Open Access Article
Open Access ArticleMonitoring the heterogeneity in single cell responses to drugs using electrochemical impedance and electrochemical noise†
Ying
Yang
abc,
Friederike M.
Mansfeld
bcde,
Maria
Kavallaris
bcd,
Katharina
Gaus
fg,
Richard D.
Tilley
 ab and
J. Justin
Gooding
ab and
J. Justin
Gooding
 *abc
*abc
aSchool of Chemistry, The University of New South Wales, Sydney, NSW 2052, Australia. E-mail: justin.gooding@unsw.edu.au
bAustralian Centre for NanoMedicine, The University of New South Wales, Sydney, NSW 2052, Australia
cThe ARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW 2052, Australia
dChildren's Cancer Institute, The University of New South Wales, Sydney, NSW 2052, Australia
eMonash Institute of Pharmaceutical Sciences, Monash University, Melbourne, VIC 3052, Australia
fEMBL Australia Node in Single Molecule Science, School of Medical Sciences, The University of New South Wales, Sydney, NSW 2052, Australia
gARC Centre of Excellence in Advanced Molecular Imaging, The University of New South Wales, Sydney, NSW 2052, Australia
First published on 28th December 2020
Abstract
Impedance spectroscopy is a widely used technique for monitoring cell–surface interactions and morphological changes, typically based on averaged signals from thousands of cells. However, acquiring impedance data at the single cell level, can potentially reveal cell-to-cell heterogeneity for example in response to chemotherapeutic agents such as doxorubicin. Here, we present a generic platform where light is used to define and localize the electroactive area, thus enabling the impedance measurements for selected single cells. We firstly tested the platform to assess phenotypic changes in breast cancer cells, at the single cell level, using the change in the cell impedance. We next show that changes in electrochemical noise reflects instantaneous responses of the cells to drugs, prior to any phenotypical changes. We used doxorubicin and monensin as model drugs and found that both drug influx and efflux events affect the impedance noise signals. Finally, we show how the electrochemical noise signal can be combined with fluorescence microscopy, to show that the noise provides information on cell susceptibility and resistance to drugs at the single cell level. Together the combination of electrochemical impedance and electrochemical noise with fluorescence microscopy provides a unique approach to understanding the heterogeneity in the response of single cells to stimuli where there is not phenotypic change.
Introduction
In cancer biology, understanding cellular heterogeneity is the key to both understanding the mechanism of disease and the response to treatment.1,2 The challenge is particularly acute because disease is often caused by aberrant cells which may be rare relative to the dominant healthy cells. Identifying, isolating and preconcentrating rare cells typically focusses on differences in cell phenotype or cell surface markers.3–5 However, even within a population of isolated cells, there may still be functional heterogeneity that is not revealed within the markers used to isolate a population.6 As such, functional assays on the single cell level are required.7,8 The majority of such single cell assays use fluorescence markers to provide functional and structural information about cells, either as endpoint assays such as flow cytometry to obtain high volumes of data,9 or live cell fluorescence microscopy based functional assays that can follow sub-cellular changes but require labelling and there are challenges with continuous monitoring over long timescales.10 The short timescale limitation with fluorescence is due to concerns over phototoxicity with continued excitation of fluorophores over time.11 What would complement the fluorescent approaches is a label free technique that can monitor live cells over many hours but with high temporal resolution.12 Electrochemical impedance spectroscopy is one such label free technique that can not only monitor cells over many hours but can do this with high sensitivity and with millisecond temporal resolution.13Since its development by Giaever and Keese in 1991,13 electrochemical impedance spectroscopy has been an ensemble technique requiring ∼2000 cells or more to enable the investigation of cell–surface interactions or drug responses. The signal from cells in electrochemical impedance comes from cells adhering to the electrode and restricting the access of ions to that electrode. Any changes in cell shape, how tightly the cells adhere to the electrode surface and membrane integrity result in a change of the dielectric properties of the cell layer, with the relative contribution of each parameter able to be determined using models.13 The result is a change in impedance. In the case of drug treatments, this means the electrochemical impedance monitors cell changes due to drug action that in most cases require the activation of signalling cascades. Such changes occur over several minutes to several hours, so this often means a lag time before an impedance signal change is observed.12 The twin challenges with impedance spectroscopy are to reduce the method down to a single cell and to rapidly monitor process that occur within a cell before any phenotype changes. The challenges with single cell measurements are predominantly electronic. In electrochemical impedance spectroscopy, measurements are equated to a circuit of resistors and capacitors. At the single cell level, the leads and circuitry of the instrument can contribute significantly to the signal, particularly at high frequencies. To address this issue, Laborde et al.14 have built all the electronics within a silicon chip to allow a microelectrode array to operate at very high frequencies to follow the migration of single cells. This approach is not generally accessible to others, however, as integrating the electronics into silicon is enormously expensive and very specialised. A second aspect of the electronic challenge is the need to connect each individual electrode to the external electrochemical circuit which limits the number of detectable recording sites.15 There are some elegant strategies to fabricate large arrays of independently addressable electrodes which again require microfabrication16 and patterning of surface chemistry to confine the cells to individual electrodes.17
In recent years, the semiconductor devices and circuits showed advantages like fast response, signal amplification, multifunctionality and multiplexity, have been widely for assessing complex dynamics in biological systems.8,18–22 The purpose of this paper is two-fold. The first is to demonstrate a far simpler strategy to achieve single cell impedance measurements of both cell phenotype changes and rapid cell responses to drugs prior to any phenotype changes. The second is introduce the concept of using electrochemical noise from single cells, and the change in that noise, to monitor uptake and efflux of drugs from single cells. Both draw on the advances in semiconductor devices for cell biology. We overcome the restriction of needing microfabrication and complicated electronics to confine and address individual cells by using unpatterned semi-conducting silicon electrodes that are in depletion. Shining light on a given location transiently activates that region of the electrode such that it becomes electrochemically active whilst the rest of the surface remains electrochemically inactive.23,24 This means that any area can be electrochemically addressed on a monolithic surface with only one connecting lead.
