Open Access Article
Open Access ArticleRecent advances in activity-based probes (ABPs) and affinity-based probes (AfBPs) for profiling of enzymes
Haixiao
Fang
a,
Bo
Peng
*b,
Sing Yee
Ong
c,
Qiong
Wu
a,
Lin
Li
 *a and
Shao Q.
Yao
*a and
Shao Q.
Yao
 *c
*c
aKey Laboratory of Flexible Electronics (KLOFE), Institute of Advanced Materials (IAM), Nanjing Tech University (NanjingTech), 30 South Puzhu Road, Nanjing, 211816, P. R. China. E-mail: iamlli@njtech.edu.cn
bFrontiers Science Center for Flexible Electronics, Xi'an Institute of Flexible Electronics (IFE), Xi'an Institute of Biomedical Materials & Engineering, Northwestern Polytechnical University, 127 West Youyi Road, Xi'an 710072, P. R. China. E-mail: iambpeng@nwpu.edu.cn
cDepartment of Chemistry, National University of Singapore, 4 Science Drive 2, 117544, Singapore. E-mail: chmyaosq@nus.edu.sg
First published on 18th May 2021
Abstract
Activity-based protein profiling (ABPP) is a technique that uses highly selective active-site targeted chemical probes to label and monitor the state of proteins. ABPP integrates the strengths of both chemical and biological disciplines. By utilizing chemically synthesized or modified bioactive molecules, ABPP is able to reveal complex physiological and pathological enzyme–substrate interactions at molecular and cellular levels. It is also able to provide critical information of the catalytic activity changes of enzymes, annotate new functions of enzymes, discover new substrates of enzymes, and allow real-time monitoring of the cellular location of enzymes. Based on the mechanism of probe-enzyme interaction, two types of probes that have been used in ABPP are activity-based probes (ABPs) and affinity-based probes (AfBPs). This review highlights the recent advances in the use of ABPs and AfBPs, and summarizes their design strategies (based on inhibitors and substrates) and detection approaches.
Introduction
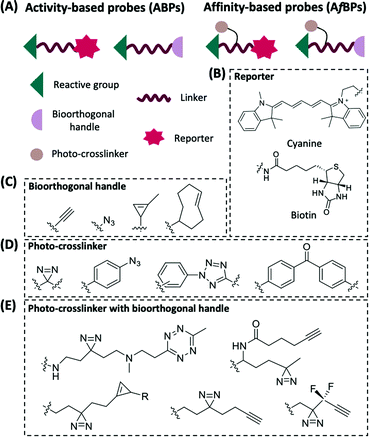
Enzymes are a specialized class of proteins characterized by their remarkable catalytic specificity and efficiency. They represent an extremely important class of biological catalysts that are indispensable for signal transduction and regulation of cellular activities.1 Therefore, understanding the roles of various enzymes involved in mammalian physiological and pathological processes is essential in providing critical information for drug discovery and disease diagnosis.2,3 In order to directly monitor enzymatic activities in cellulo and in vivo, there is an urgent need in the field of proteomics to isolate them from complex biosystems for labeling and enrichment identification purposes.4 Activity-based protein profiling (ABPP) is a chemical proteomic technique that uses small-molecule probes to directly understand the functional state of enzymes in biological systems,5,6 and is gradually becoming one of the major techniques in proteomics due to its ability to fill the gaps of other proteomic approaches.Developed by Benjamin F. Cravatt and Matthew Bogyo in the late 1990s,7,8 ABPP employs activity-based probes (ABPs) to specifically label proteins in biological samples.9 These ABPs are designed to covalently modify the active site of the target enzyme (or a family of enzymes) in a highly selective manner.10 Successful ABPP relies on the critical design and synthesis of the chemical probes. Most ABPs share a similar basic design, which usually includes a reactive group (or warhead, WH), a reporter group, and a linker (Fig. 1A). Reactive groups are often derived from known covalent inhibitors of the target enzyme. Commonly used reporter groups are fluorescent dyes and biotin (Fig. 1B) that allow labeled proteins to be visualized and enriched for subsequent studies. The linker can be a hydrophilic chain, a lipophilic chain or a peptide, in which its basic function is to provide adequate space between the reactive and the reporter groups.
Improper installation of the bulky biotin and fluorescent reporter groups, however, significantly affects the activity and cell permeability of the probes, thereby limiting their applications in living cells. The use of highly selective and reliable bioorthogonal chemistry involving small functional groups is able to overcome the above-mentioned shortcomings.11,12 Commonly used bioorthogonal reactions include copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC),13 copper-free strain-promoted azide–alkyne cycloaddition (SPAAC)14 and inverse electron-demand Diels–Alder (iEDDA)15 (Fig. 1C). This new generation of ABPs carries a suitable bioorthogonal handle, and upon covalent attachment to their targets, a reporter group is then introduced to the labeled targets via bioorthogonal reaction.
In most studies, ABPs are derived from covalent inhibitors of a target protein. For proteins that lack covalent inhibitors, photoaffinity labeling (PAL) offers a way to study their interactions. Since its development by Westheimer et al. in 1962,16 the use of PAL, or photo-crosslinking, has seen a dramatic increase in proteomic research. Upon ultraviolet (UV) irradiation, the photo-crosslinking group generates a highly reactive intermediate that reacts with its adjacent molecule, resulting in the formation of a covalent bond between the probe and the target protein (Fig. 1A). This class of PAL-based probes is also being referred to as affinity-based probes (AfBPs).17 Amongst the various types of photo-crosslinkers, arylazides,18 benzophenones,19 and diazirines20 are most commonly employed in AfBPs (Fig. 1D and E). Recently, a new type of photo-crosslinker, diaryltetrazole (Fig. 1D),21 was developed by our group that has shown a unique photo-crosslinking mechanism and could effectively reduce background labeling with excellent crosslinking efficiency.22,23
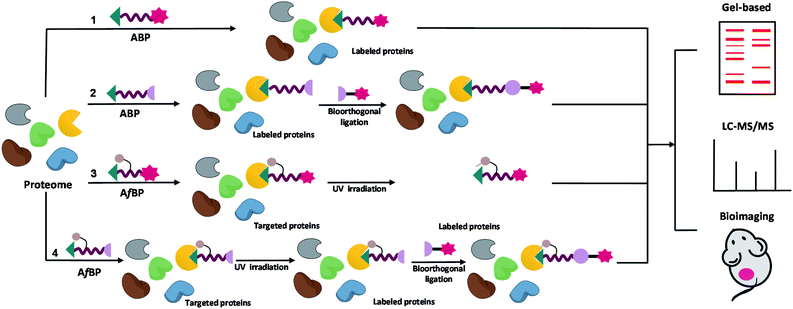
Depending on the types of functional groups on the probes, the workflow of protein profiling could proceed in different ways (Fig. 2). ABPP using ABPs is a covalent-based protein profiling method that is mainly limited by the choice of reactive groups, and therefore is difficult to profile a wide range of proteins. On the other hand, ABPP using AfBPs suffers from non-specific interferences caused by background protein labeling, and therefore is difficult to reliably identify target proteins. In order to overcome these existing limitations, we and others have developed highly effective “minimalist” photo-crosslinkers that could be conveniently introduced into probes and for subsequent use in quantitative mass spectrometry-based proteomics research and highly accurate large-scale target identification (Fig. 1E).24–28
With the rapid development of ABPP in the field of proteomics, an increasing number of advanced ABPP strategies have emerged to expand its applications. Advanced strategies such as isotopic tandem orthogonal proteolysis-ABPP (isoTOP-ABPP),29 fluopol-ABPP high-throughput screening (fluopol-ABPP HTS),30 reverse-polarity ABPP (RP-ABPP)31 and near-infrared quenched fluorescent ABPP (NIRq-ABPP)32 have since been applied for protein active-site identification, ligand discovery and tissue imaging.
In this review, we summarize the recent advances (from 2016 to present) made in the use of inhibitor- and substrate-based ABPs and AfBPs for proteome-wide enzyme profiling. Their probe designs and applications in drug target discovery and biological studies of enzyme function are also discussed. Finally, we end this review by describing our perspectives and proposing future developments of ABPP.
Inhibitor-based ABPs and AfBPs for enzymes
Enzymes play a critical role in physiological functions such as enabling intricate processes of cell metabolism to proceed in an orderly fashion and maintaining cell metabolism at normal physiological functions. Enzyme overexpression has been widely observed in many diseases.33,34 The inhibition of disease-related enzymatic activities, therefore, offers great therapeutic opportunities. For this reason, tremendous efforts have been devoted to the development of enzyme inhibitors.35,36 The application of ABPP by using probes which are designed on the basis of enzyme inhibitors could allow the rapid identification of potential off-targets, thereby providing important and valuable information for drug discovery and optimization. In addition, these probes could also help to delineate the intracellular distribution of target enzymes and discover their novel biological functions.Kinase inhibitor-based probes
Kinases are an essential class of enzymes critically involved in most signal transduction pathways. Over the years, many human malignancies have been found to be associated with the dysfunction and/or dysregulation of kinases. Consequently, many kinases have been attractive targets of drug discovery and development for years.37 The use of known kinase inhibitors as the basis in the design of kinase-based ABPs is a widely used strategy.Bruton's tyrosine kinase (BTK) plays an important role in tumorigenesis and it is essential for the survival of various B-cell malignancies including leukemia.38 The BTK inhibitor, ibrutinib, inhibits BTK by forming a covalent bond with a Cys residue in the active site of BTK.39 Chen et al. reported a BTK-selective ABP 1 based on ibrutinib (Fig. 3A).40 By using 1 in an enzyme-linked immunosorbent assay (ELISA) method, the authors were able to use the probe for quantification of endogenous levels of BTK from living cells. Similarly, Wang et al. reported a series of ABPs having different bioorthogonal handles, which were constructed by conjugating maleimide-coumarin to ibrutinib.41 Upon in vitro and in situ screenings, the authors found that one of the four ABPs (2, Fig. 3A) displayed high sensitivity and selectivity towards BTK (Fig. 3B).
