Open Access Article
Open Access ArticleTransformation of Pueraria candollei var. mirifica phytoestrogens using immobilized and free β-glucosidase, a technique for enhancing estrogenic activity†
Fonthip Makkliang a,
Wipawee Juengsanguanpornsuk
a,
Wipawee Juengsanguanpornsuk b,
Suppalak Phaisan
b,
Suppalak Phaisan c,
Attapon Sakdamas
c,
Attapon Sakdamas c,
Waraporn Putalun
c,
Waraporn Putalun b,
Seiichi Sakamoto
b,
Seiichi Sakamoto d and
Gorawit Yusakul
d and
Gorawit Yusakul *ce
*ce
aSchool of Languages and General Education, Walailak University, Nakhon Si Thammarat, Thailand
bFaculty of Pharmaceutical Sciences, Khon Kaen University, Khon Kaen, Thailand
cSchool of Pharmacy, Walailak University, Nakhon Si Thammarat, Thailand. E-mail: gorawit.yu@mail.wu.ac.th; gorawit.yu@wu.ac.th; Tel: +66 75-67-2839
dGraduate School of Pharmaceutical Sciences, Kyushu University, Higashi-ku, Fukuoka, Japan
eBiomass and Oil Palm Center of Excellence, Walailak University, Nakhon Si Thammarat, Thailand
First published on 29th September 2021
Abstract
Pueraria candollei var. mirifica (PM) has a significant beneficial effect on postmenopausal symptoms associated with estrogen deficiency. However, the estrogenic activity and intestinal absorption of isoflavonoid glycosides derived from PM, such as daidzin and genistin, are significantly lower than those of their aglycones. To enhance the estrogenic activity of the PM extract, we developed β-glucosidase and its immobilized form to increase the PM aglycone content (daidzein and genistein). The enzyme immobilization was done by alginate beads, and the resulting β-glucosidase alginate beads have a diameter of about 0.20 cm. Response surface methodology (RSM) was used to optimize certain parameters, such as the pH, temperature, and ethanol concentration. The optimal conditions of β-glucosidase for daidzein and genistein production were pH of 4.8–4.9, a temperature in the range 46.3–49.1 °C, and ethanol concentration of 10.0–11.0%. The ANOVA results indicated that the design experiment involving free and immobilized β-glucosidase was the best fit by quadratic models, which had adjusted R2 values between 0.8625 and 0.9318. Immobilized β-glucosidase can be reused up to nine times and maintained efficacy of greater than 90%. Treatment of the PM extract with β-glucosidase increased the estrogenic activity of the PM extract by 8.71- to 23.2-fold compared to that of the untreated extract. Thus, β-glucosidase has a high potential for enhancing the estrogenic activity of PM constituents, and it can be applied on an industrial scale to increase the utility of these natural products.
1. Introduction
Pueraria candollei var. mirifica (Airy Shaw & Suvat.) Niyomdham (PM) is a medicinal plant containing phytoestrogens that can be used against vasomotor symptoms.1 PM phytochemicals potentially protect against bone loss, act as antioxidants, and provide neurodegeneration protection.1 PM phytoestrogens have been divided into three groups: (i) isoflavonoids [kwakhurin, puerarin (PUE), daidzin (DZ), genistin (GT), daidzein (DZe), and genistein (GTe)], (ii) chromenes [miroestrol (MI), isomiroestrol, deoxymiroestrol (DMI), and methoxyisomiroestrol], and (iii) coumestans (coumestrol).1 DMI was identified as the most estrogenic constituent, and it is more estrogenic than MI and isomiroestrol.2 In raw materials, the glycoside isoflavones of PM phytoestrogens were found to be more abundant.3 When isoflavonoid glycosides are administered, they are hydrolyzed prior to absorption by β-glucosidase produced by the intestinal flora. In the human small intestine, aglycone products are readily absorbed and metabolized because their aglycones exhibit higher hydrophobic properties than glycosides.4,5 The presence of intestinal floras that secrete β-glucosidase varies among individuals. As a result, β-glucosidase-mediated conversion of isoflavone may be unpredictable.β-Glucosidase (β-D-glucoside glycohydrolases, EC 3.2.1.21) is a glycosyl hydrolase that belongs to the family of glycosyl hydrolases.6 It is frequently found in a variety of organisms, including microorganisms, plants, fungi, and bacteria. In the human intestine, β-glucosidase is secreted by Bifidobacterium spp. and Lactobacillus spp.7,8 Studies have revealed that β-glucosidase plays an essential role in efficiently increasing the concentration of isoflavone aglycones.9,10 β-Glucosidase may enhance the estrogenic activity of PM by converting glycosides to aglycone isoflavones, specifically DZ and GT, to DZe and GTe. The effect of PM phytoestrogen transformation on estrogenic activity and absorption has not been evaluated. Fermentation of soy isoflavonoids with Lactobacillus paracasei significantly increases DZe and GTe bioavailability.11 Additionally, DZe is capable of being metabolized to the more potent equol.12 The production of equol has a direct effect on preventing bone loss and fat accumulation as well as relieving menopausal symptoms.13,14 The biotransformation of DZ to DZe is the first step in the production of equol. However, less than half of the population is capable of producing equol endogenously,15 and Bifidobacterium, which produces β-glucosidase, is less prevalent in the elderly, particularly those with certain diseases.16 The DZe and GTe enrichment of isoflavone provides relief from menopausal symptoms and beneficial effects on C-reactive protein concentrations.17,18 The clinical outcomes for menopausal symptoms may be improved consistently among subjects because the transformation of isoflavone glycosides to the aglycone form leads to improved bioavailability.19
Free β-glucosidases are powerful and beneficial enzymes for biotransformation. However, the applications of free enzymes are limited because of their low stability, high cost, and non-reusability. The immobilized enzymes have a number of advantages over their free-β-glucosidase counterparts, including increased enzyme stability over a broad temperature and pH range, reusability, ease of separation from the reaction mixture, and applicability in a variety of systems.20 Enzyme immobilization may be classified into three types: covalent binding, adsorption on the support, and entrapment. Covalent binding and adsorption on the support have some restrictions, which may influence the binding size of the enzyme and reduce the enzyme activity.21 Entrapment is one of the best methods for immobilization, since this technique involves little enzyme loss and is not affected by the structure and chemistry of the enzyme.22 The present research focuses on the entrapment of enzymes on calcium alginate beads. This method involves simple preparation, is non-toxic (biocompatible), non-reactive with the enzyme, and affordable.23,24 In addition, calcium alginate is easily prepared in bead form and has a high surface area with greater porosity to diffuse the substrate and product.25–27 Previous studies have not focused on the effect of β-glucosidase treatment on the chemical constituents and estrogenic activity of PM phytoestrogens. In the present study, we developed and optimized both β-glucosidase and alginate-based immobilization of β-glucosidase for the production of the PM aglycone isoflavones DZe and GTe.