This activation of specific areas of a surface by light is based on light exciting photoelectrons from the valence band to the conduction band of the semi-conducting silicon electrode that is in depletion. This excitation of photoelectrons causes an increase in minority charge carriers in the silicon and hence an increase in conductivity around the site of illumination.25 We have explored fundamental aspects of this concept, referred to as light activated electrochemistry,23 including the impact of light intensity,26 orientation of the light beam27 and the thickness28 and crystallinity29 of the silicon on the spatial resolution that can be achieved and show that spatial resolutions below that needed to interrogate a single cell can be achieved.27,29 We have also shown that this technique can be used to release rare single cells captured on an unpatterned surface,3 and the detection of ion release from single cells, combined with fluorescence microscopy can be used to show how the cells respond to drug treatment.30
Herein we further show it is possible to perform conventional impedance measurements on single cells that are located anywhere on the silicon electrode surface. This is achieved by illuminate the region of a single cell and monitoring changes in electrochemical impedance as the cell phenotype changes. Furthermore, for processes occurring prior to any change in cell phenotype, or processes where there is no associated phenotypic change, we take a radical new approach of measuring the electronic noise. Changes in this noise reveal information about, for example, the uptake of drugs by a cell and the resultant efflux of these drugs. We show that for adjacent MCF-7 breast cancer cells on the silicon electrode the amount of the drug doxorubicin taken up by the cells varies and the change in impedance of individual cells correlates with the amount of drug within the cells. We demonstrate that changes in electrochemical noise occur the instance cells are exposed to drugs that the noise relates to the amount of drug within the cells. This allows us to not only differentiate drug resistant from drug sensitive cells, but explore the heterogeneity in the response of single cells from the same origin, in a label free manner, over many hours. This is an important advance for understanding single cell heterogeneity and potentially personalised medicine as it provides a method for monitoring the heterogeneity in the response of cells to drugs where the cell phenotype does not change, without necessitating the need for any labels or fluorescent species. This new capability allows the monitoring of the uptake of species into cells where there is no change in phenotype, at the single cell level, for many hours. No labels are required and the method is complementary to fluorescence microscopy. This exciting technological advance allows cell heterogeneity to be monitored and understood without concerns that the system is perturbed by any labels.
Results and discussion
Platform design and performance for light addressable single-cell impedance
The working principle for performing single cell impedance uses the concept of light activated electrochemistry and is illustrated in Fig. 1a. To localise the cell impedance to a single cell using light requires a semiconducting silicon electrode that is in depletion at the potential used for the electrochemical impedance measurement.25 When the electrode is in depletion, there is a deficiency of the minority carriers in the silicon that can drive an electrochemical reaction. In such cases, optical illumination excites electrons from the valence band of the silicon to the conduction band. This excitation increases the amount of minority carriers, and hence the conductivity. As such, the illumination allows electrochemical measurements to be performed at the site of illumination, with the rest of the surface remaining electrochemically inactive. Note, silicon was chosen as its energy band gap is sufficiently small, 1.1 eV, such that visible light, that is benign to the cells, can be used.3 The majority of other common semiconductors require ultraviolet light excitation.31 The challenge with silicon however is the easy oxidation which prevents electrochemistry occurring. To overcome this challenge the semiconducting silicon surface was firstly passivated by an alkyne-terminated organic monolayer on hydrogen terminated silicon via thermal hydrosilylation (see Fig. 1b and S1a).†32 Subsequently, anti-EpCAM IgM antibodies were attached on silicon which are specific to epithelial cell adhesion molecule (EpCAM) that is upregulated in MCF-7 breast cancer cells. The high-resolution XPS spectrums of Si 2p peaks for cell attached silicon electrodes after running a set of impedance measurements in aqueous medium were shown in Fig. S1b–d.† The incredibly minor increase in silicon oxide at 102–104 eV after incubation in cell culture medium and performing electrochemistry indicates the high quality of monolayer and its ability to prevent appreciable oxidation, which also supports the stability of the electrode during electrochemistry.33,34 Note for other applications more broadly applicable ligands such as cell adhesive ligands RGD peptides could be employed.35 The antibodies allow selective capture of a given cell type and tethers the cells to the surface for impedance measurements.36 It has been previously shown that this surface chemistry not only allows silicon electrodes to be used in biological buffers32 but that the antibody modification provides a surface where one cell type can be selectively captured despite the presence of a mixture of cells in the media.3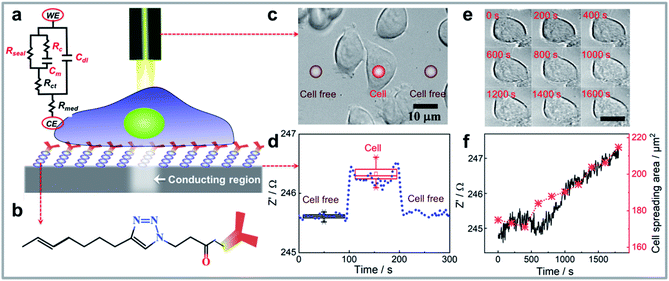 | ||
