzene (1). In a 250 mL round bottom flask a solution of morpholine (2 g, 22.95 mmol) in 100 mL of dry ethyl acetate was slowly added triethylamine (TEA) (16 mL, 114.75 mmol). Then, 3,4-difluoronitrobenzene (2.54 mL, 22.95 mmol) was added over 20 min via syringe. The solution was stirred for 72 h at RT. Solvent removal was accomplished by a rotary evaporator operating at vacuum (100-150 mbar). Crude products were purified by column chromatography over silica gel (25% ethyl acetate–75% petroleum ether) to give 4.67 g (90% yield) of 1. Yellow solid. FTIR: 3052, 2964, 2852, 1602, 1514, 1492, 1445, 1325, 1242, 1122 cm−1. 1H-NMR (CDCl3, 400 MHz): δ 7.99 (ddd, J = 9.0, 2.6, 1 Hz, 1H), 7.90 (dd, J = 13.1, 2.6 Hz, 1H), 6.91 (dd, J = 9, 8.8 Hz, 1H), 3.87 (m, 4H), 3.28 (m, 4H). 13C-NMR (CDCl3, 100 MHz): δ 153.3 (d, JC–F = 249.4 Hz), 145.6 (d, JC–F = 7.6 Hz), 141.0 (d, JC–F = 9.5 Hz), 121.1 (d, JC–F = 3.1 Hz), 117.0 (d, JC–F = 3.9 Hz), 112.7 (d, JC–F = 26.3 Hz), 66.7, 50.1 (d, JC–F = 5.0 Hz). MS (EI): m/z 226 (90, M+), 168 (100), 138 (20), 122 (10), 95 (10), 75 (5).
4.3.1.2. Synthesis of 3-fluoro-4-morpholinylaniline (2). In a 250 mL round bottom flask a solution of 1 (2 g, 8.85 mmol) and 10% Pd/C (0.018 g, 1.77 mmol) in 100 mL of dry methanol were stirred and the system was alternately evacuated and filled with Ar. Then H2 atmosphere was introduced via a balloon system and allowed to react for 4 h at RT. The mixture was filtered, and the solvent was evaporated. Finally, the crude obtained was purified by silica column chromatography (60% ethyl acetate–40% petroleum ether) to obtain 2 (1.18 g, 68% yield). Purple solid. FTIR: 3417, 3052, 2939, 1638, 1514, 1492, 1449, 1335, 1220, 1110 cm−1. 1H-NMR (CDCl3, 400 MHz): δ 6.79 (dd, J = 9.1, 8.6 Hz, 1H), 6.41 (m, 2H), 3.84 (m, 4H), 3.55 (sa, 2NH), 2.96 (m, 4H). 13C-NMR (CDCl3, 100 MHz): δ 156.9 (d, JC–F = 245.4 Hz), 143.0 (d, JC–F = 10.3 Hz), 131.9 (d, JC–F = 9.6 Hz), 120.3 (d, JC–F = 4.4 Hz), 110.6 (d, JC–F = 3.0 Hz), 103.9 (d, JC–F = 23.8 Hz), 67.1, 51.8 (d, JC–F = 2.5 Hz). MS (EI): m/z 196 (90, M+), 149 (15), 138 (100).
4.3.1.3. Synthesis N-carboethoxy-3-fluoro-4-morpholinylaniline (3). In a 100 mL round bottom flask a solution of 2 (0.2 g, 1.02 mmol) in 30 mL of dry THF under Ar atmosphere and TEA (0.71 mL, 5.1 mmol) was added dropwise over 15 min at 0 °C. Then, ethyl chloroformate (0.107 mL, 1.12 mmol) was added dropwise over 15 min. The mixture was stirred and kept into an ice bath at 0 °C for 4 h. The reaction mixture was treated with water (150 mL) to precipitate the product, the solid was filtered and allowed to dry in an oven at 70 °C for 4 h to obtain 3 (0.232 g, 85% yield). Brown crystal solid. FTIR: 3324, 2995, 1692, 1115 cm−1. 1H-NMR (CDCl3, 400 MHz): δ 7.27 (d, J = 12.0 Hz, 1H), 6.97 (dd, J = 8.6, 1.6 Hz, 1H), 6.87 (dd, J = 9.0, 8.8 Hz, 1H), 6.51 (s, 1NH), 4.22 (q, J = 7.1 Hz, 2H), 3.86 (m, 4H), 3.03 (m, 4H), 1.30 (t, J = 7.1 Hz). 13C-NMR (CDCl3, 100 MHz): δ 155.7 (d, JC–F = 245.7 Hz), 153.7, 135.7 (d, JC–F = 9.1 Hz), 133.5 (d, JC–F = 10.9 Hz), 119.2 (d, JC–F = 4.1 Hz), 114.6 (d, JC–F = 9.3 Hz), 108.0 (d, JC–F = 33.0 Hz), 67.2, 61.5, 51.3 (d, JC–F = 3.0 Hz), 14.7. MS (EI): m/z 268 (100, M+) 240 (10), 222 (15), 210 (60), 195 (15), 182 (20), 164 (20), 137 (30).