β-Glucosidase activity and stability are influenced by environmental conditions, such as temperature, pH, and ethanol. However, the ethanol concentration is essential to keep the substrates solubilized. Response surface methodology (RSM) was applied using a central composite design (CCD), and the pH, temperature, and ethanol concentration were optimized to achieve the best activity of β-glucosidase for the production of PM aglycone isoflavones. Increased temperature and ethanol levels have a combined detrimental effect on enzyme activity and stability,28 therefore, the interaction between these factors is expected. The optimal temperature has a considerable influence on the pH range in which the enzyme mixture exhibits high activity.29 RSM is the method of choice for addressing these interactions. Finally, we evaluated the estrogenic activity of the resulting PM extracts using the MCF-7 cell model. The process was shown to increase the estrogenic activity of the PM extract significantly and could be applied in the botanical extract industry.
2. Materials and methods
2.1. Chemicals and reagents
p-Nitrophenol (≥99%), p-nitrophenyl-β-D-glucopyranoside (p-NPG) (98%), and daidzin (DZ) were purchased from Sigma-Aldrich, Inc. (MO, USA). Daidzein (DZe), puerarin (PUE), and genistein (GTe) were purchased from LKT Laboratories Inc. (MN, USA). Genistin (GT, 99%) was from Fujicco (Tokyo, Japan). Calcium chloride dihydrate (CaCl2·2H2O, 97%) was purchased from Rankem (RFCL Limited, New Delhi, India). Sodium alginate was purchased from Loba Chemie Pvt Ltd. (91%, Mumbai, India). β-Glucosidase of Trichoderma reesei was purchased from Xian Lyphar Biotech Co., Ltd. (Shaanxi, China). We isolated miroestrol (MI) and deoxymiroestrol (DMI) from PM roots.302.2. Immobilization of β-glucosidase
Sodium alginate solution (3.8% w/v) was prepared by dissolving sodium alginate powder in hot ultrapure water and then stirring at a rate of 500 rpm. The sodium alginate solution and β-glucosidase were mixed in various proportions to obtain a homogeneous solution containing 2000 U mL−1 β-glucosidase in a final sodium alginate concentration of 2.0–4.0% (w/v). Subsequently, drops of this mixture solution were produced with the tip of the transfer pipette into a 0.20 M CaCl2 solution (20 mL) under continuous stirring at 500 rpm for two hours. Finally, these alginate beads were washed with ultrapure water and kept at 4 °C until use. An equation was used to determine the immobilization yield of β-glucosidase. Aadd denotes the enzyme's initial activity, while Afree denotes the enzyme's remaining free activity following immobilization. The ESI (ESI†, Analysis of β-glucosidase activity) details the procedure for determining enzyme activity.A scanning electron microscope (SEM) was used to examine the surface morphology of alginate beads, as described in the ESI† (Analysis of the surface morphology using a scanning electron microscope).
2.3. Reaction between β-glucosidase and an extract of Pueraria candollei var. mirifica
Free β-glucosidase and immobilized β-glucosidase were reacted with isoflavonoids of the PM extract, which was prepared by macerating 500 g of dry PM powder in 2.5 L of 80% EtOH for two days at room temperature. The extract was collected and then concentrated using a rotary evaporator and a lyophilizer under vacuum, and then 37.5 g of the product was obtained. The powdered PM extract (10 g) was dissolved in 20% EtOH (100 mL). The clear extract was collected and kept at −20 °C. Previous research established that 20% EtOH extracts could recover between 89–99% of the phytoestrogens found in PM.31 The PM isoflavonoids were reacted with immobilized or free β-glucosidase on alginate beads. A 2.5 mL solution of PM extract was mixed with an equal volume of 50 mM sodium citrate phosphate buffer (pH 5) containing β-glucosidase. The reaction mixture was shaken in a temperate incubator. At specific time intervals, the sample solution (100 μL) was collected. Next, alkaline ethanol (300 μL of 50 mM sodium carbonate buffer pH 9.6 and 200 μL of EtOH) was added to the sample solution to terminate the reaction. Finally, this solution was injected into an HPLC-UV system for isoflavone analysis (ESI,† Instrumental and high-performance liquid chromatography for isoflavonoid analysis). Then, the appropriate enzyme amount was selected for optimization of the extraction parameters, including the solution pH, temperature, and EtOH concentration.2.4. Response surface methodology (RSM)