| Fig. 1 (a) Schematic representation of the single cell impedance sensing platform consisting of electrochemical measuring and imaging unit. Also shown is the simplified electrical equivalent circuit for a living cell attached on modified silicon electrode. The sealing resistance (Rseal) from cell adhesion is connected with the membrane capacitance (Cm) and resistance of cytoplasm (Rc) in parallel. The charge transfer resistance (Rct), double layer capacitance (Cdl) at the electrode/electrolyte interface, and resistance for cell culture medium (Rmed) between counter electrode (CE) and working electrode (WE) are also presented. (b) A customized silicon surface was designed to selectively attach the cells of interest onto the surface and perform light activated electrochemistry at the illumination area, the chemical structure of the surface is shown. (c) As a demonstration of the platform working principle, bright-field images of cells can be acquired using an optical microscope, subsequently, the platform design that has an integrated laser beam that serves as a contactless wire for recording electrochemical impedance data on distinct spots (red circles indicate the laser beam on cell or cell free region) on cells attached to the monolayer-modified silicon surface. The surface placed in an electrochemical chamber is connecting with the electrochemical workstation, therefore, the impedance data for these illuminated spots could be collected simultaneously. (d) The raw data shows the impedance change when moving laser beam at one single cell or adjacent cell free region as indicated in panel (c) using the platform. E = 0.1 V, f = 400 Hz. (e) Representative microscopy images of an MCF-7 cell attached on silicon electrode showing the spreading area change during the impedance monitoring process, (f) shows the positive correlation between the impedance change and cell spreading area (analysed cell-cross area in (e) using ImageJ, with detailed tutorial in the ESI†). | ||
The modified silicon surface with captured MCF-7 cells was inserted into a custom designed chamber that allowed simultaneous impedance and microscopy measurements to be performed.30 Whilst under the microscope, a laser beam was moved over the surface. According to previous work, the photo-generated charge carrier diffusing inside of the bulk silicon will make the electrochemical active region larger than the beam size.23,24 Therefore, a laser beam with optimized full width at half maximum (FWHM) of 5 μm, which makes the spatial resolution of light activated electrochemistry around 18 μm,3,24,30 was utilized to perform single cell impedance measurement. The impedance of individual cells, and cell free regions, were measured as the laser beam moved across the surface (Fig. 1c). Using this setup, the impedance for each individual cell can be addressed sequentially by moving the light beam to each cell of interest. Optical microscopy was used to observe the cell morphology, either under bright field or fluorescent imaging. The increase in impedance as the light source scanned past the cell is attributed to the cell limiting the access of ions to the electrochemically active portion of the silicon electrode. Fig. 1d shows an example of the change in actual impedance data when the laser beam was illuminated on a single attached cell or at adjacent cell-free regions. The impedance data without attached cells was used as the reference,37 and the normalized impedance in terms of 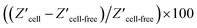 was used to compare the difference in impedance for cell and cell-free regions. In this work, such difference is readily visible with the normalized impedance typically lying in the range of 0.30–0.60%. This result demonstrates that the system can determine the impedance from attached cells. Importantly, over the time frames of the experiments presented herein, there was no change in cell viability (ESI Table 1†). This was shown using a trypan blue exclusion tests after impedance measurement.38 These results demonstrate that the use of light to electrochemically activate regions of the surface to perform measurements has no significant effect on cell viability.
was used to compare the difference in impedance for cell and cell-free regions. In this work, such difference is readily visible with the normalized impedance typically lying in the range of 0.30–0.60%. This result demonstrates that the system can determine the impedance from attached cells. Importantly, over the time frames of the experiments presented herein, there was no change in cell viability (ESI Table 1†). This was shown using a trypan blue exclusion tests after impedance measurement.38 These results demonstrate that the use of light to electrochemically activate regions of the surface to perform measurements has no significant effect on cell viability.
We examined the spreading characteristics of one single MCF-7 cell after capturing cells on the silicon electrode (Fig. 1e) and monitored its impedance at frequency of 400 Hz for 30 min (Fig. 1f). Fig. 1f showed with the increase of attachment and spreading of the MCF-7 cell, there is a concomitant increase in the real part of the impedance signal. This is because the cell membrane is insulating and the increased cell spreading on surface will restrict the current flow underneath the cell so the seal resistance (Rseal, Fig. 1a) is increased. The real part of the impedance signal at a frequency of 400 Hz was sensitive to this change, therefore, we see there is a positive correlation between the real part impedance change and cell spreading area increase over 30 min.