4.3.1.4. Synthesis (R)-[N-3-(3-fluoro-4-morpholinylphenyl)-2-oxo-5-oxazolidinyl]methanol (4). In a 50 mL round bottom flask a solution of 3 (0.2 g, 0.75 mmol) was dissolved in 25 mL of dry THF under Ar atmosphere at −78 °C and n-BuLi was added dropwise (0.33 mL, 0.82 mmol) over 15 min. Then, glycidyl butyrate (0.155 mL, 0.82 mmol) was added dropwise over 15 min via syringe. After 1 h at −78 °C the reaction was kept at RT for 12 h. Finally, the reaction was then quenched with saturated ammonium chloride solution and 20 mL of water were added, the organic phase was extracted with ethyl acetate (3 × 50 mL). Solvent was removed under vacuum and the crude product was purified by column chromatography (80% ethyl acetate-20% petroleum ether) to obtain 4 (0.11 g, 50% yield). Brown solid. FTIR: 3404, 3246, 2933, 2840, 1732, 1514, 1451, 1418, 1227 cm−1. 1H-NMR (400 MHz, CDCl3): δ 7.43 (dd, J = 14.4, 2.6 Hz, 1H), 7.11 (ddd, J = 9.0, 2.6, 1.2 Hz, 1H), 6.91 (t, J = 9.0 Hz, 1H), 4.72 (m, 1H), 3.96 (m, 3H), 3.86 (m, 4H), 3.74 (m, 1H), 3.03 (m, 4H), 2.96 (m, 1H). 13C-NMR (CDCl3, 100 MHz): δ 155.5 (d, JC–F = 246.1 Hz), 154.8, 136.4 (d, JC–F = 9.1 Hz), 133.2 (d, JC–F = 10.4 Hz), 118.9 (d, JC–F = 4.3 Hz), 114.0 (d, JC–F = 3.3 Hz), 107.5 (d, JC–F = 26.4 Hz), 72.95, 67.0, 62.7, 51.0 (d, JC–F = 3.0 Hz), 46.4. MS (EI): m/z 296 (100, M+) 238 (50), 164 (13), 149 (40).
4.3.1.5. Synthesis (R)-[N-3-(3-fluoro-4-morpholinylphenyl)-2-oxo-5-oxazolidinyl]methyl methanesulfonate (5). In a 50 mL round bottom flask a solution of 4 (0.1 g, 0.338 mmol) in 25 mL of dry DCM under Ar atmosphere and TEA (0.1 mL, 0.67 mmol) was added dropwise. Then, methanesulfonyl chloride (0.04 mL, 0.47 mmol) was added slowly to the solution over 15 min and the reaction was kept at 0 °C for 40 min. The mixture obtained was treated with water (150 mL) and 150 mL of DCM were added, and the organic phase was extracted with DCM (2 × 150 mL). The combined organic portions were dried with MgSO4 and evaporated to obtain 5 (0.124 g, 98% yield). White solid. 1H-NMR (400 MHz, DMSO-d6): δ 7.50 (dd, J = 15.0, 2.4 Hz, 1H), 7.21 (dd, J = 9.1, 2.4 Hz, 1H), 7.07 (t, J = 9.1 Hz, 1H), 4.99 (m, 1H), 4.48 (m, 2H), 4.16 (t, J = 9.3 Hz, 1H), 3.80 (dd, J = 9.3, 6.2 Hz, 1H), 3.78 (m, 4H), 3.24 (s, 3H), 2.97 (m, 4H).