Response surface methodology (RSM) is an effective statistical technique for experimental design and optimization. This technique is used to examine the responses (dependent variables) as a result of the interaction of independent variables. Based on the optimization of single parameters (pH, temperature, and EtOH concentration of the reaction mixture), the optimal ranges of the mentioned parameters were selected for RSM. The central composition design (CCD) method was used in this study to optimize the interaction between the independent variables pH (3, 5, and 7, X1), temperature (30, 50, and 70 °C, X2), and EtOH concentration (5, 10, and 15% v/v, X3). The DZe (Y1) and GTe (Y2) responses were obtained using immobilized β-glucosidase, whereas the DZe (Y3) and GTe (Y4) responses were obtained using free β-glucosidase. The effects of independent factors on the responses were randomized and fitted to the appropriate model. Design-Expert® software was used to perform the statistical analyses. As illustrated in Table S1,† the CCD is composed of axial points (−1.68 and +1.68), center points (0), and factorial points (−1 and +1). Twenty sets of experiments with six center points were conducted in this study, as shown in Table S2.†2.5. Evaluation of estrogenic activity by an MCF-7 proliferation assay
After optimizing the conditions using RSM, the PM extract was prepared and analyzed for isoflavonoid, MI, and DMI contents. Then, estrogenic activity was compared between the PM extracts and extracts treated with β-glucosidase. For the control sample, a solution of PM extract (25 mL) was mixed with an equal volume of 50 mM sodium citrate phosphate buffer pH 5 and incubated. In the case of enzymatic treatment, one hundred beads of immobilized β-glucosidase or 0.2 U mL−1 free β-glucosidase were used. Following the reaction, the treated extracts were purified using octadecylsilyl silica resin. The resin (30 mL volume) was initially packed and equilibrated with 5% EtOH. The resulting reaction mixture was diluted one-to-one with water, resulting in a final concentration of EtOH of 5%. Contaminants, such as buffer salts, were eluted with 150 mL water. The bound compounds were eluted with EtOH at a concentration of 80%. The extracts were dried using a vacuum rotary evaporator and a freeze dryer. The target compounds were identified, and the total MI and DMI were determined using an indirect competitive enzyme-linked immunosorbent assay (ELISA) using a previously described procedure.32The culture and treatment of MCF-7 human breast cancer cells were performed in accordance with our previous study.31 MCF-7 cells were cultured in DMEM/F12 supplemented with charcoal-treated FBS (10%, v/v) and antibiotics. Cells were seeded in 96-well plates at a density of 7 × 103 cells per well. After 48 hours of growth, the cells were treated with 10−10 M estradiol (E2) as a control estrogen. PM extracts at various concentrations (0.01–10 ng mL−1) were diluted in estrogen-free medium containing 0.1% (v/v) EtOH. The treatment lasted six days, with the medium being replaced every three days. The effects of E2 and PM extract on cell proliferation were determined using the tetrazolium 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, which was incubated at 0.45 mg mL−1 for 2 hours at 37 °C. The resulting formazan was dissolved in 100 μL solubilizing solution containing 2% (v/v) glacial acetic acid, 40% (v/v) dimethylformamide, and 16% (w/v) sodium dodecyl sulfate. A microplate reader (595 nm) was used to determine the formazan concentration. The proliferative effect of the PM extract treatment was quantified as the percentage of cells that proliferated (% RPE) relative to that of the 10−10 M E2. RPE (%) = (PME/E2) ×100, where PME and E2 refer to the cell proliferation induced by the PM extract and 10−10 M E2, respectively.
2.6. Software and statical analysis
Design-Expert® software version 13 was used to conduct the RSM (Stat-Ease Inc. MN, USA). Statistically significant differences were determined by an ANOVA, which was performed using SPSS ver. 26.3. Results and discussion
3.1. Immobilization of β-glucosidase on alginate beads
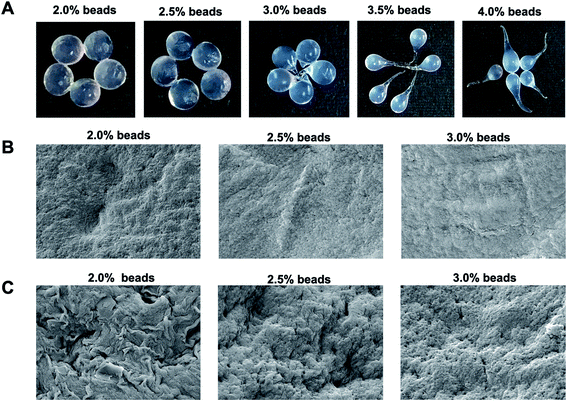
Different shapes of β-glucosidase beads immobilized in 2.0, 2.5, 3.0, 3.5, and 4.0% w/v alginate solutions were obtained (Fig. 1). Higher alginate concentrations resulted in higher immobilized yields, and a drop-like shape was obtained with alginate concentrations of 3.0–4.0% w/v. We obtained spherical alginate beads at concentrations of 2.0 and 2.5% w/v alginate solution. The immobilized yields of β-glucosidase on alginate beads were 72 ± 0.18, 74 ± 0.24, 75 ± 0.12, 75 ± 0.28, and 76 ± 0.12% using alginate solutions at 2.0, 2.5, 3.0, 3.5, and 4.0% w/v, respectively. At higher concentrations of alginate, the enzyme could be trapped in the cross-linked structure with a smaller pore size.33 The surface morphology of alginate beads immobilized with β-glucosidase (2.0, 2.5, and 3.0% w/v) was revealed. Beads of β-glucosidase alginate had a diameter of approximately 0.20 cm. The smooth surface of enzyme-free alginate beads was observed (Fig. 1). A porous structure can be observed in immobilized β-glucosidase alginate beads (Fig. 1), where the substrate reached the immobilized β-glucosidase, and the products were released into the reaction solution. The results are consistent with a previous report in which the highest immobilized yield was obtained using 3% sodium alginate and 0.2 M CaCl2.343.2. β-Glucosidase reaction toward PM isoflavonoid