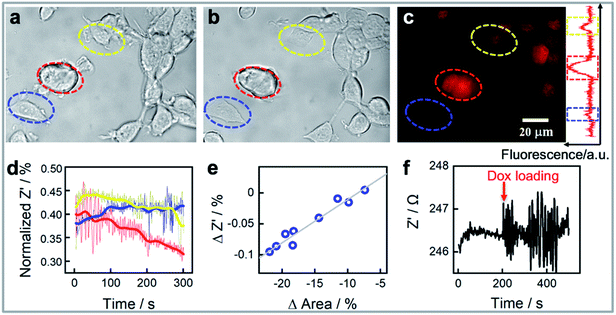
We next demonstrate that our platform has the capability to monitor the response of the cells to drugs in real time. As an effective chemotherapy medication to treat breast cancer, doxorubicin was chosen as the model drug.39Fig. 2 shows an example of three MCF-7 cells monitored at the single cell level before (Fig. 2a) and 30 minutes after exposure to 1.0 μM of doxorubicin (Fig. 2b). It is apparent from the intensity of the fluorescence from doxorubicin in fluorescent microscopy images that the three cells accumulate different amounts of doxorubicin (Fig. 2c). We also observe that the normalized impedance change of these cells after exposure to doxorubicin (Fig. 2d) varies, integrating the area covered by each single cell in Fig. 2a and b during the measurement reveals that the method provides information on the cell adhesion and changes in cell spreading at a single cell level. The reduction in cell area for the cells identified by the red and yellow circles correlated well with the decreases in normalized impedance. By interpreting more cell area and normalized impedance data, Fig. 2e shows a linear relationship between the decrease in impedance and a reduction in the cell contact area determined from the bright field images. Note however that the impedance change is not only dependent on the cell contact area, but also on the cell–surface adhesion.13 A change in cell–surface adhesion could be the explanation for why the cell (circled in blue colour) in Fig. 2a and b has a reduced cell contact area of ∼6% but the impedance increased slightly. These results highlight the need for methods that can explore the single cell heterogeneity, as in this case, three immortal cells from a single population exhibit differences in how the impedance changes after exposure to doxorubicin. Further highlighting the heterogeneity of the response of cells to the drug is that although the change in impedance of the nine cells shown in Fig. 2e was directly correlated with the change in area of the cells, thus showing the single cell impedance provides a means to monitor cellular morphology and cell–substrate adhesion interactions change during drug treatment, the change in area of the cells was not directly correlated with the amount of drug uptake (Fig. S2†). When conducting this experiment, we observed that when the cell is exposed to the drug the impedance measurement suddenly shows much larger fluctuations, or noise, than prior to the addition of the drug (Fig. 2f). Such impedance fluctuations are clearly noted in Fig. 2d for the three cells listed in Fig. 2a–c. Such as the red circled cell, which showed remarkable morphological change, consistent change in normalized impedance change, and the significant impedance noise (Fig. 2a–d). Meanwhile, three cells not only showed various normalized impedance change, but also showed different impedance fluctuations. The impedance noise and how it can be used for quantifying the heterogeneity in cell responses to drugs will be discussed further in the next section.
Doxorubicin induced cellular response detection: cell impedance noise levels indicate the drug uptake levels for doxorubicin treated MCF-7 cell line
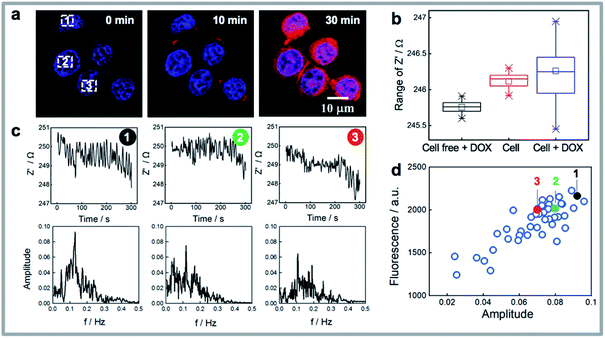
Cellular uptake is an important determinant of the pharmacodynamic properties of drugs and drug candidates. The cytotoxic effect of doxorubicin on human breast cancer cells (MCF-7) has been reported to be positively correlated with the amount of doxorubicin taken up by cells.40 As the rapid uptake of drugs precedes any downstream signalling events, it is not detected using conventional impedance measurements. Generally, optical imaging is the main method to visualize the drug accumulation levels in living cells. The question being asked here is, can the sudden increase in noise observed in the single cell impedance data for the MCF-7 cells after doxorubicin loading be used to quantify the uptake of the drug by the cells? This sudden increase in noise is evident in a single cell trace shown in Fig. 2f upon the injection of the drug. As doxorubicin is also fluorescent, the onset of noise can clearly be correlated with fluorescence microscopy showing that the noise commences as the drug enters the cell. Note in the cell free regions there was no obvious increase in noise on addition of doxorubicin (Fig. S3a and d†).To clarify whether the impedance fluctuations were specific to doxorubicin or not, we next exposed the cells to the antibiotic monensin, which is also an Na+/K+ antiporter (Fig. S5†). As with doxorubicin, exposure of the cell to 10 μM monensin, a sudden increase in the electrochemical noise is observed (Fig. S5a†). Note over the 30 minutes of this experiment, after the addition of the drug, there is an increase in impedance which is associated with the cells spreading. This can be seen in the bright field images before and after monensin loading for 30 min (Fig. S5b†). What is evident in Fig. S5b† is after the drug treatment, more filopodia spreading from the cell are observed. The 10 nM monensin did not lead to MCF-7 cell death41 although a decrease in intracellular K+ concentration was noted from fluorescence images (Fig. S5c†). This decrease in K+ concentration is because monensin causes rapid transfer of ions across the cell membrane.42 This gives a clue that the impedance fluctuations were due to ion transport through the cell membrane on exposure to the drugs used herein. Doxorubicin (Adriamycin) and many other antibiotics are known to facilitate the translocation of ions across biological membranes.43 If one considers a living cell in terms of electronic components, as is typically done in impedance spectroscopy, electrical models of a cell consists of both resistance and capacitance (a simplified equivalent circuit is inserted in Fig. 1a). Ion flux across the cell membrane will change both the resistance of cytoplasm and membrane capacitance, and hence reflected as increased impedance noise.