4.3.1.6. Synthesis (R)-[N-3-(3-fluoro-4-morpholinylphenyl)-2-oxo-5-oxazolidinyl]methyl azide (6). In a 50 mL round bottom flask a solution of 5 (0.1 g, 0.267 mmol) in 25 mL of DMF was treated with sodium azide (0.07 g, 1.069 mmol) under Ar atmosphere and it was heated at 85 °C for 16 h. The obtained product was extracted with 50 mL of a saturated aqueous solution of sodium chloride and DCM (3 × 50 mL). The combined organic portions were dried with MgSO4 and evaporated in a vacuum oven to remove all possible solvent to obtain 6 (0.021 g, 25% yield). Bright red solid. 1H-NMR (400 MHz, CDCl3): δ 7.44 (dd, J = 14.4, 2.6 Hz, 1H), 7.13 (ddd, J = 9.0, 2.6, 1.2 Hz, 1H), 6.93 (t, J = 9.0 Hz, 1H), 4.87 (m, 1H), 4.12 (t, J = 9.0 Hz, 1H), 3.90 (dd, J = 9.0, 5.5 Hz, 1H), 3.62 (m, 4H), 3.77 (d, J = 5.5 Hz, 2H), 3.01 (m, 4H). 13C-NMR (CDCl3, 100 MHz): δ 155.5 (d, JC–F = 246.5 Hz), 153.8, 136.6 (d, JC–F = 9.0 Hz), 132.9 (d, JC–F = 10.5 Hz), 118.9 (d, JC–F = 4.2 Hz), 114.0 (d, JC–F = 3.4 Hz), 107.6 (d, JC–F = 26.3 Hz), 70.9, 66.9, 51.0 (d, JC–F = 3.0 Hz), 48.2, 44.6. MS (EI): m/z 321 (100, M+) 235 (5), 209 (10), 191 (35), 151 (11), 150 (30), 149 (25).
4.3.1.7. Synthesis (R)-[N-3-(3-fluoro-4-morpholinylphenyl)-2-oxo-5- oxazolidinyl] methanamine (7). In 50 mL round-bottom flask a mixture of 6 (0.02 g, 0.061 mmol) and 10% Pd/C (0.06 g, 0.563 mmol) in 15 mL of dry methanol was stirred, and the system was alternately evacuated and filled with Ar. Then H2 atmosphere was introduced via a balloon system and allowed to react for 24 h at RT. The reaction was evacuated and more H2 atmosphere was added and kept for a further 12 h at RT. The reaction mixture was filtered and washed with methanol (3 × 20 mL), and the solvent was evaporated to obtain 7 (0.005 g, 28% yield). White solid. 1H-NMR (400 MHz, CDCl3): δ 7.36 (dd, J = 14.4, 2.6 Hz, 1H), 7.00 (ddd, J = 9.0, 2.6, 1.1 Hz, 1H), 6.84 (t, J = 9.0 Hz, 1H), 4.86 (m, 1H), 4.07 (t, J = 9.0 Hz, 1H), 3.81 (dd, J = 9.0, 5.8 Hz, 1H), 3.73 (m, 6H), 3.01 (m, 4H). NMR-13C (100 MHz, DMSO-d6): δ 154.6 (d, JC–F = 243.9 Hz), 154.0, 135.5 (d, JC–F = 8.9 Hz), 133.4 (d, JC–F = 10.5 Hz), 119.2 (d, JC–F = 4.3 Hz), 114.2 (d, JC–F = 3.1 Hz), 106.7 (d, JC–F = 26.1 Hz), 72.0, 66.1, 50.7 (d, JC–F = 3.0 Hz), 47.2, 43.0. MS (EI): m/z 321 (100, M+) 235 (5), 209 (22), 164 (13), 151 (21), 150 (9), 149 (6).
4.4 Synthesis of multiple-walled carbon nanotubes
4.5 Antibacterial activity of MWCNTs
Antibacterial activity of the nanomaterials was evaluated by a radial diffusion assay and the Mueller Hinton broth microdilution assay.For the assay, 96-well U-bottom ELISA type plates with lids were used and filled as follows. 50 μL of the nanomaterial suspension at a concentration twice the concentration to be evaluated (0.78 to 50 μg mL−1) were added in all columns of rows A to G. In comparison, 50 μL of the solvent used in the NTC dispersions were added in all wells of row H. Subsequently, 50 μL of the inoculum of the bacterial strain was added in all rows in columns 2 to 12, while 50 μL of Mueller Hinton broth was added in column 1. Column 1 was used as blank since it contained no inoculum, and row H was used as growth control because it did not contain the nanomaterial to be evaluated. Once the plates were filled, they were covered and sealed with parafilm and incubated at 37 °C for 18–20 h. Finally, the plates were read. The presence of turbidity and/or a bacterial growth button at the bottom of the wells indicated bacterial growth, while the absence of this indicated antibacterial activity of the nanomaterial evaluated. The antibacterial experiments were performed twice by triplicate.