The reactivity of immobilized β-glucosidase (2.0, 2.5, and 3.0% w/v alginate beads) and free β-glucosidase (0.04, 0.20, and 1.0 U mL−1) with PM extract was first examined. After the reaction, sample solutions for free β-glucosidase were collected at 0, 30, and 60 minutes, whereas sample solutions for immobilized β-glucosidase reaction were collected at 0, 60, and 120 minutes. The enzyme reaction was stopped with alkaline ethanol before the HPLC-UV analysis of the isoflavonoids. The maximum response of DZe and GTe generation was obtained using immobilized β-glucosidase on 2.0% w/v alginate beads (ESI, Fig. S1†). Low alginate concentrations (2.0% w/v) produced higher porosity than 2.5 and 3.0% w/v, thus allowing the substrate and product to flow in and out of beads (Fig. 1). For further experiments, β-glucosidase immobilized in 2.0% w/v alginate beads was used. The target compound conversion in the case of free β-glucosidase were dependent on the enzyme concentration (ESI, Fig. S2†). For the following experiment, 0.2 U mL−1 β-glucosidase was used since it gave a medium response of DZe and GTe synthesis, which suggests that the optimization could be improved further. For comparison, immobilized β-glucosidase (210 U, 21 U per bead) and free β-glucosidase (1 U) were added to the reaction volume (5 mL). The immobilized approach necessitated a substantially higher enzyme amount and a slightly longer reaction time. The increased β-glucosidase quantity required in the immobilized enzyme system was due to the limited flow of the substrate and product into and out of the matrix.3.3. Reaction optimization and RSM analysis
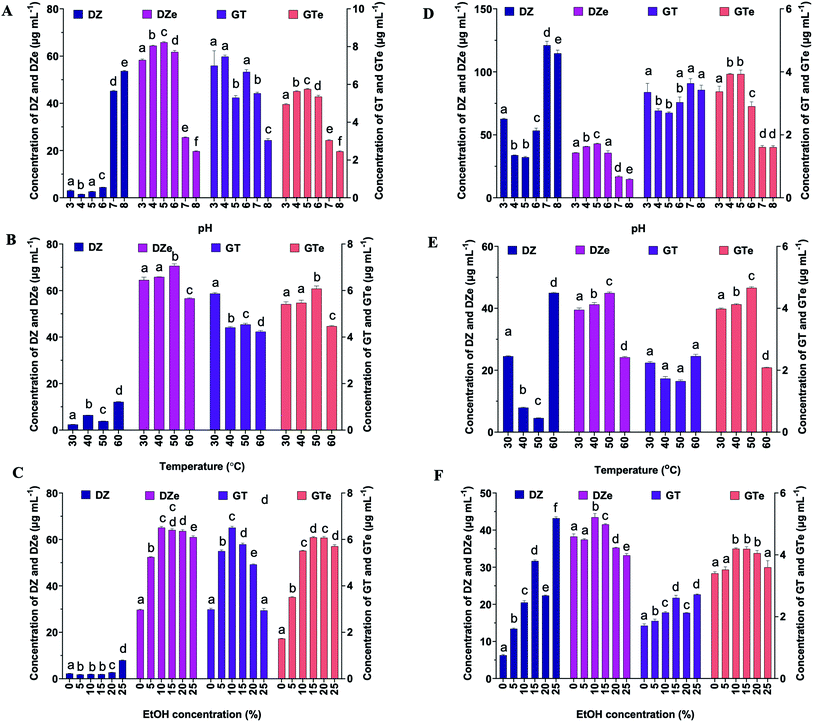
Each factor was optimized individually prior to RSM optimization. Sodium citrate phosphate buffer (50 mM) was used to optimize both the free and immobilized reaction solutions of β-glucosidase at pH 3, 4, 5, and 6. A 50 mM Tris–HCl buffer solution was used as a buffer solution at pH 7 and 8. For both the free and immobilized β-glucosidase on alginate beads, the greatest response of DZe and GTe was achieved with 50 mM sodium citrate phosphate buffer (pH 5) (Fig. 2A and D). The optimum pH of free-β-glucosidase ranged from 4 to 5, whereas the immobilized β-glucosidase exhibited a broader pH range (4–6). The result was similar to that obtained for β-glucosidase from Aspergillus fumigatus ABK9 entrapped into alginate beads, in which the immobilized enzyme exhibited a broader pH range (pH 4.5–6.5) than that exhibited by the free enzyme (pH 5–6).35 The results indicated that at pH 6, free β-glucosidase was lost at 18% and 28% for DZe and GTe production, respectively, whereas the immobilized β-glucosidase exhibited lower loss of DZe and GTe production at approximately 7%, where the losses were compared to that those productions at pH 5. The results suggest that β-glucosidase immobilized on alginate beads may protect the enzyme. Trichoderma β-glucosidases mainly exhibit optimal reactivity at pH values over 4.0 to 5.5 and in the temperature range of 35 to 80 °C,36 and this study obtained similar results. Therefore, a suitable buffer was chosen to optimize the temperature of free and immobilized β-glucosidase.The temperature of the reaction mixture influences the catalytic activity. The optimal reaction temperature for β-glucosidase was investigated between 30 and 60 °C at intervals of 10 °C. The reactions of DZe and GTe increased, resulting in a temperature increase from 30 to 50 °C. The production of DZe and GTe decreased at temperatures above 50 °C (Fig. 2B and E). For example, at 60 °C, the immobilized β-glucosidase was more stable than the free β-glucosidase. The immobilized β-glucosidase retained DZe and GTe productivities at 80% and 74%, respectively, while the productivities by free β-glucosidase remained at 54% and 45%, respectively, compared to 50 °C. High temperatures are likely to denature enzymes. Thermal energy can disrupt the weak non-covalent interactions in the conformation of the native enzyme protein, leading to denaturation.28 Thus, for both the immobilized and free β-glucosidase, the reaction temperature of 50 °C was optimal for converting DZ and GT to DZe and GTe, respectively. To dissolve PM isoflavones and retain enzyme activity, the EtOH concentration had to be adjusted. The reaction mixture was examined at various concentrations of EtOH (5, 10, 15, 20, and 25% (v/v) final concentrations) and water. The reaction of DZe and GTe increased as the EtOH concentration increased from 0% to 10% (Fig. 2C and F) and decreased steadily as the EtOH concentration increased above 10% because higher EtOH concentrations denature the enzyme. Ethanol disrupts or loosens the compact structure of the enzyme by disrupting the tertiary hydrophobic interactions.37 Therefore, 10% EtOH was chosen as the best medium for both immobilized and free β-glucosidase reactivity.