Fast Fourier Transform (FFT) is a widely used mathematical tool to convert a signal from its time domain to frequency domain,44,45 hence FFT is applied here to analyze the impedance noise data by varying the impedance fluctuations with different frequencies and amplitudes. Fig. 3a shows three cells where the uptake of drug increases with time but where the fluorescence intensity for the three cells, from the fluorescent doxorubicin (red colour in the images), is quite different among the cells. Note from Fig. 3b it can be observed that the median impedance of the cell covered region does not change significantly on exposure to the drug, while the interquartile range, reflective of the amplitude of the impedance fluctuations, increases dramatically. The impedance curves and box plots in Fig. S3a–d† summarized the impedance differences for cell free region, a dead cell (where there is membrane lysis), and a living cell, before and after exposure to doxorubicin. The absence of the significant increased noise in these cases further confirms the impedance noise is the main response of a living cell to the drug. To analyze the raw impedance data in Fig. 3c, the impedance signal was converted from its original time domain to a single side amplitude spectrum in the frequency domain. In the obtained Fourier amplitude spectrum, the frequency with the maximum amplitude in the FFT spectrum was identified as the dominant frequency which typically occurred at ∼0.1 Hz. Note in FFT spectrum for cell free region or a dead cell after treating with doxorubicin or a living cell prior to addition of doxorubicin (see Fig. S3e–g†), there was no dominant frequency. By comparing the amplitude spectra in Fig. 3c for three cells, it shows the amplitude of the dominant frequency tends to change with the amount of drug that enters the cell. This is especially the case at the dominant frequency at 0.11 Hz where the more drug that enters the cell, the higher is the amplitudes at the fundamental frequencies (Fig. 3d). To further verify this observation, single cell impedance data from 36 cells from 6 different silicon electrodes were collected and processed using FFT analysis, and the relationship between fluorescence intensity of doxorubicin in MCF-7 cells and amplitudes is given in Fig. 3d. It can be seen the doxorubicin uptake level correlates well with the amplitude from FFT spectrum. The results indicate the electrochemical noise could provide quantitative information for drug uptake at the single cell level at early time points, which does not require any pre-labelling. This capability is particularly meaningful to quantify the uptake and effect on cells of drugs that are not fluorescent as we showed with monensin above.
Influence of the cell type and doxorubicin exclusion level on the impedance noise
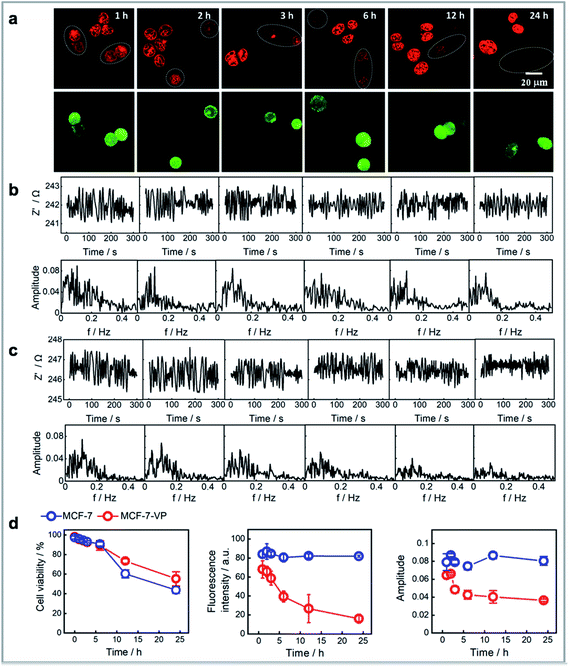
We next sought to show that the FFT amplitude signal from single cells could be used as a measure of the tolerance of different cells to the drug. Although doxorubicin has been a valuable clinical antineoplastic agent in recent years, cell resistance to the drug can limit its efficacy. Thus, we compared the response of normal drug sensitive MCF-7 cells to doxorubicin over time versus that from multidrug-resistant MCF-7 cell line (MCF-7–VP).46 The MCF-7–VP cells were developed by stepwise selection of MCF-7 cells to etoposide (VP) and are cross-resistant to doxorubicin. The MCF-7–VP cells were analysed using the single cell impedance sensing system before and after loading with doxorubicin. Fig. S6† summarizes a set of comparisons between the two cell lines that include the fluorescence images, the fluorescence intensities from doxorubicin on these cells, the amplitudes at the dominant frequencies from the FFT spectrum of impedance signals, and the relationships between fluorescence intensity and amplitude. We can see the accumulation levels for the two MCF-7 cell lines are quite similar immediately for the two cell types after a 30 minute exposure to doxorubicin (Fig. S6a†). There was a similar linear dependence trend between the fluorescence intensity and amplitude observed for the two cell lines (Fig. S5b†). The box plots for the fluorescence intensity and amplitudes (Fig. S5c†) clearly represent these similarities, which suggests the magnitude of the impedance or the noise cannot identify a doxorubicin resistant MCF-7 cell from drug sensitive cells over the short time scales of this measurement. Note, in the case of both cell types, the cells are exposed to the concentration of doxorubicin, and as the uptake of doxorubicin is known to be similar in resistant and sensitive cells, the noise is expected to be similar.The mechanism for the drug resistance by MCF-7 cells is complex. It is accepted that the mechanism of doxorubicin resistance involves ABCB1 (MDR1, Pgp),47 ABCC1 (MRP1), and other transporters (ABCC2, ABCC3, ABCG2, and RALBP1).48,49 These identified transmembrane pumps are responsible for doxorubicin efflux from cells. Thus, it is in the efflux of doxorubicin that differentiates the drug resistant from the non-resistant cells. Consequently, we designed the experiments to see whether the impedance can reflect the effects of these transporters. The doxorubicin sensitive MCF-7 and resistant MCF-7–VP cells were treated with doxorubicin and left in doxorubicin free cell media for 1 h, 2 h, 3 h, 6 h, 12 h and 24 h after exposure to the doxorubicin followed by plating both cell lines onto the same modified silicon electrode surface. Subsequently single cell impedance was performed on these cells. To differentiate the two cell lines on the same surface, we labelled MCF-7–VP using a green-fluorescent