4.6 Automated molecular docking studies
Automated molecular docking studies were performed with the crystallographic structure of ribosomal RNA (rRNA) from E. coli in the MOE 2018 program. The crystal structure of ribosomal rRNA was obtained from the Protein Data Bank (PDB) for docking with the compounds using PDB structure ID: 4V4Q. All ligands (e.g., magnesium ion, water molecules, and ribosomal proteins) and a repeat strand of ribosomal RNA were removed to prepare the crystallographic structure. Once the E. coli ribosomal RNA structure was arranged, the geometry of the models was optimized with the QuickPrep application implemented in MOE that adds hydrogen atoms and sets the protonation states, fixes the receptor atoms away from the ligand and leaves the receptor atoms close to the ligand.In docking of linezolid with MOE, the interaction indicated by the program (hydrogen bonding) between the hydrogen of the linezolid amino group and the 5′ oxygen of the phosphate group of residue G2505 of ribosomal RNA (rRNA) was observed. In this docking, the score was good, with a value of −7.21. A second linezolid docking was also performed in PTC of the same E. coli rRNA fragment, but now with the LEDOCK 2018 program, obtaining a pose similar to that obtained by MOE.
Finally, the score, conformational energy and placement energy values for structures 7, Sn-7 and Sn-7-H+ were determined. These values were assigned by the program after several calculations and were related to the coupling quality, where lower scores (more negative values) indicate more favorable couplings. Finally, the structures with the best values were selected and displayed.
Author contributions
José A. Alatorre-Barajas – investigation, methodology, formal analysis, writing – original draft. Eleazar Alcántar-Zavala – investigation, methodology. M. Graciela Gil-Rivas – investigation, methodology. Edgar Estrada-Zaval – investigation, methodology. Adrián Ochoa-Terán – conceptualization, formal analysis, funding acquisition, writing – review & editing. Y. Gochi-Ponce – conceptualization, funding acquisition, writing – review & editing. Julio Montes-Ávila – resources, formal analysis. Alberto Cabrera – investigation, methodology. Balter Trujillo-Navarrete – resources, formal analysis, writing – review & editing. Yazmin Yorely Rivera-Lugo – investigation, methodology. Gabriel Alonso-Núñez – resources, formal analysis. Edgar A. Reynoso-Soto – resources. J. L. Medina-Franco – resources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors thank to Tecnológico Nacional de México (Grant 5366.19-P and 10716.21-P) and Consejo Nacional de Ciencia y Tecnología (CONACYT) for the financial support through the Research Grant SALUD-2015-1-261324, ITT NMR facilities Grant INFR-2011-3-173395 and Scholarship Program, which supported to José A. Alatorre Barajas and Eleazar Alcántar-Zavala for their graduate studies. The docking software MOE was support by Instituto de Investigaciones Biomédicas at the UNAM, through the programs “Novel alternatives for the Treatment of Infection Deseases (NUATEI-IIB-UNAM)” and “Apoyo a la Investigación y el Posgrado (PAIP)” Grant 5000-9163 in the Chemistry Faculty of the UNAM.References
- A. Despotovic, B. Milosevic, I. Milosevic, N. Mitrovic, A. Cirkovic, S. Jovanovic and G. Stevanovic, Am. J. Infect. Control, 2020, 48, 1211–1215 CrossRef.
- Y. Li, L. Ren and J. Zou, Can. J. Infect. Dis. Med. Microbiol., 2019, 2019, 6417959, DOI:10.1155/2019/6417959.
- N. Manchal, M. R. S. Mohamed, M. Ting, H. Luetchford, F. Francis, J. Carrucan and R. Norton, Infect. Dis. Heal., 2020, 25, 175–180 CrossRef PubMed.
- V. T. Anju, P. Paramanantham, S. L. Sb, A. Sharan, M. H. Alsaedi, T. M. S. Dawoud, S. Asad and S. Busi, Photodiagnosis Photodyn. Ther., 2018, 24, 300–310 CrossRef PubMed.
- S. J. Brickner, M. R. Barbachyn, D. K. Hutchinson and P. R. Manninen, J. Med. Chem., 2008, 51, 1981–1990 CrossRef CAS PubMed.
- H. Houri, H. Kazemian, H. Sedigh Ebrahim-Saraie, A. Taji, Z. Tayebi and H. Heidari, J. Glob. Antimicrob. Resist., 2017, 10, 200–203 CrossRef PubMed.