Based on the abovementioned ideal circumstances, the CCD was used. The reduced quadratic models of all responses were determined from the CCD (Table 1), where the model F-values were 44.28, 20.87, 33.06, and 22.99, with all p-values of <0.0001 for Y1, Y2, Y3, and Y4, respectively. The lack of fit values for all models were nonsignificant. Design models can be used to forecast the best values for dependent variables. This proposed model has an excellent correlation between the experimental data and the fitted model, as seen by the adjusted R2 values of 0.9318, 0.8625, 0.9101, and 0.8741 for Y1, Y2, Y3, and Y4, respectively. The difference between the adjusted R2 and predicted R2 of Y1 and Y3 was less than 0.2, indicating that Y1 and Y3 had good model prediction abilities. The prediction efficacy of the Y2 and Y4 models was lower than that of the Y1 and Y3 models. Because PM extract had a higher concentration of DZ than the GT extract, it is possible that the enzyme velocity for DZ is more than those of GT.
| Source of variation | Y1 | Y2 | Y3 | Y4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sum of squares | F-value | P-value probability | Sum of squares | F-value | P-value probability | Sum of squares | F-value | P-value probability | Sum of squares | F-value | P-value probability | |
| Model1 | 3161.88 | 44.28 | <0.0001 | 11.25 | 20.87 | <0.0001 | 2384.13 | 33.06 | <0.0001 | 12.12 | 22.99 | <0.0001 |
| X1 | 22.04 | 1.85 | 0.1967 | 0.1330 | 1.48 | 0.2454 | 41.65 | 3.47 | 0.0854 | 0.0414 | 0.4715 | 0.5044 |
| X2 | 141.85 | 11.92 | 0.0043 | 0.0537 | 0.5979 | 0.4532 | 20.86 | 1.74 | 0.2105 | 0.0496 | 0.5645 | 0.4658 |
| X3 | 2.17 | 0.1827 | 0.6760 | 0.6871 | 7.65 | 0.0161 | 0.0136 | 0.0011 | 0.9737 | 0.3319 | 3.78 | 0.0739 |
| X1X2 | — | — | — | — | — | — | — | — | — | — | — | — |
| X2X3 | — | — | — | — | — | — | — | — | — | — | — | — |
| X1X3 | — | — | — | — | — | — | — | — | — | — | — | — |
| X12 | 1206.97 | 101.41 | <0.0001 | 2.79 | 31.03 | <0.0001 | 1019.29 | 84.81 | <0.0001 | 4.44 | 50.48 | <0.0001 |
| X22 | 1122.77 | 94.34 | <0.0001 | 6.92 | 76.98 | <0.0001 | 1275.62 | 106.14 | <0.0001 | 7.14 | 81.25 | <0.0001 |
| X32 | 1260.34 | 105.89 | <0.0001 | 2.51 | 27.93 | <0.0001 | 450.38 | 37.47 | <0.0001 | 2.22 | 25.24 | 0.0002 |
| Residual | 154.72 | 1.17 | 156.24 | 1.14 | ||||||||
| Lack of fit | 101.93 | 1.21 | 0.4368 | 0.6682 | 0.8353 | 0.6099 | 102.02 | 1.18 | 0.4487 | 0.6135 | 0.7250 | 0.6739 |
| Pure error | 52.79 | 0.5000 | 54.22 | 0.5289 | ||||||||
| Cor total | 3316.60 | 12.42 | 2540.37 | 13.27 | ||||||||
| Std dev. | 3.45 | 0.2998 | 3.47 | 0.2964 | ||||||||
| Mean | 25.00 | 1.44 | 21.20 | 1.90 | ||||||||
| C.V. (%) | 13.80 | 20.85 | 16.35 | 15.61 | ||||||||
| PRESS | 528.75 | 3.73 | 470.44 | 3.35 | ||||||||
| R2 | 0.9533 | 0.9059 | 0.9385 | 0.9139 | ||||||||
| Adj R2 | 0.9318 | 0.8625 | 0.9101 | 0.8741 | ||||||||
| Pre R2 | 0.8406 | 0.6993 | 0.8148 | 0.7478 | ||||||||
The coded equation shows the regression model of independent variables and the responses of Y1, Y2, Y3, and Y4.
| Y1 = 43.67 − 1.27X1 − 3.22X2 + 0.3991X3 − 9.15X12 − 8.83X22 − 9.35X32 |
| Y2 = 3.42 − 0.0845X1 − 0.0967X2 + 0.0661X3 − 0.6336X12 − 0.7348X22 − 0.5995X32 |
| Y3 = 37.18 − 1.75X1 − 1.24X2 − 0.0315X3 − 8.41X12 − 9.41X22 − 5.59X32 |
| Y4 = 3.03 − 0.0551X1 − 0.0603X2 + 0.1559X3 − 0.5548X12 − 0.7039X22 − 0.3923X32 |
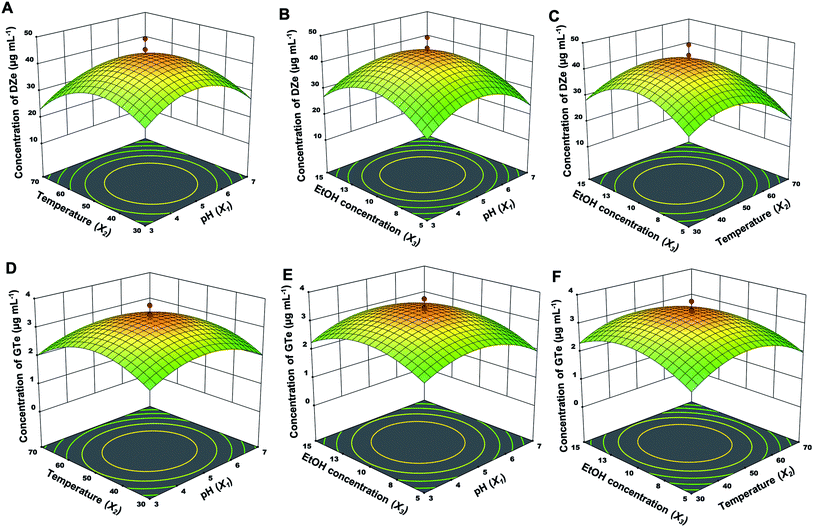
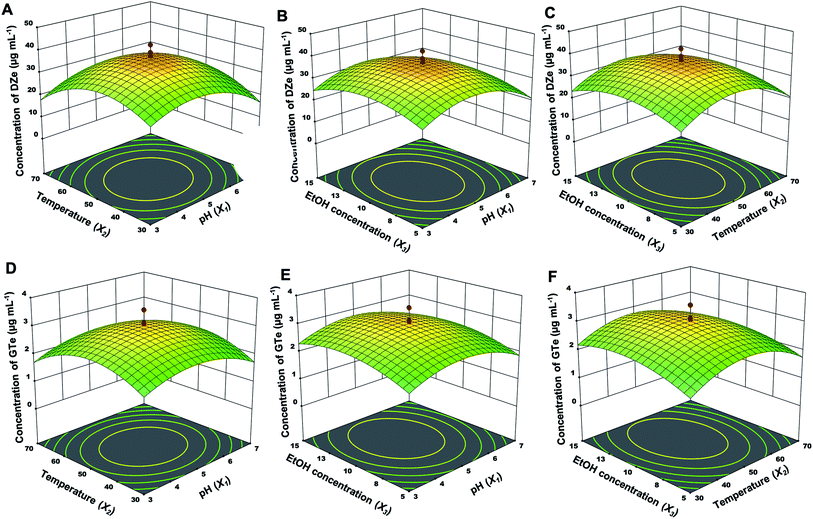
The independent factor interactions of each parameter were explained using a three-dimensional (3D) response, including pH and temperature (X1 and X2) interactions, pH and EtOH concentration (X1 and X3) interactions, and temperature and EtOH concentration (X2 and X3) interactions, as shown in Fig. 3 (immobilized enzyme) and Fig. 4 (free enzyme). Regarding the reduced quadratic models, the interaction between factors did not have a significant impact on all responses. Moreover, the influence order of the single factors was X2 > X1 > X3 for DZe production. The optimal conditions of all factors were almost the same between the free and immobilized β-glucosidase. In the case of the immobilized β-glucosidase reaction, the optimal conditions for Y1 and Y2 were obtained at pH values of 4.9 and 4.9, temperatures of 46.3 and 48.7 °C, and EtOH concentrations of 10.1 and 10.3%, respectively. In the same manner, free β-glucosidase optimally produced Y3 and Y4 at pH values of 4.8 and 4.9, temperatures of 48.6 and 49.1 °C, and EtOH concentrations of 10.0 and 11.0%, respectively. Glutaraldehyde-based enzyme immobilization indicated improved thermal stability.38 While calcium alginate immobilization physically traps β-glucosidase; thus, the active site and structure of the enzyme were not altered by immobilization. The optimal condition of free and immobilized β-glucosidase were almost the same.