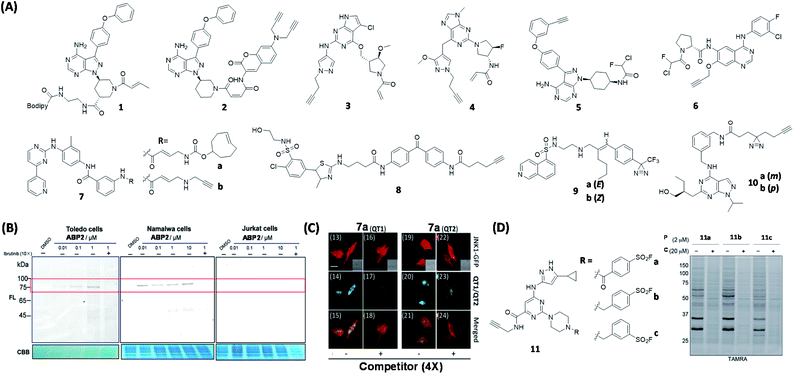 | ||
| Fig. 3 Kinase inhibitor-based ABPs and AfBPs and their in cellulo applications. (A) Reported kinase inhibitor-based ABPs and AfBPs. (B) Gel-based labeling of BTK with 2 in BTK-positive (Toledo, Namalwa) and BTK-negative cells (Jurkat), only positive cells showed a band around 75 kDa which demonstrated the successful labeling of cellular BTK. Reproduced from ref. 41 with permission from Royal Society of Chemistry, copyright 2019. (C) Imaging of JNK1-GFP-transfected HeLa cells using 7a with two TP reporters, both of which successfully labeled JNK1 in living cells. Reproduced from ref. 48 with permission from Royal Society of Chemistry, copyright 2019. (D) Reported sulfonyl fluoride-based ABPs for kinases (left). In-gel fluorescence analysis of Jurkat cells treated with either DMSO or the probe competitor, followed by treatment with 11a to 11c. 11 was successfully used for broad-spectrum kinase labeling in cells. P = probe, C = competitor. Reproduced from ref. 53 with permission from American Chemical Society, copyright 2017. | ||
Epidermal growth factor receptor (EGFR) is a member of the receptor tyrosine kinase (RTK) family. Mutations and overexpression of EGFR are closely associated with tumor pathogenesis.42 In 2017, Planken et al. developed a new EGFR inhibitor (PF-06747775)43 based on PF-06459988, an irreversible EGFR T790M mutant inhibitor. ABPs 3 and 4 (Fig. 3A) were designed on the basis of two EGFR inhibitors, PF-06747775 and PF-06459988. The proteome-wide reactivity of 3 was demonstrated to be lower than that of 4 by spectroscopic experiments, highlighting the superior specificity of 3. The results also indicated that, in addition to the T790M mutant of EGFR, 3 displayed a higher inhibitory effect on both exon19-deletion (Del) and L858R mutants when compared to the wild-type EGFR.
A number of proteomic studies have shown that several types of acrylamide-based kinase inhibitors exhibited off-target protein labeling in the submicromolar to micromolar concentration ranges.44 For example, ibrutinib was found to inhibit other members of the Tec kinase family that contain a critical Cys residue in the kinase domain.45 This off-target activity often results in unwanted toxicity. Ojida's group synthesized a series of ABPs by attaching α-chlorofluoroacetamide (CFA) as a WH, together with a handle, to two kinase inhibitors, ibrutinib and afatinib (EFGR inhibitor), giving probes 5 and 6 (Fig. 3A), respectively.46 Subsequent proteome-wide protein profiling experiments indicated that 5 and 6 were able to inhibit BTK and EGFR, respectively, with low off-target activity. These results thus demonstrate the potential of using the highly specific CFA as a new covalent inhibitor WH.
c-Jun N-terminal kinase (JNK) is critically involved in many important stress signaling pathways, and its dysregulation is closely associated with many pathological states.47 Inspired by the discovery of the first irreversible JNK inhibitor (JNK-IN-8) having nanomolar inhibitory activity and high selectivity, our group developed JNK ABPs 7a & 7b (Fig. 3A) based on the structure of JNK-IN-8.48 We subsequently carried out proteome-wide reactivity profiling, as well as live-cell bioimaging (Fig. 3C), with these probes together with different two-photon (TP) fluorescence reporters, to detect endogenous JNKs from live mammalian cells.
By using a benzophenone photo-crosslinker, Desrochers et al. synthesized AfBP 8 (Fig. 3A) for phosphatidylinositol 4-kinases B (PI4KB) based on the PIK inhibitor, PIK93.49 PI4KB, which belongs to the PIK family, can be recruited to the Golgi membrane for activation and subsequent phosphorylation through protein–protein interactions.50 The authors used 8 to study the role of PI4KB in hepatitis C virus (HCV) infection by simultaneously tagging PI4KB-interacting Golgi recruitment protein, ACBD3. This work demonstrates the use of AfBP as a tool to study the role of ribonucleic (RNA) viruses via the disruption of PIK regulation.
In another study, Overkleeft's group reported AfBPs 9a & 9b (Fig. 3A) based on the protein kinase A (PKA) inhibitor, H89.51 Gel-based experiments revealed that 9a having a (E)-configuration (same as that in H89) could effectively label both PKA and protein kinase B-α (AKT1). In contrast, 9b having a (Z)-configuration had a diminished AKT1 inhibition/labeling effect. This study shows that there is a peripheral difference between the two kinases, hence providing useful information for the future design of AKT1-selective molecules.
Recently, Bush et al. developed a set of pan-/cyclin-dependent kinases (CDK) AfBPs based on the pan-CDK inhibitor, roscovitine.52 The optimal probes 10a & 10b (Fig. 3A) containing an alkyl diazirine were modified to include an alkyne bioorthogonal handle, and 10b was found to enrich 5 CDKs as well as 12 other kinases.