calcein that causes no change in the impedance or noise. From the fluorescence images in Fig. 4a, one obvious observation is that the doxorubicin accumulation levels for MCF-7–VP cells are lower than for the MCF-7 cells, and this difference becomes more pronounced with the time after exposure to doxorubicin from 1 h to 24 h. The results show that the MCF-7–VP cell line has the ability to transport doxorubicin out of the cell more effectively than the MCF-7 cells, as expected, such that there are lower amounts of the drug in the MCF-7–VP cells.50 The FFT amplitude spectrum from the impedance signal correlates with these fluorescence microscopy results (Fig. 4b and c). For the MCF-7–VP cells, the amplitude at the dominant frequency decreased with increased doxorubicin exposure time, reflecting the doxorubicin effluxing ability of MCF-7–VP cells and the lower amounts of doxorubicin being retained in the cell (Fig. 4c). This decrease in amplitude was particularly evident between hours 2 to 5, but as with the fluorescence from the doxorubicin, it continued to decline over the next 20 h. Note, neither the fluorescence intensity, nor the amplitude changed significantly over the same time period for the MCF-7 cells. The magnitude of the amplitude from MCF-7–VP cells was lower than that from MCF-7 cells 2 h after drug loading (Fig. 4d). These results suggested the electrochemical noise signal as a measure of the transport of doxorubicin across the plasma membrane of cells and hence quantify the exclusion ability for single MCF-7–VP cells. Thus, the noise measurements can distinguish the drug susceptibility from cells at the single cell levels after only short duration of drug exposure.
Conclusions
We introduced a photo-electrochemical technology for directly assessing single cancer cell activity in real time based on impedance measurement. The system is based on functionalized silicon, on which light can localise electrochemistry to a single cell without any need for electrode patterning or predetermined surface architecture. The light addressable resolution for single-cell level impedance offers important capability that is not readily achieved with current cell–substrate impedance techniques unless using microfabrication. We firstly validated its fundamental capability to follow the attachment and spreading of individual cells. We show the ability to explore variations in the uptake of doxorubicin by single MCF-7 cells, as determined by simultaneous fluorescence microscopy, and how the doxorubicin influenced the spreading and adhesion changes of the cells using single cell impedance spectroscopy. The variation in individual drug uptake and response to this drug, when there is no phenotypic change in the cell is a new capability that could not be resolved by using traditional cell impedance assay that is performed on an ensemble cells level.51 These initial results demonstrate the potential of light addressable single cell impedance to explore cell heterogeneity.As powerful as the single cell impedance measurement that we have developed is, with this method, in common with all impedance methods, the response of the cell to the drug doxorubicin is a downstream response from the drug uptake.12 That is, there is a lag time between the drug uptake and the impedance change which is frequently 30 minutes or more.12 We noticed however in this measurement that the drug uptake was associated with a sudden increase in noise in the impedance signal. This noise was attributed to the flux of ions through the cell membrane as the drug passed into the cell with similar increases in noise when the cells were exposed to the ion antiporter, monensin. Further support for the suggestion that the noise is related to ion-transfer come from the similarity between the noise we observe and that from electrophysiology experiments where ion-channels are voltage stimulated.52,53 In biosensing, electrochemical noise has hardly been explored although it has gained some interest in looking at organic films on electrodes as anti-corrosion coatings where the noise arises from naturally occurring fluctuations in potential and current around a mean value in electrochemical cell.54
By converting this noise from the time domain to the frequency domain, using a fast Fourier transform, the amplitude at the dominant frequency was shown to correlate with the amount of doxorubicin that was taken up by the cell. That is, the FFT signal from the noise in the impedance served as a new type of cell electrochemical impedance measurement that did not rely on changes in cell phenotype but rather was an instantaneous measure of the amount of drug being transported into the cell. Furthermore, the noise signals provide additional information to conventional impedance measurements as we show that the signals are obtained regardless of whether the impedance changes. We compared the noise profiles for MCF-7 cells versus doxorubicin resistant MCF-7–VP cells when there was no change in the real impedance at the single cell level. Initially we observed very similar noise profiles for both cell types. However, within 3 h the amplitude at dominant frequency in the FFT spectrum for the MCF-7–VP cells began to decrease as the doxorubicin was effluxed from the drug resistant MCF-7–VP cells. As expected for the resistant cells, where the transporters to efflux the drug from the cells are upregulated,48,50 the amplitude at the dominant frequency continued to decline as more doxorubicin was effluxed from the cells as determined using fluorescence microscopy. The FFT spectra thus provide complementary and supporting evidence to other experimental studies like imaging results.