The investigated values of the three parameters from the experimental design were used for further experiments. These values confirm that the experimental models are reliable and applicable for product production. Validations of the established models were evaluated, and the reaction was performed under the optimal conditions for immobilized β-glucosidase (X1, 4.9; X2, 46.3; X3, 10) and its free form (X1, 4.8; X2, 48.6; X3, 10). The resulting prediction accuracy with immobilized β-glucosidase was 98 and 95% for Y1 and Y2, respectively, whereas the prediction accuracy of the free enzyme models was 91 and 79% for Y3 and Y4, respectively (Table 2).
| Immobilized β-glucosidase | Prediction accuracy (%) | Free β-glucosidase | Prediction accuracy (%) | ||||
|---|---|---|---|---|---|---|---|
| Optimum condition | Predicted values | Validated values | Optimum condition | Predicted values | Validated values | ||
a 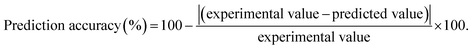 |
|||||||
| X1 = 4.9 | Y1 = 44.00 | Y1 = 43.3 ± 0.88 | 98 | X1 = 4.8 | Y3 = 37.31 | Y3 = 34.3 ± 1.47 | 91 |
| X2 = 46.3 | Y2 = 3.41 | Y2 = 3.59 ± 0.12 | 95 | X2 = 48.6 | Y4 = 3.02 | Y4 = 3.81 ± 0.16 | 79 |
| X3 = 10.0 | X3 = 10.0 | ||||||
3.4. Reusability of immobilized β-glucosidase on alginate beads
Immobilized β-glucosidase was established to enhance the reusability of the product. The reaction of immobilized β-glucosidase (10 beads) and PM extract (5 mL) was performed under the optimum conditions, and the first round of reaction produced 49.6 ± 1.11 and 3.37 ± 0.29 μg mL−1 Y1 and Y2, respectively, which was set as 100% activity. Before the subsequent reaction, β-glucosidase alginate beads were washed by stirring for 5 min. The next round of reactions followed the abovementioned procedure. At the 9th cycle of the immobilized glucosidase process, the yields for Y1 and Y2 were above 90%. After the 9th cycle, the response of Y1 and Y2 gradually decreased (Fig. S3†), which may have been due to enzyme loss from the alginate beads under the continuous shaking stress and activity loss due to instability at the late reaction cycle. Genetic engineering of β-glucosidase for specific immobilization on regenerated amorphous cellulose showed more than 96% efficiency after 30 rounds of recycling.39 Immobilized β-glucosidase on magnetic chitosan microspheres retained high efficacy over eight batches.403.5. MCF-7
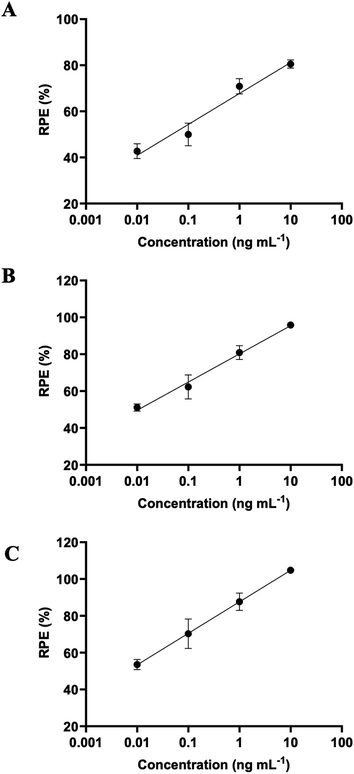
The contents of each PM isoflavone and total MI and DMI are shown in Table 3. The contents of PUE did not seem to be much changed by the β-glucosidase treatment, although the conversion of DZ and GT to DZe and GTe, respectively, was significant. The total contents of MI and DMI slightly increased after the β-glucosidase treatment. Previously, miroestrol-3-O-β-D-glucopyranoside was isolated at a yield of 23 mg/40 kg PM root.41 Thus, the compound may be cleaved to yield MI. The DZ content in the PM extract was more abundant than that in GT, which corresponded to the higher DZe yield than GTe. Although GTe exhibited more binding potency toward the estrogen receptor (ER) of MCF-7 cells, DZe was reported to be a more efficient substance against ovariectomized bone loss than GTe.42 The PM extract exhibited estrogen activity in a concentration-dependent manner (Fig. 5). The concentration of PM extract, which produced 80% RPE compared to 10−10 M E2, was defined as EC80. Less EC80 indicated more estrogenic potency (Table 3). The immobilized and free β-glucosidase treatments showed enhanced estrogenic potency by 8.71- and 23.2-fold, respectively, compared to the non-β-glucosidase-treated extract. Free β-glucosidase enhanced estrogenic activity more than the immobilized form. The free β-glucosidase reaction provided more DZe, and the increased DZe and GTe contents may correspond to increased estrogen potency. The PM extract treated with β-glucosidase could be further developed as a functional food against estrogen deficiency disorders, such as menopause, osteoporosis, Alzheimer's disease, and systemic inflammation. Further clinical trials of PM may lead to more efficacious and safe treatments for menopause-related symptoms. Based on current knowledge, the controversial results of clinical trials are associated with a lack of PM extract standardization and interpersonal variations in isoflavone absorption. This β-glucosidase-derived PM extract (standardized) may lead to more consistent clinical outcomes.