- B. M. Valle-Ramírez, Master thesis, Instituto Tecnologico de Tijuana-Centro de Graduados e Investigación, 2017.
- G. Mittal, V. Bhandari, R. Gaind, V. Rani, S. Chopra, R. Dawar, R. Sardana and P. K. Verma, BMC Infect. Dis., 2019, 19, 717, DOI:10.1186/s12879-019-4368-6.
- F. Román, C. Roldán, P. Trincado, C. Ballesteros, C. Carazo and A. Videl, Antimicrob. Agents Chemother., 2013, 57, 2428–2429 CrossRef PubMed.
- J. Feng, D. S. Billal, A. Lupien, G. Racine, E. Winstall, D. Légaré, P. Leprohon and M. Ouellette, J. Proteome Res., 2011, 10, 4439–4452 CrossRef CAS.
- R. Bi, T. Qin, W. Fan, P. Ma and B. Gu, J. Glob. Antimicrob. Re., 2018, 13, 11–19 CrossRef PubMed.
- J. Simon, E. Flahaut and M. Golzio, Materials, 2019, 12, 624, DOI:10.3390/ma12040624.
- C. J. Serpell, K. Kostarelos and B. G. Davis, ACS Cent. Sci., 2016, 2, 190–200 CrossRef CAS.
- H. He, L. A. Pham-Huy, P. Dramou, D. Xiao and P. Zuo, Biomed Res. Int., 2013, 2013, 578290, DOI:10.1155/2013/578290.
- H. Zare-Zardini, M. Davarpanah, M. Shanbedi, A. Amiri, M. Maghrebi and L. Ebrahimi, J. Biomed. Mater. Res. A, 2014, 6, 1774–1781 CrossRef.
- K. Chen and S. Mitra, Colloid. Surface. B, 2019, 173, 386–391 CrossRef CAS PubMed.
- A. William, in Fullerens, Graphenes and Nanotubes: A Pharmaceutical Approach, ed. A.M. Grumezescu, Elsevier Inc., 1st edn, 2018, vol. 1, ch. 2, pp. 31–65 Search PubMed.
- S. Merum, J. B. Veluru and R. Seeram, Mater. Sci. Eng. B Solid-State Mater. Adv. Technol., 2017, 223, 43–63 CrossRef CAS.
- R. Alshehri, A. M. Ilyas, A. Hasan, A. Arnaout, F. Ahmed and A. Memic, J. Med. Chem., 2016, 59, 8149–8167 CrossRef CAS PubMed.
- M. Sireesha, V. J. Babu and S. Ramakrishna, Mater. Sci. Eng. B-Adv., 2017, 223, 43–63 CrossRef.
- R. Alshehri, A. M. Ilyas, A. Hasan, A. Arnaout, F. Ahmed and A. Memic, J. Med. Chem., 2016, 59, 8149–8167 CrossRef CAS.
- S. Sharmeen, A. F. M. M. Rahman, M. M. Lubna, K. S. Salem, R. Islam and M. A. Khan, Bioact. Mater., 2018, 3, 236–244 CrossRef.
- A. Takagi, A. Hirose, T. Nishimura, N. Fukumori, A. Ogata, N. Ohashi, S. Kitajima and J. Kanno, J. Toxicol. Sci., 2008, 33, 105–116 CrossRef CAS PubMed.
- D. Bains, G. Singh, J. Bhinder, P. K. Agnihotri and N. Singh, ACS Appl. Bio. Mater., 2020, 3, 2092–2103 CrossRef CAS.
- D. Zhang, M. A. Kandadai, J. Cech, S. Roth and S. A. Curran, J. Phys. Chem., 2006, 110, 12910–12915 CrossRef CAS PubMed.
- C. Liu, H. Shi, H. Yang, S. Yan, S. Luan, Y. Li, M. Teng, A. F. Khan and J. Yin, Mat. Design, 2017, 120, 128–134 CrossRef CAS.
- G. Guzel Kaya, S. Medaglia, V. Candela-Noguera, M. A. Tormo-Mas, M. D. Marcos, E. Aznar, H. Deveci and R. Martínez-Máñez, Pharmaceutics, 2020, 12, 1126, DOI:10.3390/pharmaceutics12111126.
- E. J. Antony, A. Shibu, S. Ramasamy, M. S. Paulraj and I. V. M. V. Enoch, Mat. Sci. Eng. C, 2016, 65, 194–198 CrossRef CAS PubMed.