Sulfonyl fluoride is an electrophilic functional group that is resistant to hydrolysis and can react with nucleophilic sites found in a variety of proteins. Taunton's group designed a series of probes (11, Fig. 3D) based on a pyrimidine 3-aminopyrazole scaffold bearing different phenylsulfonyl fluoride substituents.53 By performing proteome-wide screening, the authors found that 11 could covalently label a broad range of intracellular kinomes with high efficiency. The optimized ABP 11b was able to efficiently compete with high intracellular concentrations of adenosine triphosphate (ATP) in kinase binding, and covalently modify up to 133 endogenous kinases (Fig. 3D). More recently, other sulfonyl fluoride-based probes have also been reported, capable of Lys-selective covalent labeling of proteins such as eIF4E (eukaryotic initiation factor 4E) and Hsp90 (a heat shock protein).54,55
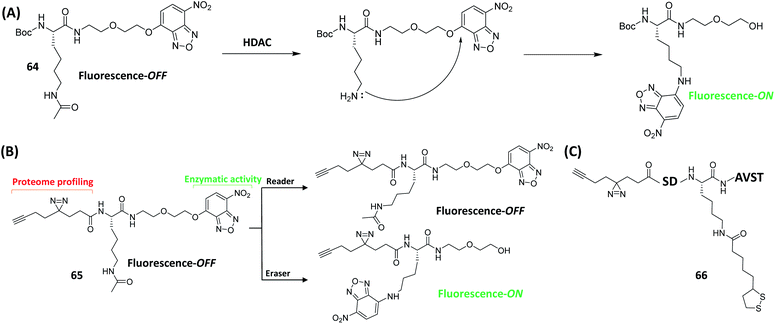
Protease inhibitor-based probes
Abnormal protease activities have been implicated in the pathogenesis of many human diseases such as cancer, osteoporosis and arthritis.56 As a result, proteases represent another class of biological targets that have received much attention in recent years. The very nature of the enzymatic reaction (i.e. substrate cleavage) endowed by proteases provides unique opportunities to the development of probes for various purposes, including proteome-wide profiling and imaging of protease activities. Based on this, the concept of quenched fluorescent activity-based probes (qABPs) was first introduced by Bogyo in 2005 (Fig. 4A).57qABPs belong to a sub-class of ABPs, and are usually composed of three parts: a fluorophore, a fluorescence quencher and a protease inhibitor. We designed another series of modular qABPs, which contains a mandelic acid core surrounded by a fluorophore, a quencher and an enzyme substrate WH (Fig. 4B).58 When the probe is introduced into the cell, its WH is enzymatically cleaved leading to the release of the quenched moiety that can sensitively report endogenous enzyme activity through three possible pathways of achieving fluorescent signal amplification. Among them, pathway 3 is the most efficient way (Fig. 4B). As pathway 3 causes the release of a large amount of quencher and highly fluorescent intermediate to diffuse out of the active site of the enzyme, while allowing the labeling of nearby available nucleophiles, such as proteins that are in the same subcellular organelle as the target. The unique feature of such probes is that they emit fluorescent Turn-ON signals only upon the covalent attachment to the target enzyme via specific enzymatic reaction. Therefore, this design allows no-wash bioimaging of the activity and cellular localization of target enzyme both in vitro and in vivo, and with high resolution.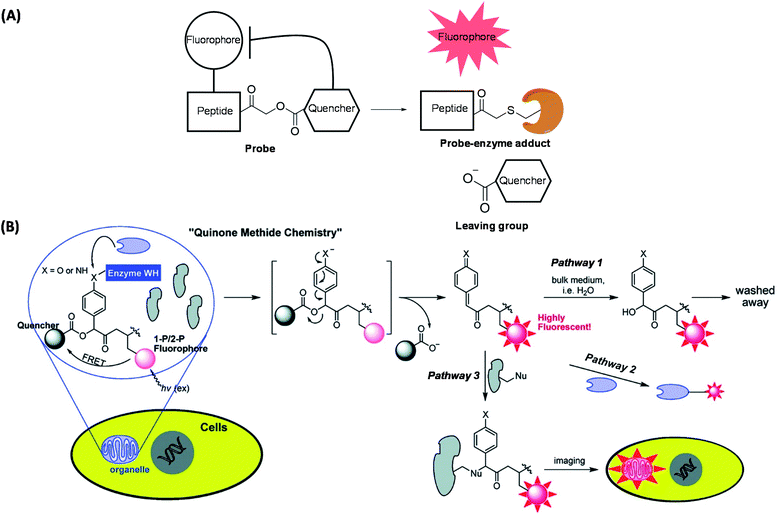 | ||
| Fig. 4 Two qABP working strategies. (A) Schematic illustration of activity-dependent fluorescent labeling of Cys proteases by using qABP. (B) Another strategy of the enzyme activity-dependent qABP reaction. The structure of qABP contains a mandelic acid core which was surrounded by a fluorophore (red), a quencher and an enzyme WH (blue). The resulting intermediate of enzymatic reaction has three possible pathways within the cells: (pathway 1) hydration/quenching by water; (pathway 2) covalent attachment to the target enzyme; (pathway 3) accumulation in the organelle where the enzymatic reaction occurs. Reproduced from ref. 58 with permission from American Chemical Society, copyright 2011. | ||
Recently, Bogyo's group extended the concept of qABP by using protease substrates as parts of the probe design (Fig. 5A);59 the authors developed multivariate “AND-gate” probes containing specific protease substrate sequences which were shown to improve the overall tumor selectivity. These probes (12 & 13, Fig. 5B) will only be activated to produce a fluorescent signal upon the cleavage of both substrate sequences by their respective proteases and were subsequently used to highlight tumor margins in multiple mouse models of cancer.
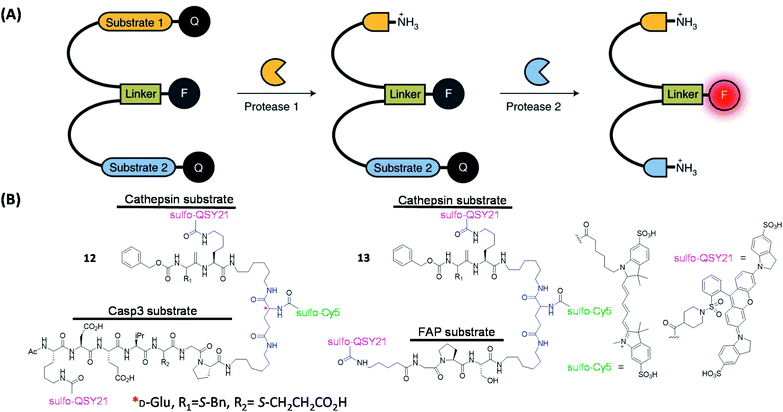 | ||
| Fig. 5 The working strategies and structures of the “AND-gate” probes. (A) Schematic illustration of an AND-gate fluorescent probe. Q = quencher, F = fluorophore. Reproduced from ref. 59 with permission from Springer Nature, copyright 2020. (B) Structures of reported AND-gate probes. | ||
Several protease-targeting ABPs have recently been used successfully in animal models for various biological and medical applications including in image-guided surgical removal of tumors. Caspases and cathepsins are important proteases found in all animals and other organisms. Caspase-3 (Casp3, a Cys protease) is one of the key caspases that play a major role in cell apoptosis.60 Cathepsin B, is another key Cys protease known to be closely associated with tumor infiltration.61 Previously reported Casp3 probes having a DEVD-based sequence interacted with both cathepsin B and legumain (also a Cys protease).62,63 In an effort to improve target specificity amongst these three Cys proteases, Blum's team performed further probe optimization by using sequential screening to identify highly specific Casp3 ligands.64 Trp and Phe were reported to have weak legumain interactions,65 hence peptides were synthesized to include EWD (Glu–Trp–Asp), EPD (Glu–Pro–Asp) and EFD (Glu–Phe–Asp) sequences. Different lengths of diamino linkers and various quenchers were conjugated in the prime site of LE28.63In vitro inhibitory activity assays indicated that the replacement of P2 Pro with Phe decreases the binding affinity to legumain but retains the activity against Casp3. Subsequently, the EFD sequence was used to develop qABPs. The bulky quencher (BBQ) was attached to the position close to acyloxymethyl ketone group (AOMK, a well-known Cys protease-targeting WH) to cause a spatial conflict with the main site of cathepsin B. Upon proteome-wide profiling, the resulting qABP 14 (Fig. 6A) was found to be virtually free of labeling by both cathepsin B and legumain (Fig. 6B).
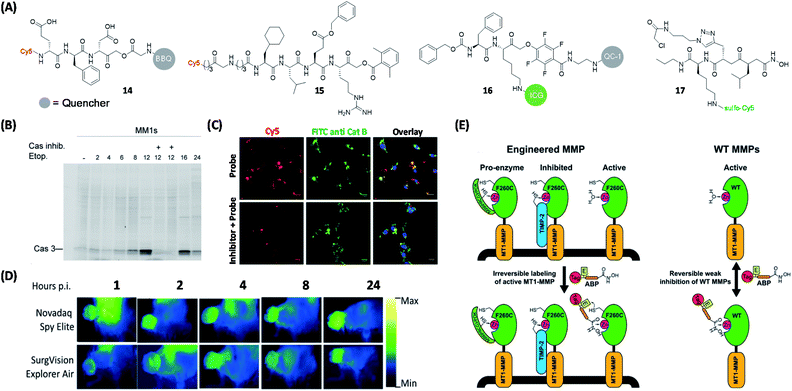 | ||
| Fig. 6 Protease qABPs for visual labeling of different proteases in live cells and mouse tumors. (A) Structures of reported ABPs to study proteases. (B) In-gel fluorescence analysis of Casp3 in MM1s cells treated with 14. The addition of Casp3 inhibitor blocked the labeling of Casp3 by the probe, which demonstrated the specificity of the probe to Casp3. Reproduced from ref. 64 with permission from Royal Society of Chemistry, copyright 2016. (C) Labeling of cathepsin B in non-small lung cancer cells from patient using 15 and anti-cathepsin B FITC antibody. The labeling of cathepsin B using 15 was blocked by the addition of cathepsin B inhibitor, but the inhibitor did not influence the labeling efficiency of the antibody. Reproduced from ref. 66 with permission from Royal Society of Chemistry, copyright 2019. (D) Still images from Novadaq Spy Elite and SurgVision Explorer Air of 4T1 tumors at different time points after injection of 16, which showed the potential of 16. Reproduced from ref. 69 with permission from Springer, copyright 2020. (E) Specific labeling of active MMP-14 using protein engineering coupled with 17. The ABP only labeled active MMPs and cannot label enzymes that are inaccessible to zinc or their inhibitory forms. Reproduced from ref. 75 with permission from American Chemical Society, copyright 2018. | ||
Protease substrate screening in the design of selective probes is a powerful tool for many targets of interest. In 2019, Salvesen et al. employed two peptide-based chemical libraries for dissecting the preferences of cathepsin B at P1, P2, P3 and P4 positions.66 One of the AOMK-based inhibitors that exhibited the highest selectivity in the screen was further functionalized with a Cy5 dye to generate a highly selective ABP for cathepsin B (15, Fig. 6A). The authors further demonstrated that 15 could selectively label cathepsin B in an array of 18 human cancer cell lines and non-small lung cancer cells from patients (Fig. 6C).