| Extract treatment | Content (μg g−1 PM extract) | EC80 (ng mL−1) | |||||
|---|---|---|---|---|---|---|---|
| PUE | DZ | GT | DZe | GTe | MI and DMI | ||
| Control | 1063 ± 17.5 | 50.0 ± 1.01 | 8.32 ± 0.19 | 34.9 ± 0.87 | 2.74 ± 0.11 | 43.9 ± 1.48 | 8.80 ± 3.81 |
| Immobilized β-glucosidase | 954 ± 8.95 | 4.07 ± 0.07 | 5.25 ± 0.36 | 82.6 ± 4.45 | 4.42 ± 0.23 | 49.8 ± 3.34 | 1.01 ± 0.32 |
| Free β-glucosidase | 975 ± 3.74 | 10.6 ± 0.34 | 3.46 ± 0.01 | 88.0 ± 0.67 | 4.47 ± 0.05 | 49.7 ± 1.05 | 0.38 ± 0.12 |
4. Conclusion
The β-glucosidase enzyme was successfully immobilized in alginate beads, which retained the enzyme's reactivity and could be reused. β-Glucosidase effectively converts the PM isoflavone glycoside to an aglycone. Immobilized β-glucosidase could be reused for up to 9 cycles. In addition, the immobilized enzymes can be conveniently separated from the product. The response surface methodology (RSM) central composite design (CCD) established reliable response models, and the prediction accuracy for DZe and GTe was 79–98%. The estrogenic potency of the PM extracts improved by 8.71- to 23.2-fold after treatment with β-glucosidase. Thus, β-glucosidases were efficient for PM isoflavone conversion and enhanced estrogenic activity.Authorship contributions
Fonthip Makkliang: data curation, formal analysis, investigation, methodology, doftware, visualization, writing – original draft. Wipawee Juengsanguanpornsuk: investigation. Suppalak Phaisan: data curation. Attapon Sakdamas: investigation. Waraporn Putalun: resources. Seiichi Sakamoto: resources. Gorawit Yusakul: conceptualization, funding acquisition, methodology, project administration, resources, supervision, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was financially supported by the Agricultural Research Development Agency (Public Organization) (grant number: CRP6305030030). The authors would also like to thank the Center for Scientific and Technological Equipment of Walailak University for providing all facilities and equipment. Partially, this research was financially supported by the new strategic research project (P2P), Walailak University, Thailand.References
- S. Malaivijitnond, Front. Med., 2012, 6, 8–21 CrossRef PubMed.
- S. Chansakaow, T. Ishikawa, H. Seki, H. Seki, M. Okada and C. Chaichantipyuth, J. Nat. Prod., 2000, 63, 173–175 CrossRef CAS PubMed.
- N. Peerakam, P. Sirisa-Ard, N. Q. Huy, T. V. On, P. T. Long and A. Intharuksa, Asian J. Chem., 2018, 30, 2086–2090 CrossRef CAS.
- K. D. Setchell, N. M. Brown, L. Zimmer-Nechemias, W. T. Brashear, B. E. Wolfe, A. S. Kirschner and J. E. Heubi, Am. J. Clin. Nutr., 2002, 76, 447–453 CrossRef CAS PubMed.
- Y. Okabe, T. Shimazu and H. Tanimoto, J. Sci. Food Agric., 2011, 91, 658–663 CrossRef CAS PubMed.
- J. R. Ketudat Cairns and A. Esen, Cell. Mol. Life Sci., 2010, 67, 3389–3405 CrossRef CAS PubMed.
- S. Yan, P. C. Wei, Q. Chen, X. Chen, S. C. Wang, J. R. Li and C. Gao, Biochem. Biophys. Res. Commun., 2018, 496, 1349–1356 CrossRef CAS PubMed.
- S. Delgado, L. Guadamuro, A. B. Flórez, L. Vázquez and B. Mayo, Innovative Food Sci. Emerging Technol., 2019, 51, 148–155 CrossRef CAS.
- X. Pei, J. Zhao, P. Cai, W. Sun, J. Ren, Q. Wu, S. Zhang and C. Tian, Protein Expression Purif., 2016, 119, 75–84 CrossRef CAS PubMed.
- M. Matsuura, J. Sasaki and S. Murao, Biosci., Biotechnol., Biochem., 2014, 59, 1623–1627 CrossRef.
- F. de Oliveira Silva, T. C. Lemos, D. Sandora, M. Monteiro and D. Perrone, J. Sci. Food Agric., 2020, 100, 2991–2998 CrossRef CAS PubMed.
- S. E. Mustafa, S. Mustafa, A. Ismail, F. Abas, M. Y. Abd Manap, O. A. Ahmed Hamdi, S. Elzen, L. Nahar and S. D. Sarker, Heliyon, 2020, 6, e05298 CrossRef PubMed.