The key advance of the experimental design is that many single cells can be interrogated by simply moving the light source from one cell to the next. Hence the heterogeneity of the single cell responses to external stimuli can be investigated. In addition, there is no need to label the cells with a biomarker, which means the approach is not specified to a specific target or cellular pathway, and maximal information including cell spreading and attachment, drug uptake and exclusion behaviors could be gathered to evaluate the effects of drugs. In addition, the changes in the cell recorded by impedance can also be correlated with light microscopy, and even fluorescence microscopy if the drug is fluorescent like doxorubicin. A further advantage is that the cells remain viable throughout the non-destructive measurement, and thus it allows the cells to be studied using other single cell assays, such as single cell genomic analysis. This ability allows for a correlation between single-cell impedance changes and genomic profile, and hence it will be possible to derive a deep understanding of the relationships between cellular responses and genotypes.
Hence the single cell impedance measurement provides a means to monitor differences in changes of cell phenotype as a result of drug exposure of single cells whilst the associated noise in the impedance responses can be used to correlate this change in phenotype with drug uptake. Shown here for a fluorescent drug that can also be visualized via light microscopy, this approach has the potential to be used with non-fluorescent drugs, and hence applied for cell screening prior to the downstream molecular analysis.
Author contributions
Y. Y. performed all experiments, developed the analysis method for the noise data and interpreted the data. F. M. M. provided the idea of the drug resistant cell lined and assisted with developing these cells. M. K. and K. G. helped design the biological experiments and assisted in data interpretation. R. D. T. helped with data interpretation, experimental design and writing of the paper. J. J. G. led the project, was involved in all aspects of the project design and data interpretation. Y. Y. and J. J. G. wrote the manuscript with assistance from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Australian Research Council (ARC) Centre of Excellence in Convergent Bio-Nano Science and Technology (CE14100036), the ARC Australian Laureate Fellowship (FL150100060) and the NHMRC Investigator award (APP1091261 and APP1196648).Notes and references
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 CrossRef CAS.
- I. J. Fidler, Cancer Res., 1978, 38, 2651–2660 CAS.
- S. G. Parker, Y. Yang, S. Ciampi, B. Gupta, K. Kimpton, F. M. Mansfeld, M. Kavallaris, K. Gaus and J. J. Gooding, Nat. Commun., 2018, 9, 2288 CrossRef.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy and A. Muzikansky, Nature, 2007, 450, 1235–1239 CrossRef CAS.
- S. L. Stott, C.-H. Hsu, D. I. Tsukrov, M. Yu, D. T. Miyamoto, B. A. Waltman, S. M. Rothenberg, A. M. Shah, M. E. Smas and G. K. Korir, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18392–18397 CrossRef CAS.
- J. Calbo, E. van Montfort, N. Proost, E. van Drunen, H. B. Beverloo, R. Meuwissen and A. Berns, Cancer Cells, 2011, 19, 244–256 CrossRef CAS.
- N. Navin, J. Kendall, J. Troge, P. Andrews, L. Rodgers, J. McIndoo, K. Cook, A. Stepansky, D. Levy and D. Esposito, Nature, 2011, 472, 90 CrossRef CAS.
- M. A. Colicos, B. E. Collins, M. J. Sailor and Y. Goda, Cell, 2001, 107, 605–616 CrossRef CAS.
- H. M. Davey and D. B. Kell, Microbiol. Mol. Biol. Rev., 1996, 60, 641–696 CrossRef CAS.
- A. Sakaue-Sawano, H. Kurokawa, T. Morimura, A. Hanyu, H. Hama, H. Osawa, S. Kashiwagi, K. Fukami, T. Miyata and H. Miyoshi, Cell, 2008, 132, 487–498 CrossRef CAS.
- J. W. Lichtman and J.-A. Conchello, Nat. Methods, 2005, 2, 910–919 CrossRef CAS.
- M. Parviz, K. Gaus and J. J. Gooding, Chem. Sci., 2017, 8, 1831–1840 RSC.
- I. Giaever and C. R. Keese, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 7896–7900 CrossRef CAS.
- C. Laborde, F. Pittino, H. A. Verhoeven, S. G. Lemay, L. Selmi, M. A. Jongsma and F. P. Widdershoven, Nat. Nanotechnol., 2015, 10, 791–795 CrossRef CAS.
- D. J. Bakkum, U. Frey, M. Radivojevic, T. L. Russell, J. Müller, M. Fiscella, H. Takahashi and A. Hierlemann, Nat. Commun., 2013, 4, 1–12 Search PubMed.
- D. Tsai, D. Sawyer, A. Bradd, R. Yuste and K. L. Shepard, Nat. Commun., 2017, 8, 1–11 CrossRef CAS.
- Y. Wu, J. Lian, V. R. Goncales, R. Pardehkhorram, W. Tang, R. D. Tilley and J. J. Gooding, Langmuir, 2020, 36, 5243–5250 CrossRef CAS.
- Y. Jiang, R. Parameswaran, X. Li, J. L. Carvalho-de-Souza, X. Gao, L. Meng, F. Bezanilla, G. M. Shepherd and B. Tian, Nat. Protoc., 2019, 14, 1339 CrossRef CAS.
- Y. Jiang and B. Tian, Nat. Rev. Mater., 2018, 3, 473–490 CrossRef.
- Y. Jiang, X. Li, B. Liu, J. Yi, Y. Fang, F. Shi, X. Gao, E. Sudzilovsky, R. Parameswaran and K. Koehler, Nat. Biomed. Eng., 2018, 2, 508–521 CrossRef CAS.