Based on a previously reported synthesis of depalmitoylase-specific fluorogenic peptide library,67 Bogyo and coworkers recently reported a similar method that could be used to screen for tumor extract-specific sequence by using a hybrid combinatorial substrate library called HyCoSuL.68 By identifying the different combinations of natural and non-natural amino acid residues, “hits” peptide sequences were subsequently taken to construct the corresponding ABPs for tumor-associated enzymes (e.g. proteases).
Recently, Popkin's group also developed a tumor-visualization platform by using fluorescent probes in human keratinocyte carcinoma excision specimens.69 By using a near-infrared protease-targeting qABP that was previously reported by Bogyo et al.,70 the authors successfully applied the platform for rapid and convenient global assessment of margins during skin cancer resection. Suurs et al. successfully utilized cathepsin qABP for surgical guidance (16, Fig. 6A).71 This new qABP was synthesized by modifying a previously reported cathepsin qABP;72 by replacing the original fluorophore and the quencher with an FDA-approved indocyanine green (ICG) dye and QC-1, respectively. The resulting probe was found to be specifically activated by cathepsin X, B/L and S. With the help of a combination of different optical fluorescence imaging camera systems, probe 16 was shown to be able to delineate tumor tissues with high precision during surgery (Fig. 6D).
As a type of zinc-dependent endopeptidases, matrix metalloproteinases (MMPs) play a critical part in cancer pathology.73 Among them, MMP-14 (MT1-MMP) is considered the main protease involved in the transformation of tumor cells to invasive carcinoma. Based on a previous work,74 Bogyo's team designed ABP 17 based on a new isobutylsuccinylhydroxamic acid motif (Fig. 6A). ABP 17 was engineered to orthogonally bind to an engineered MMP that contained a Cys residue near the active site of the enzyme (having an active site-bound zinc; Fig. 6E).75 In addition, the probe has a reversible weak inhibition of WT MMPs. From protein profiling and cell imaging experiments, the authors found that, in the tumor tissue microenvironment, activation of MMP14 was mainly associated with advanced tumors, their surrounding and activated stromal cell populations.
Carbohydrate inhibitor-based probes
Glycosidases (or glycoside hydrolases) are a class of enzymes that hydrolyze glycosidic bonds and play important roles in protein post-translational glycosylation in various organisms.76 The main hydrolysis product of glucosidases is glucose, which is an indispensable part of glucose metabolism. Depending on the type of glycosidic bonds they hydrolyze, glucosidases can be classified as α- or β-glucosidases. Based on the configuration of the heterotrimeric carbon in the hydrolyzed glucosyl group, glucosidases can also be divided into retaining and inverting glucosidases.77 Most glucosidases are retaining glucosidases and their catalysis usually involves a two-step mechanism mediated by key catalytic residues including a nucleophilic residue and a pair of general acid/base.78 Their catalytic reaction proceeds through a covalent glycosyl–enzyme intermediate, in which the carbon configuration of the glycosyl molecule is flipped twice. On the other hand, such conformational secondary flipping does not occur in inverting glucosidases, leading to formation of products with different configurations.79 To target retaining glycosidases, capturing the covalent intermediates of glycosidic reactions thus became the basis in the design of most inhibitors and ABPs (18–22, Fig. 7A), with suitable reporter groups that have been successfully used in various probe designs (Fig. 7B).80–86 Cyclophellitol-derived epoxides and aziridines are common inhibitors of retaining glucosidases. Upon binding to the retaining glucosidases, their catalytic nucleophile attacks the epoxide/aziridine and generates covalent enzyme-inhibitor adducts.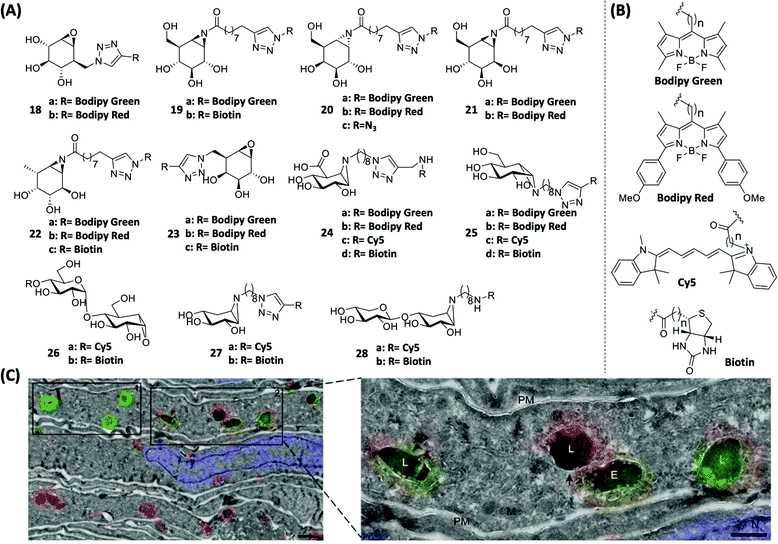 | ||
| Fig. 7 Inhibitor-based ABPs for glycosidases and labeling of GBA and hrGBA in human tissues by using ABPs. (A) Structures of cyclophellitol-derived epoxides- and aziridines-based ABPs. (B) Structures of various reporter groups used in ABPs. (C) Overlay of electron micrographs and confocal fluorescence images of NHDF ultrathin cryosections expressing Man-R. The metabolic processes of hrGBA and endogenous GBA in vivo were traced using 18. 18a-labeled hrGBA (green), 18b-labeled endogenous GBA (red) and nuclei labeled with DAPI (blue). E = endosome; L = lysosome. M = mitochondria; PM = plasma membrane. Reproduced from ref. 90 with permission from Wiley-VCH GmbH, copyright 2019. | ||
In 2010, Witte et al. reported ABPs 18 that could label glucocerebrosidase (GBA) in situ for the first time by linking reporter groups to cylcophellitol.86 Such a class of ABPs exhibited superior properties over the previously widely used tagged deoxyfluorosugars.87,88 Therefore selective labeling of a specific type of glycosidase was achieved. The loss of enzymatic activity in GBA is known to cause lysosomal storage impairment, eventually leading to Gaucher disease.89 Human recombinant GBA (hrGBA) is currently an approved protein-based drug for the treatment of Gaucher disease. Recently, Meel et al. successfully labeled hrGBA and endogenous GBA by using 18a and 18b, respectively, in normal human dermal fibroblasts (NHDFs) which express the mannose receptor (Man-R) (Fig. 7C).90 Galactosylceramidase (GALC) is a glycosidase mainly located in the lysosome and is responsible for galactose ceramide hydrolysis. A deficiency in GALC causes Krabbe disease.91 In 2017, Overkleeft's group reported ABPs 23 (Fig. 7A) for the specific labeling of GALC.92 These ABPs consist of the β-galactopyranose-configured cyclophellitol-epoxide core that is responsible for conferring specificity to GALC. ABP 23b was used in the in situ imaging of active GALCs in mouse brains. Based on a previous finding that substituting the C8 position of cyclophellitol with bulky groups was effective in generating GBA-selective inhibitors,93 Wu et al. recently reported β-glucuronidase-specific ABPs 24 (Fig. 7A), which allowed rapid and quantitative visualization of exo-acting β-glucuronidase (GUSB) and endo-acting heparanase (HPSE) from human tissue lysates.94 One of the best-performing probes, 24c, was developed by introducing a spacer between the Cy5 dye and aziridine nitrogen, and oxidizing the C6-equivalent position of cyclophellitol to mimic the carboxylate group in glucuronide (GlcUA).
Lysosomal α-glucosidase (GAA) is a retaining α-glucosidase, low levels of GAA expression are known to cause glycogen storage disease type II, also known as Pompe disease.95 Overkleeft's group found that α-glucose-configured nitrogen-substituted cyclophellitol aziridines could specifically label GH31 retaining α-glucosidases.96 Therefore, a set of fluorophore- or biotin-modified ABPs (25, Fig. 7A) were designed on the basis of α-glucose-configured N-alkyl cyclophellitol aziridines for successful monitoring of GH31 retaining α-glucosidase. Amongst the various ABPs, 25a was shown to be able to quantitatively report the endogenous level of GAA in cell lysates of patients diagnosed with Pompe disease. Recently, the same group also found that ABPs derived from maltose analogues (26, Fig. 7A) showed excellent labeling specificity towards retaining α-amylases. These probes thus could potentially be used for the screening of α-amylase inhibitors.97
In 2019, Schröder et al. detected and differentiated β-xylosidases and endo-β-1,4-xylanases in the secretome of A. niger by using ABPs 27 and 28 (Fig. 7A), respectively.98 Fluorescence labeling and proteomic analysis revealed that A. niger secreted different catabolic enzymes according to the carbon source used in its growth process.