- F. E. Akram, T. El-Tayeb, K. Abou-Aisha and M. El-Azizi, Ann. Clin. Microbiol. Antimicrob., 2016, 15, 48, DOI:10.1186/s12941-016-0164-y.
- H.-W. Han, K. D. Patel, J.-H. Kwak, S.-K. Jun, T.-S. Jang, S.-H. Lee, J. Campbell Knowles, H.-W. Kim, H.-H. Lee and J.-H. Lee, Biomolecules, 2021, 11, 1028, DOI:10.3390/biom11071028.
- S. Makled, N. Boraie and N. Nafee, Drug Deliv. and Transl. Res., 2021, 11, 1037–1055 CrossRef CAS.
- P. Guo, B. A. Buttaro, H. Y. Xue, N. T. Tran and H. L. Wong, Eur. J. Pharm. Biopharm., 2020, 151, 189–198 CrossRef CAS.
- K. D. Patil, S. B. Bagade and S. C. Bonde, Materials Technology, 2020 DOI:10.1080/10667857.2020.1816021.
- S. Shah, D. Christipher, S. Sharma, M. Soniwala and J. Chavda, J. Drug Deliv. Sci. Technol., 2020, 60, 102013, DOI:10.1016/j.jddst.2020.102156.
- M. H. Teaima, M. K. Elasaly, S. A. Omar, M. A. El-Nabarawi and K. R. Shoueir, Saudi Pharm. J., 2020, 28, 859–868 CrossRef CAS PubMed.
- O. I. Parisi, M. Fiorillo, A. Caruso, A. R. Cappello, C. Saturnino, F. Puoci, A. Panno, V. Dolce, H. El-Kashef and M. S. Sinicropi, Int. J. Pharm., 2014, 461, 163–170 CrossRef CAS PubMed.
- F. De Maio, V. Palmieri, G. Santarelli, G. Perini, A. Salustri, I. Palucci, M. Sali, J. Gervasoni, A. Primiano, G. Ciasca, M. Sanguinetti, M. De Spirito, G. Delogu and M. Papi, Nanomaterials, 2020, 10, 1431, DOI:10.3390/nano10081431.
- B. Moreno-Valle, J. A. Alatorre-Barajas, Y. Gochi-Ponce, E. Alcántar-Zavala, Y. Y. Rivera-Lugo, J. Montes-Ávila, B. Trujillo-Navarrete, G. Alonso-Núñez, E. A. Reynoso-Soto and A. Ochoa-Terán, J. Nanoparticle Res., 2020, 22, 315, DOI:10.1007/s11051-020-05044-w.
- M. Bukowski, B. Wladyka and G. Dubin, Toxins, 2010, 2, 1148–1165 CrossRef CAS PubMed.
- N. Stach, A. Karim, P. Golik, R. Kitel, K. Pustelny, N. Gruba, K. Groborz, U. Jankowska, S. Kedracka-Krok, B. Wladyka, M. Drag, A. Lesner and G. Dubin, Int. J. M. Sci., 2021, 22, 2220, DOI:10.3390/ijms22042220.
- J. Feng, S. Michalik, A. N. Varming, J. H. Andersen, D. Albrecht, L. Jelsbak, S. Krieger, K. Ohlsen, M. Hecker, U. Gerth, H. Ingmer and D. Frees, J. Proteome Res., 2013, 12, 547–558 CrossRef CAS PubMed.
- L. Shaw, E. Golonka, J. Potempa and S. J. Foster, Microbiology, 2004, 150, 217–228 CrossRef CAS PubMed.
- E. Alcántar-Zavala, E. Hernández-Guevara, A. Ochoa-Terán, J. Montes-Ávila, E. A. Estrada-Zavala, A. J. Salazar-Medina, E. Alday, A. Cabrera, G. Aguirre, V. Miranda-Soto, C. Velazquez, S. P. Díaz-Camacho and J. L. Medina-Franco, Bioorg. Chem., 2020, 105, 104359, DOI:10.1016/j.bioorg.2020.104359.
- G. D. García-Olaiz, E. Alcántar-Zavala, A. Ochoa-Terán, A. Cabrera, R. Muñiz-Salazar, J. Montes-Ávila, A. J. Salazar-Medina, E. Alday, C. Velazquez, J. L. Medina-Franco and R. Laniado-Laborín, Bioorg. Chem., 2020, 95, 103483, DOI:10.1016/j.bioorg.2019.103483.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04691h |
| This journal is © The Royal Society of Chemistry 2021 |