- H. J. Jou, S. C. Wu, F. W. Chang, P. Y. Ling, K. S. Chu and W. H. Wu, Int. J. Gynaecol. Obstet., 2008, 102, 44–49 CrossRef CAS PubMed.
- J. Wu, J. Oka, J. Ezaki, T. Ohtomo, T. Ueno, S. Uchiyama, T. Toda, M. Uehara and Y. Ishimi, Menopause, 2007, 14, 866–874 CrossRef PubMed.
- Y. Ideno, K. Hayashi, J. Nakajima-Shimada, Y. Onizuka, M. Kishi, T. Ueno and S. Uchiyama, PLoS One, 2018, 13, e0201318 CrossRef PubMed.
- S. Arboleya, C. Watkins, C. Stanton and R. P. Ross, Front. Microbiol., 2016, 7, 1204 CrossRef PubMed.
- L. Khaodhiar, H. A. Ricciotti, L. Li, W. Pan, M. Schickel, J. Zhou and G. L. Blackburn, Menopause, 2008, 15, 125–132 CrossRef PubMed.
- W. L. Hall, K. Vafeiadou, J. Hallund, S. Bugel, C. Koebnick, M. Reimann, M. Ferrari, F. Branca, D. Talbot, T. Dadd, M. Nilsson, K. Dahlman-Wright, J. A. Gustafsson, A. M. Minihane and C. M. Williams, Am. J. Clin. Nutr., 2005, 82, 1260–1268 CrossRef CAS PubMed.
- T. Izumi, M. K. Piskula, S. Osawa, A. Obata, K. Tobe, M. Saito, S. Kataoka, Y. Kubota and M. Kikuchi, J. Nutr., 2000, 130, 1695–1699 CrossRef CAS PubMed.
- S. B. Jadhav and R. S. Singhal, Carbohydr. Polym., 2014, 105, 49–56 CrossRef CAS PubMed.
- H. U. Rehman, A. Aman, A. Silipo, S. A. Qader, A. Molinaro and A. Ansari, Food Chem., 2013, 139, 1081–1086 CrossRef PubMed.
- F. A. Gourdani, P. Ghadam, M. M. Heravi and M. Malmir, J. Iran. Chem. Soc., 2021, 18, 1471–1478 CrossRef.
- Z. Ölçer and A. Tanriseven, Process Biochem., 2010, 45, 1645–1651 CrossRef.
- H. Tumkurk, G. Demirel, H. Altinok, S. Aksoy and N. Hasiroi, J. Food Biochem., 2008, 32, 234–246 CrossRef.
- S. Talekar and S. Chavare, Recent Res. Sci. Technol., 2012, 4, 1–5 CAS.
- G. Dey, B. Singh and R. Banerjee, Braz. Arch. Biol. Technol., 2003, 46, 167–176 CrossRef CAS.
- A. Blandino, A. Macias and D. Cantero, Appl. Biochem. Biotechnol., 2003, 110, 53–60 CrossRef CAS PubMed.
- P. A. Skovgaard and H. Jorgensen, J. Ind. Microbiol. Biotechnol., 2013, 40, 447–456 CrossRef CAS PubMed.
- J. Herlet, P. Kornberger, B. Roessler, J. Glanz, W. H. Schwarz, W. Liebl and V. V. Zverlov, Biotechnol. Biofuels, 2017, 10, 234 CrossRef CAS PubMed.
- G. Yusakul, W. Juengsanguanpornsuk, B. Sritularak, S. Phaisan, T. Juengwatanatrakul and W. Putalun, Nat. Prod. Res., 2020, 1–5, DOI:10.1080/14786419.2020.1727473.
- W. Juengsanguanpornsuk, G. Yusakul, W. Kraithong and W. Putalun, J. Herb. Med., 2021, 29, 100463 CrossRef.
- G. Yusakul, T. Kitisripanya, T. Juengwatanatrakul, S. Sakamoto, H. Tanaka and W. Putalun, J. Nat. Med., 2018, 72, 641–650 CrossRef CAS PubMed.
- Y. Gong, G. T. Han, Y. M. Zhang, J. F. Zhang, W. Jiang, X. W. Tao and S. C. Gao, J. Polym. Eng., 2016, 36, 363–370 CAS.
- Keerti, A. Gupta, V. Kumar, A. Dubey and A. K. Verma, ISRN Biochem., 2014, 2014, 178498 CAS.
- A. Das, T. Paul, P. Ghosh, S. K. Halder, P. K. Das Mohapatra, B. R. Pati and K. C. Mondal, Waste Biomass Valorization, 2014, 6, 53–61 CrossRef.
- P. Tiwari, B. N. Misra and N. S. Sangwan, BioMed Res. Int., 2013, 2013, 203735 Search PubMed.
- H. Yoshikawa, A. Hirano, T. Arakawa and K. Shiraki, Int. J. Biol. Macromol., 2012, 50, 865–871 CrossRef CAS PubMed.
- D. de Andrades, N. G. Graebin, M. K. Kadowaki, M. A. Z. Ayub, R. Fernandez-Lafuente and R. C. Rodrigues, Int. J. Biol. Macromol., 2019, 129, 672–678 CrossRef CAS PubMed.
- S. L. Hu, D. M. Wang and J. Hong, Biotechnol. Bioprocess., 2018, 23, 39–48 CrossRef CAS.
- P. Zheng, J. Wang, C. Lu, Y. Xu and Z. Sun, Process Biochem., 2013, 48, 683–687 CrossRef CAS.
- M. H. Bang, D. G. Lee, Y. S. Baek, J. G. Cho, M. W. Han, K. S. Choi, D. K. Chung, S. K. Ko, C. H. Oh, S. Y. Cho, K. Y. Chai, J. H. Kim and N. I. Baek, Chem. Nat. Compd., 2013, 49, 443–445 CrossRef CAS.
- C. Picherit, V. Coxam, C. Bennetau-Pelissero, S. Kati-Coulibaly, M. J. Davicco, P. Lebecque and J. P. Barlet, J. Nutr., 2000, 130, 1675–1681 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05109a |
| This journal is © The Royal Society of Chemistry 2021 |

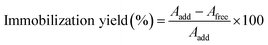
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00×.
00×.