ABPP has also been used as a tool to study gut flora. Whidbey et al. developed a platform based on β-glucuronidase-selective ABPs to detect and identify subsets of microorganisms in the gut that are responsible for heterologous biological metabolism.99
Proteasome inhibitor-based probes
Proteasomes degrade unwanted and damaged proteins, therefore they are the key factor of cellular protein concentration regulation, and the function of the proteasomes is closely related to the development of cancer and neurodegenerative diseases.100 In 1997, Bogyo and Ploegh developed radioactive ABPs that can label proteasomes.101 Later, Crews and coworkers developed an ABP by attaching the anti-cancer drug eponemycin with biotin. The proteome study revealed that proteasomes were the main targets of eponemycin.102 In 2003, Ovaa et al. reported a two-step labeling strategy for proteasomes in living cells.103 By attaching three proteasome subunit inhibitors with different fluorescent groups, the same group reported three proteasome ABPs that target different subunits (29, 30 and 31, Fig. 8A).104 Using a mixture of the ABPs (ABP-cocktail), they achieved simultaneous gel-based detection of human constitutive core particles (cCP) and immunoproteasome core particles (iCP) proteasomes (β1c, β1i, β2c, β2i, β5c and β5i), which can also be used to distinguish immune cells from non-immune cells.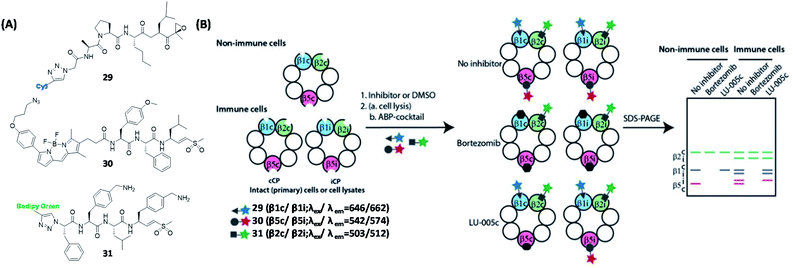 | ||
| Fig. 8 Inhibitor-based ABPs for labeling of six human proteasome subunits and their working strategies. (A) Structures of ABPs that target different subunits of human proteasomes. (B) Schematic representation of ABPP using proteasome ABP mixture. Visual gel analysis of six human proteasome subunits was achieved using three ABP mixtures. Reproduced from ref. 104 with permission from Wiley-VCH GmbH, copyright 2016. | ||
This work, together with studies of glycosidase ABPs, have shown that sometimes it is possible to derive probes selective for a single enzyme or a small closely related family of enzymes.
Other enzyme inhibitor-based probes
Deubiquitinases (DUBs) are a large group of proteases that catalyze the cleavage of ubiquitin (Ub) from proteins. Due to its important biological functions in the regulation of protein degradation, various ABPs have been developed to study the function of DUBs. In 2016, Tate's team discovered a specific inhibitor of the ubiquitin-specific protease (USP) via high-throughput screening. Following this lead, 32 was subsequently designed (Fig. 9A), and shown to be able to identify 12 endogenous USPs from pull-down experiments.105 Recently, the same group also discovered a specific covalent inhibitor of the ubiquitin carboxy-terminal hydrolase L1 (UCHL1) and subsequently synthesized the corresponding ABP 33 (Fig. 9A).106 By using proteome profiling, the authors demonstrated that 33 could block the pro-fibrotic response in a cellular model of idiopathic pulmonary fibrosis. Poly (ADP-ribose) polymerase (PARP) is a family of enzymes present in most eukaryotic cells that are involved in multifunctional protein post-translational modification (PTM).107 Howard et al. developed an AfBP 34 (Fig. 9A) based on olaparib (a potent inhibitor of PARP1) and showed that 34 could effectively label PARP1/2 in live cells.108 They also found that 34 enabled successful and selective labeling and inhibition of recombinant PARP6 in vitro, therefore providing a potential tool for differentiating PARP6 activities from endogenous sources.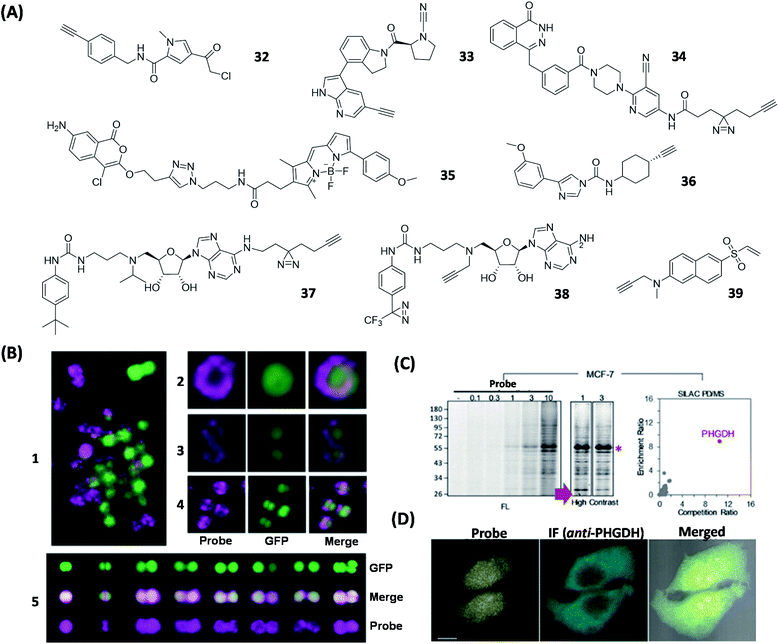 | ||
| Fig. 9 ABPs and AfBPs designed based on specific enzyme inhibitors and their labeling of enzymes in cells. (A) Structures of reported enzyme inhibitor-based ABPs. (B) Confocal images of S. aureus Newman labeled with 35 (1). Confocal images of S. aureus Newman-GFP cell labeled with 35 during exponential phase (2 and 3) and stationary phase (4 and 5). 35 successfully demonstrated that FphB is concentrated in the isolated membrane of bacterial cells. Reproduced from ref. 109 with permission from Springer Nature, copyright 2018. (C) Proteome profiles of MCF-7 cells labeled with 39, with a visible FL band around 55 kDa. These demonstrated the specific labeling of PHGDH by 39 in living cells. (D) Confocal images of MCF-7 cells treated with 39 and anti-PHGDH. Reproduced from ref. 115 with permission from Wiley-VCH GmbH, copyright 2018. | ||
Serine hydrolases play various roles in the regulation of host–pathogen interactions in many organisms.7 In 2018, Bogyo's group successfully identified 10 previously uncharacterized S. aureus serine hydrolases by using competitive ABPP. The new hydrolases were named fluorophosphonate-binding hydrolases (Fph A-J).109 FphB was selected for the use in the next-stage study due to its overall enzymatic activity in cells and high specificity toward a previously reported inhibitor called JCP251. The authors found that, by substituting the C3 methoxyl group in JCP251 with Bodipy-TMR fluorophore to generate ABP (35, Fig. 9A), the resulting probe was able to retain the original inhibitory activity, allowing it to be used as an effective FphB-targeting probe. The proteome reactivity labeling profile of FphB with 35 further showed that FphB activity was mainly concentrated in the separating septum located on the bacterial cell surface (Fig. 9B).
Fatty acid amide hydrolase (FAAH) is an endosomal enzyme that metabolizes many fatty acid amides (FAAs), which are messenger molecules in various living organisms. Severe neurotoxicity was reported in the clinical study of BIA 10-2474 (a FAAH inhibitor). Upon further investigation, Stelt et al. revealed that BIA 10-2474's toxicity was caused by its poor target specificity, which resulted in substantial alterations in the human cortical neuron lipid network.110 To further understand the exact mechanism-of-action that caused BIA 10-2474's toxicity, Cravatt's group employed 36 (Fig. 9A), an ABP designed on the basis of the demethylation metabolite of BIA 10-2474.111 The authors found 36 was able to irreversibly react with the catalytic Cys present in several aldehyde dehydrogenases, including aldehyde dehydrogenases A2 (ALDHA2). Their findings thus suggested that BIA 10-2474's severe toxicity was possibly caused by the off-target inhibition of this compound as well as its metabolites on these important human enzymes, which are required to properly maintain the physiological stability of the nervous system.
DOT1L is a Lys-methylating enzyme. Overexpression of DOT1L leads to cell cycle arrest, as well as promoting differentiation and apoptosis of tumor cells.112 For this reason, DOT1L has become an increasingly important therapeutic target for cancer treatment. Based on a potential DOT1L inhibitor, FED1, which showed good in vitro inhibitory but poor in vivo activity, clickable AfBPs, including 37 and 38 (Fig. 9A), were generated to better understand the cellular activity of FED1 in targeting endogenous DOT1L.113 It was found that 38 displayed higher fluorescence signals in live-cell imaging experiments, whereas 37 showed better performance in most in vitro experiments, indicating that the primary amine in the adenine moiety of FED1 may be important for target binding. Further live-cell imaging studies showed that both 37 and 38 were mostly trapped in the cytoplasm of A431 cells and failed to localize to the nucleus where endogenous DOT1L normally resides. These data serve as the basis to potentially explain the relatively poor cellular activities of FED1 and its derivatives, thus providing clues for future improvement of DOT1L inhibitors.