- Y. Goda and M. A. Colicos, Nat. Protoc., 2006, 1, 461–467 CrossRef CAS.
- A. Starovoytov, J. Choi and H. S. Seung, J. Neurophysiol., 2005, 93, 1090–1098 CrossRef.
- M. H. Choudhury, S. Ciampi, Y. Yang, R. Tavallaie, Y. Zhu, L. Zarei, V. R. Gonçales and J. J. Gooding, Chem. Sci., 2015, 6, 6769–6776 RSC.
- Y. Yang, S. Ciampi, Y. Zhu and J. J. Gooding, J. Phys. Chem. C, 2016, 120, 13032–13038 CrossRef CAS.
- Y. B. Vogel, J. J. Gooding and S. Ciampi, Chem. Soc. Rev., 2019, 48, 3723–3739 RSC.
- Y. Yang, S. Ciampi, M. H. Choudhury and J. J. Gooding, J. Phys. Chem. C, 2016, 120, 2874–2882 CrossRef CAS.
- Y. Yang, S. Ciampi, Y. Zhu and J. J. Gooding, J. Phys. Chem. C, 2016, 120, 13032–13038 CrossRef CAS.
- M. H. Choudhury, S. Ciampi, X. Lu, M. B. Kashi, C. Zhao and J. J. Gooding, Electrochim. Acta, 2017, 242, 240–246 CrossRef CAS.
- S. Gautam, V. R. Goncales, R. N. P. Colombo, W. X. Tang, S. I. C. de Torresi, P. J. Reece, R. D. Tilley and J. J. Gooding, Chem. Commun., 2020, 56, 7435–7438 RSC.
- Y. Yang, M. Cuartero, V. R. Gonçales, J. J. Gooding and E. Bakker, Angew. Chem., Int. Ed., 2018, 57, 16801–16805 CrossRef CAS.
- W. Wu, C. Jiang and V. A. Roy, Nanoscale, 2015, 7, 38–58 RSC.
- S. Ciampi, P. K. Eggers, G. Le Saux, M. James, J. B. Harper and J. J. Gooding, Langmuir, 2009, 25, 2530–2539 CrossRef CAS.
- J. J. Gooding and S. Ciampi, Chem. Soc. Rev., 2011, 40, 2704–2718 RSC.
- S. Ciampi, M. James, P. Michaels and J. J. Gooding, Langmuir, 2011, 27, 6940–6949 CrossRef CAS.
- J. J. Gooding, S. G. Parker, Y. Lu and K. Gaus, Langmuir, 2014, 30, 3290–3302 CrossRef CAS.
- M. Fahlman, S. Fabiano, V. Gueskine, D. Simon, M. Berggren and X. Crispin, Nat. Rev. Mater., 2019, 4, 627–650 CrossRef CAS.
- M. Varshney and Y. Li, Biosens. Bioelectron., 2007, 22, 2408–2414 CrossRef CAS.
- W. Strober, Curr. Protoc. Immunol., 1997, 21, A.3B.1–A.3B.2 Search PubMed.
- T. Aas, A.-L. Børresen, S. Geisler, B. Smith-Sørensen, H. Johnsen, J. E. Varhaug, L. A. Akslen and P. E. Lønning, Nat. Med., 1996, 2, 811–814 CrossRef CAS.
- S. Aroui, S. Brahim, M. De Waard and A. Kenani, Biochem. Biophys. Res. Commun., 2010, 391, 419–425 CrossRef CAS.
- B. C. Pressman and M. Fahim, Annu. Rev. Pharmacol. Toxicol., 1982, 22, 465–490 CrossRef CAS.
- A. M. Tartakoff, Cell, 1983, 32, 1026–1028 CrossRef CAS.
- P. Henderso, J. D. McGivan and J. B. Chappell, Biochem. J., 1969, 111, 521–535 CrossRef.
- P. Welch, IEEE Trans. Audio Electroacoust., 1967, 15, 70–73 CrossRef.
- D. E. Smith, Anal. Chem., 1976, 48, 221A–240A CAS.
- E. Schneider, J. K. Horton, C. H. Yang, M. Nakagawa and K. H. Cowan, Cancer Res., 1994, 54, 152–158 CAS.
- U. A. Germann, Eur. J. Cancer, 1996, 32, 927–944 CrossRef.
- L. Suman, M. Anupama, C. Wei Ning and C. Balram, Curr. Drug Metab., 2010, 11, 115–128 CrossRef.
- S. Awasthi, S. S. Singhal, J. Singhal, Y. Yang, P. Zimniak and Y. C. Awasthi, Int. J. Oncol., 2003, 22, 721–732 CAS.
- J. D. Allen, S. C. Jackson and A. H. Schinkel, Cancer Res., 2002, 62, 2294–2299 CAS.
- C. Xiao, B. Lachance, G. Sunahara and J. H. T. Luong, Anal. Chem., 2002, 74, 5748–5753 CrossRef CAS.
- M. A. Messerli, L. P. Collis and P. J. S. Smith, Electroanalysis, 2009, 21, 1906–1913 CrossRef CAS.
- A. J. W. Hartel, P. J. Ong, I. Schroeder, M. H. Giese, S. Shekar, O. B. Clarke, R. Zalk, A. R. Marks, W. A. Hendrickson and K. L. Shepard, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E1789–E1798 CrossRef CAS.
- S. S. Jamali, D. J. Mills and J. M. Sykes, Prog. Org. Coat., 2014, 77, 733–741 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc05489e |
| This journal is © The Royal Society of Chemistry 2021 |