3-Phosphoglycerate dehydrogenase (PHGDH) is an essential enzyme involved in the serine biosynthetic pathway and has been found to be upregulated in numerous rapidly proliferating cancer cells.114 In order to develop effective ABPs suitable for in situ profiling and live-cell imaging of endogenous PHGDH, molecular docking experiments were first carried out to identify potential small-molecule PHGDH binders. We found that several vinyl sulfone-containing electrophilic phosphotyrosine mimics, including phenyl vinyl sulfone (PVS) and phenyl vinyl sulfonate (PVSN), were well-fitted in the active site of PHGDH, thus presenting a potential electrophilic trap to covalently react with the nearby Cys residue in PHGDH. Further development of these “leads” by taking advantage of the vinyl sulfone's ability to serve as an effective fluorescence quencher,37 led to the discovery of 39 (Fig. 9A), the first-ever fluorescence Turn-ON ABP that could selectively and covalently label endogenous PHGDH from numerous cancer cells.115 This dual-purpose probe was able to simultaneously image and profile endogenous enzymatic activities from live mammalian cells (Fig. 9C and D).
Substrate-based ABPs and AfBPs for enzymes
In addition to designing ABPs and AfBPs based on the inhibitors, another widely used approach employed in the design of enzyme-targeting ABPs and AfBPs involves the chemical modification of natural substrates of the target enzymes. These probes have been dominantly used for the biological function studies of enzymes, while inhibitor-based probes are more often used for drug target discovery. Some bifunctional probes that are composed of two binding units could allow rapid enzymatic activity detection and spectroscopic analysis at the same time.Steviol-based probes
In plants, UDP-glycosyltransferases (UGTs) utilize UDP glucose (UDPG) as a donor to transfer part of the glucose to secondary metabolites, in an event known as glycosylation.116 Steviol glycoside (SG) is a natural sweetener biosynthesized from steviol through a series of glycosylation reactions catalyzed by multiple UGTs.117Xiao's group tagged the “minimalist” photo-crosslinker diazine-alkyne to steviol and synthesized a steviol-derived AfBP 40 (Fig. 10A) to identify various UGTs involved in steviol catalysis. The proteomic profiling by using 40 confirmed that UGT73E1 was involved in the glycoside biosynthesis of steviol.118 To improve the selectivity and sensitivity for the identification and labeling of UGTs in biosynthesis, Wong et al. developed another method to profile UGTs by using a dual-substrate probe constructed from an alkyne-labeled acceptor 41 and a diazirine-labeled donor 42 (Fig. 10A).119 Because none of them have the essential properties of the fully functionalized probes, they will not capture the proteins in proteomics experiments. Upon being catalyzed by specific UGTs, the diazirine-modified moiety in donor 42 was transferred to acceptor 41, thus generating the steviol glycoside-derived probe (Fig. 10B), which, upon subsequent photo-crosslinking, could be used to capture and identify the corresponding UGTs. Using this approach, the authors successfully labeled two glycosyltransferases, UGT73C1 and SrUGT85C2, and showed that both enzymes displayed the same activity against steviol. On the other hand, other UGTs such as SrUGT91D2 and SrUGT76G1, which are involved in steviol glycoside biosynthesis but only catalyze non-steviol glycoside substrates, were not labeled. Compared to previous studies, ABPs generated from this newly developed method showed significantly improved selectivity and sensitivity towards endogenous UGTs.
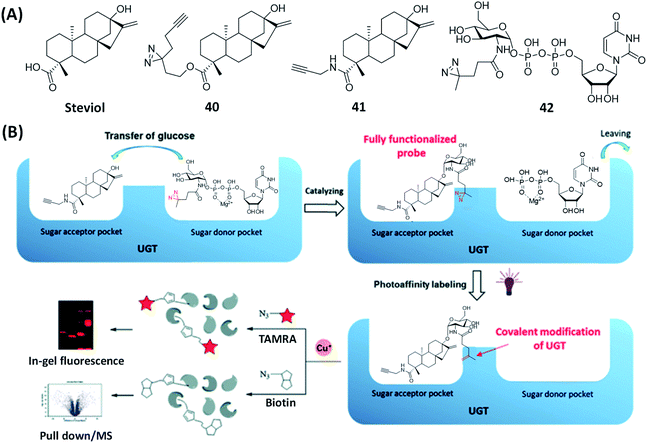 | ||
| Fig. 10 A bisubstrate probe strategy for specific labeling of UGTs. (A) The structures of steviol and the reported substrate-based ABP and AfBPs. (B) Bisubstrate probe strategy for the identification of UGT cellular localization and activities by steviol-based ABPs. Reproduced from ref. 119 with permission from Royal Society of Chemistry, copyright 2020. | ||
Furthermore, they found that this modular dual-substrate-probe labeling strategy had an impact on the catalytic efficiency of natural enzymes. By optimizing the probe structure and improving the combination of modules, it may help to broaden the application of the dual-substrate probe labeling strategy for other enzymes.
Nicotinamide adenine dinucleotide (NAD+)-based probes
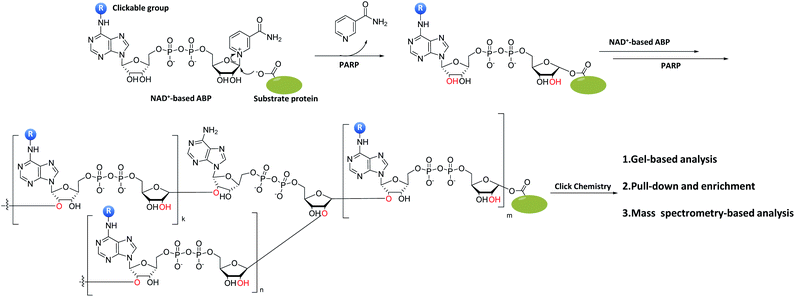
Protein ADP-ribosylation is a PTM in which members of the PARP enzymes covalently attach ADP-ribose moieties from NAD+ to target proteins.120 PARPs are ADP-ribosylating enzymes that comprise at least 18 members. As the first identified and best-studied PAPR family member, PARP1 regulates a variety of biological functions including DNA repair, chromatin reorganization, transcriptional regulation, apoptosis and mitosis.121 Under severe and/or continuous stress, PARP1 is activated to promote the synthesis of poly (ADP-ribose) (PAR) chains.107 The over-activation of PARP leading to rapid depletion of NAD+ in cells could affect various biological processes. It is therefore necessary to be able to precisely monitor this process in biological cells, preferably in real time. One example is the monitoring and study of the biological functions of NAD+-associated enzymes by using synthetic NAD+-based ABPs (Fig. 11). The mechanism of these ABPs is similar to that of NAD+. First, an ADP-ribosyl group binds to the carboxylate side chain of the substrate protein residues, and the process is then repeated with the addition of more ADP-ribosyl groups to the 2′-OH groups of both ribose rings, resulting in the formation of PAR, followed by the enrichment and analysis of PARP substrate proteins by click chemistry. The NAD+ analogue 43 was successfully synthesized by Jiang et al., and subsequently used to identify 79 proteins as potential PARP1 substrates in a large-scale proteome-wide study.122 In another study, Buntz et al. reported two NAD+-based ABPs to study protein PARylation.123 In order to allow PAR activity to be directly monitored in vivo and in real-time, the probes were made to be fluorescently active by conjugating them to a fluorophore (tetramethylrhodamine, TMR) (44, Fig. 12). SDS-PAGE analysis showed that only 44b could fluorescently label PARP1 auto-modification. Due to the low cell permeability of NAD+ analogues, carrier peptide Pep-1 was co-incubated with 44b to enhance the cellular uptake.124 Labeling results obtained using flow cytometry demonstrated that more than 90% of the cells were successfully labeled. The formation of PAR in cells was further confirmed by confocal microscopy; a strong fluorescence increase was observed within a few seconds after the induction of DNA damage. Wallrodt et al. also synthesized a series of NAD+ analogues by modifying the adenine moiety at different sites (45a–d; Fig. 12);125 their results indicated that modifications at position 7 (45d) and 8 (45a) of adenine interfered with the recognition of the probes by their target enzymes. Substitution with a small group at position 6 (45b), on the other hand, appeared to be tolerable. Amongst the various probes developed, 2-modified NAD+ analogues (45c, f, g), which were resistant to chemical modifications (with long and bulky chains), were shown to be the most effective. To explore the function of these probes in live cells, 45g was taken as a representative to label endogenous PAR from HeLa cells. By further extending the studies on the effects of chemical modification of adenine to other PARPs,126 the authors showed that 6-modified NAD+ analogues (45b, b′) could be reasonably recognized by PARP1/2/5/6, while 2-modified analogues (45c, c′) exhibited overall better PARP-targeting activities. Further studies revealed that both PARP1 and PAPR2 were able to process 2-modified NAD+ analogues (45c, c′), but not the 7- or 8-modified ones (45d and 45a, respectively). Collectively, these studies demonstrated that 2-modified NAD+ analogues are the best choices in the construction of PAPR1- and PAPR2-specific ABPs. Tankyrases (e.g., PARP5 and PARP6), on the other hand, displayed a preference for 6-modified NAD+ analogues (45b, b′), making them preferred choices in future design of ABPs for the study of these PARPs.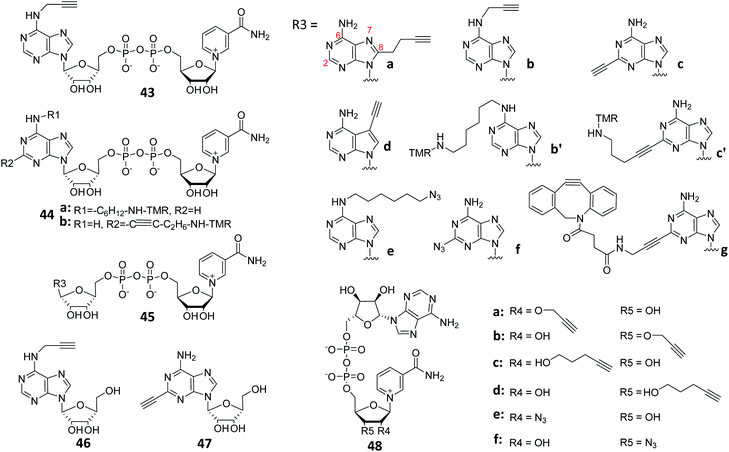 | ||
| Fig. 12 Structures of reported ABPs based on NAD+ by modifying adenine or nicotinamide ribose-OH at different sites. | ||
Multiple metabolic precursors of NAD+ (e.g., NR, NA, NAM and NMN) are known to be well-tolerated by numerous human cells to sustain endogenous NAD+ production,127 suggesting that it is possible to also metabolically generate NAD+-based ABPs in live cells that may be used for subsequent proteomic analysis of ADP-ribosylated proteins. With this in mind, Hang et al. used a previously reported adenine 6-modified NAD+ precursor 46 (Fig. 12),128 and fed it to HeLa cells. Upon successful incorporation into ADP-ribosylated proteins, the authors were able to find that oxidative stress-induced ADP ribosylation of V-Ha-Ras (HRas, a member of Rho GTPases) occurs at the C-terminal hypervariable regions, Cys181 and Cys184.129 This suggests that not only bacterial toxin, mammalian cells alone were able to regulate the ADP-ribosylation of endogenous GTPases.
Recently, Kalesh et al. reported a dual metabolic marker approach for the labeling of substrate proteins in PARylation (Fig. 12).130 By using ABP 46 and adenine 2-modified NAD+ precursor ABP 47, and in combination with tandem mass tag (TMT) isobaric mass spectrometry and hierarchical Bayesian modeling, the authors quantified the response of over a few thousand proteins upon treatment with clinical PARP inhibitors (olaparib and rucaparib).
Different from the aforementioned studies, Zhang et al. investigated the effect of substitution at nicotinamide ribose (NR)-OH and synthesized an array of NAD+ analogues (48, Fig. 9).131 Co-incubation of PARP1 with the NR 3′-OH (48f) caused a strong auto-PARylation of PARP1, and the addition of veliparib (an inhibitor of PARP1/4) effectively inhibited this process. In contrast, NR 2′-OH analogues (48a, c, e) did not show any substrate activity for PARP1. Meanwhile, 48b and 48d exhibited different activities toward PARP1. Kinetic data showed that the Km of 48f was slightly higher than that of NAD+. The Kcat of 48b was significantly lower than those of NAD+ and 48f. Successful monitoring of PARylation in living cells was achieved by using 48f with the assistance of transient permeabilization of cell membranes.132 When compared to the previously reported adenine-modifying NAD+ analogues (45b and 45c), 48f possessed a significantly higher substrate activity against both PARP1 and PARP2 (Fig. 13A and B). Collectively, these results indicate that the NR 3′-OH NAD+ analogue (48f) showed higher activity and selectivity towards PARP1 and PARP2 (Fig. 13C and D).
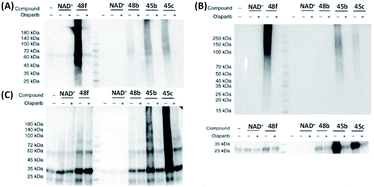 | ||
| Fig. 13 Gel-based analysis of substrate activities of NAD+, 48b, f and 45b, c for (A) PARP1; (B) PARP2; (C) catalytic domain of PARP5a; and (D) catalytic domain of PARP10. All the probes were successfully recognized by the corresponding enzymes, and the addition of PARP inhibitor olaparib efficiently inhibited the interactions between the probes and PARPs. Reproduced from ref. 131 with permission from Springer Nature, copyright 2019. | ||
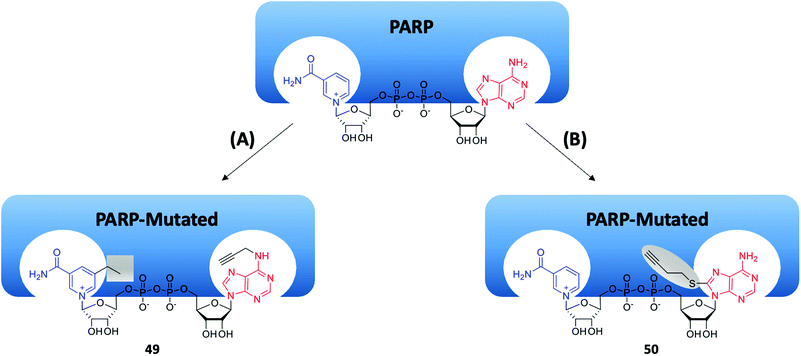
Cohen's group reported a “bump-hole” strategy, which is an additional method for recognizing the enzymatic target protein of PARP1.133 This strategy created an orthogonal pair of NAD+ analogues (49, Fig. 14A) and mutated PARP1 by attaching a bulky motif at the nicotinamide of NAD+ while mutating the “gatekeeper” residue of the PARP1 nicotinamide binding pocket (Fig. 14A), which did not interfere with the normal biological functions of wide-type PARP1. However, this approach diminished the PARylation activity of PARP1 and PARP2, while preserving the MARylation activity. A similar but more elegant strategy was reported in 2016. Instead of modifying the nicotinamide portion, Kraus's team attached an alkyl stalk to the adenine portion of NAD+ (50, Fig. 14B), which served as both a “bump” and a clickable handle for subsequent pull-down experiments (Fig. 14B).134 They successfully identified hundreds of ADP ribosylation sites for PARP1, PARP2 and PARP3, as well as thousands of PARP1-mediated ADP ribosylation sites using this approach.
Recently, Zhang's group designed and synthesized another series of NAD+ analogues (51, Fig. 15A) containing both diazirine-modified adenine and 3-azide-modified ribose.135 These bifunctional probes served as good substrates of PARylation and were able to capture PARylation-interacting proteins, including both readers and erasers, via PAL (Fig. 15B). With probe 51c alone, the authors were able to capture up to 247 possible interacting proteins from cell lysates. Among these proteins, VCP and RBBP7 were selected for further biological validations; in ELISA-binding assays and immunoblotting experiments, both VCP and RBBP7 were shown to specifically interact with PARP1. Using a similar strategy, Cohen's group developed clickable AfBPs (52 and 53, Fig. 15C) based on benzamide adenine dinucleotide (BAD, another NAD+ analogue).136 They identified hundreds of both known and unknown NAD+/NADH binding proteins by using 52 and 53 (261 and 141 proteins were identified from the 52- and 53-treated samples, respectively), which primarily labeled different proteins but also share some common targets. These clickable NAD+-based AfBPs provide new tools for analyzing proteins that interact with NAD+ in different diseases.
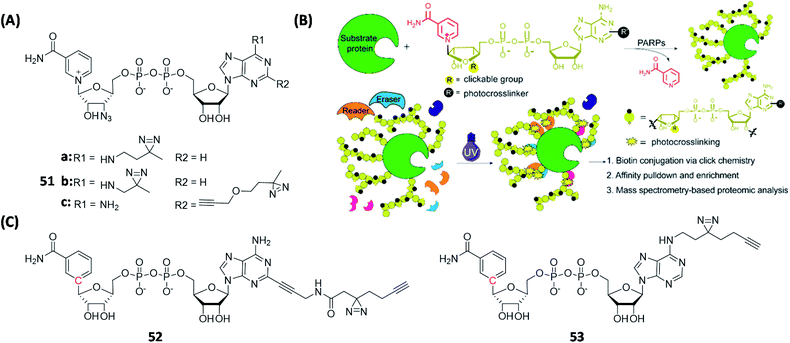 | ||
| Fig. 15 Use clickable AfBPs to capture proteins associated with PARylation. (A) Structures of reported bifunctional NAD+ probes. (B) Schematic illustration of the process for capturing PARylation-related interacting proteins using a bifunctional NAD+ probe. Reproduced from ref. 135 with permission from American Chemical Society, copyright 2021. (C) Structures of reported BAD-based clickable AfBPs. | ||
Ubiquitin (Ub)-based probes
Ubiquitination is an important reversible PTM in eukaryotic cells. It plays important roles in protein localization, metabolism, function, regulation, and degradation.137 Ubiquitination occurs via a three-enzyme-catalyzed reaction which involves E1, E2 and E3 proteins. Ub molecules can be coupled to other Ubs to form poly-Ub chains. By hydrolyzing the isopeptide linkage in ubiquitinated proteins, DUBs remove Ub molecules from their substrates as well as Ub-processing precursors.138 Therefore, Ub-based ABPs and AfBPs are effective tools to monitor the biological functions of key enzymes that are involved in the ubiquitination cycle.Most of the probes used in the characterization of DUBs were designed on the basis of mono- or diUb. Since most DUBs have active Cys residues in their catalytic centers, most reported Ub-based probes contain a thiol-reactive functional group (e.g. a Michael acceptor) that allows for covalent reactions with the Cys residues in DUBs. Strictly speaking, such probes may be considered “inhibitors”, but we simply group them herein as “substrate-based ABPs” for convenience, as all of them were derived from natural substrates of DUBs (i.e. Ub).
DUBs employ multiple Ub-binding pockets to break down polyubiquitin (poly-Ub) chains. Ovaa's team reported a series of non-hydrolyzable protease-resistant diUb ABPs (i.e.54, Fig. 16A)139 for the identification and investigation of specific linkage reactions of DUBs with diverse Ub-binding pockets. These probes covered seven different linkage positions (connected at seven Lys positions of Ub, i.e. K6/K11/K27/K29/K33/K48/K63) and contained protease-resistant triazole bonds as well as a propargyl group at the C-terminus of the adjacent Ub part. In vitro DUB cleavage experiments were used to determine the linkage specificity of the S2 pocket in DUBs. Results showed that OTUD2 was conjugated with K11- and K33-linked diUb, and OTUD3 was bonded with K11-linked diUb in the S1–S2 pocket. Whedon et al. developed two selenoCys (SeCys)-based approaches to introduce DUB-reactive dehydroalanine (DHA) at the C-terminus of Ub.140 Through efficient oxidation or alkylative β-elimination of SeCys, DHA could be selectively conjugated at the C-terminus of Ub where several Cys residues in the target of Ub were present. ABP 55 was designed to target TRIM-25, an E3 ligase tripartite sequencing protein (Fig. 16A). Upon positive interaction between 55 and DUBs, the Cys sulfhydryl group in 55 formed a covalent bond through DHA with the active site of DUBs. The captured DUBs were next identified in a pull-down experiment. These so-called alkylation–elimination DUB-targeting strategies effectively resolved the interference of other Cys residues in the target of Ub, thus providing new chemical biology tools for future design of novel DUB ABPs.
Liang et al. reported two diUb-based AfBPs, 56 and 57 (Fig. 16A), which were able to selectively analyze ubiquitin-binding proteins from cell lysates.141 Different Ub–Ub linkages were found to exhibit practical variations. For example, Lys63-conjugated poly-Ub was the signal for transduction.142 Based on this knowledge, the diUb core in the two designed AfBPs (i.e.56 and 57) were conjugated via Lys48 and Lys63 linkage, respectively, resulting in probes that exhibited different protein selection preferences. In addition, the Ala46 residues of two Ub units were mutated to Cys, which was then attached to photo-crosslinkers including diazine and aryl-azide. With such probes, the authors discovered that at least two Ub units were required to efficiently capture Ub-binding proteins via PAL.
Meledin et al. constructed a ubiquitinated α-globin ABP (58, Fig. 16A), through a semisynthetic strategy based on sequential DHA formation on the expressed proteins.143 α-Globin can be ubiquitinated at multiple sites to facilitate their proteasomal degradation.144 By using large-scale proteomic analysis, the authors discovered that USP15 acted as a potential DUB for the regulation of α-globin, and overexpression of UPS15 exacerbated the thalassemia symptoms.
The Lys27-linked poly-Ub plays a vital role in autoimmunity and DNA repair.145 Based on previous studies,141,146 Tan et al. reported a diUb probe 59 (Fig. 16A) for the identification of DUBs that regulate the synthesis and degradation of K27-linked poly-Ub.147 The probe carried an aryl-azide group as the photo-crosslinker at the isopeptide position, and a biotin tag was attached to the C-terminus of the probe. When compared with the earlier-discussed, DHA-based probes,140,14359 exhibited higher crosslinking activity for Lys27-linkage-targeting DUBs. Pull-down experiments demonstrated that the probes could recognize K27-targeting DUBs present in the crude proteome, a key property which was not possible with earlier DHA-based probes.
Most Ub-based DUB-targeting ABPs, however, had poor cell permeability due to their macromolecular nature. This has severely limited their in cellulo and in vivo applications. In 2018, Zhuang's group reported a cell-permeable DUB-targeting ABP (60, Fig. 16A) that enabled intracellular DUB profiling.148 An HA tag and a cyclic polyarginine (cR10) peptide were conjugated to Ub via a disulfide bond, the HA tag was then attached to the N-terminus of the probe. As a cell-permeabilizing peptide, cR10 enabled the cellular uptake of 60, which was then removed from the probe upon successful cell entry and subsequent disulfide bond cleavage caused by endogenous GSH.14960 was successfully used to monitor endogenous DUBs present in various organelles by fluorescence imaging.
Recently, a tetrazole group which is a new type of photo-crosslinker,150 was attached to the C-terminus of Ub, producing probe 61 (Fig. 16A).151 This probe successfully pulled down and identified fifteen known DUBs, and upon further optimizations, the probe was used to identify specific DUBs proteomes in cells that were grown during G1/S and G2/M phases. By comparing enriched DUBs from the two cell growth phases, five DUBs in G1 phase and one DUB in G2 phase were specifically identified, respectively. Amongst them, BAP1 and USP36 were closely related to the regulation of ubiquitination in the G1 phase as reported previously.152 The doubly enriched DUB USP16, previously known to have a function in regulating protein localization in the G2 phase,153 was also discovered. Using a time-gradient identification assay, the function of this DUB during cell mitosis was further demonstrated.
Mulder et al. developed a cascading ABP 62 (Fig. 16A),154 based on a previous study that the mutation Ub-G76A still retained the ability of being processed through the E1–E2–E3 cascade.155 With this knowledge, the authors hypothesized that partial replacement of this C-terminal Ala of this Ub mutant further with the electrophilic DHA (giving 62) may also preserve the activity of Ub (Fig. 16B). Indeed, 62 was found to be conjugated to the corresponding enzyme in each step during the E1–E2–E3 cascade reaction by two different ways either forming thioether adducts by covalently binding to enzymes (pathway 1), or generating thioester compounds with enzymes via the natural linkage pathway (pathway 2). Therefore, 62 was an excellent ABP that could be used in the study of each step in the E1–E2–E3 cascade reaction.
Virdee's team deduced that the E2–Ub complex could serve as a starting substrate for the analysis of trans-thiolation activities in E3 ligases, i.e. RING-in-between-RING (RBR) and homologous to E6AP carboxy-terminus (HECT). Therefore, ABP 63 (Fig. 16A) was prepared as a non-hydrolytic mimic of E2–Ub, in which the unstable thioester bond was suitably stabilized. An appropriate and dynamically tuned electrophilic reagent was further installed between Ub and E2 as well.156 The resulting 63 was able to successfully monitor the trans-thiolation activity of parkin (a RBR E3 ligase) in vitro from cell lysates, thereby providing a potential tool to directly and quantitatively detect endogenous parkin activity. With the same strategy, the authors were able to further identify that the neural-associated E3 ligase, MYCBP2 (PHR1), was a novel class of cyclic E3 ligases capable of selective esterification of Thr over Ser in their substrates.
Other substrate-based probes
For enzymes that lack effective ABPs or AfBPs, rational substrate design or high-throughput screening may generate effective initial leads for subsequent construction of specific chemical probes. Moreover, by modifying existing enzyme-targeting probes with different functional groups while retaining their enzyme specificity, some probes may be repurposed that enable simultaneous single-step fluorescence-based enzyme detection and proteome-wide profiling.Histone deacetylases (HDACs) are a class of enzymes that play an imperative part in the structural modification of chromosomes and in gene expression regulations.157 In 2016, Sun's group combined a simple HDAC substrate, e.g. the acylated Lys (Kac), with a fluorogenic dye, O-NBD, to develop the corresponding activity-based fluorescence Turn-ON probe 64 (Fig. 17A).158 Upon enzymatic cleavage of Kac in the probe by HDAC, the resulting uncaged Lys intramolecularly attacked the O-NBD moiety, subsequently turning on the probe fluorescence. The enhanced fluorescence intensity was shown to be up to 50 folds, which made probe 64 an excellent HDAC-based fluorogenic ABP. Based on these results, the authors further introduced a diazirine-alkyne photo-crosslinker group into the probe, generating a dual-purpose fluorescent probe 65 (Fig. 17A). The probe 65 was shown to not only function as a fluorogenic substrate probe, but also recognize and capture target proteins from cell lysates. By combining fluorescence assays and in-gel fluorescence scanning, 65 successfully identified and differentiated epigenetic readers BRD4-1 and erasers Sirt2 (Fig. 17B).
Lys lipoylation (Klip) is a conserved Lys PTM, and plays an important role in the regulation of cellular metabolism.159 K105 (QSDKlipASVT) is a peptide sequence of the branched-chain α-ketoacid dehydrogenase (BCKDH), which is one of several metabolic multi-complexes where Klip is known to occur.160 By utilizing a similar strategy as in the design of the earlier-discussed HDAC probe, the same group developed an AfBP 66 (Fig. 17C), which was based on the peptide sequence QSDKlipASVT with replacement of the N-terminal Gln residue with a photo-crosslinker.161 Gel-based profiling and pull-down experiments demonstrated that Sirt2 (an NAD+-dependent deacetylase) caused significant delipoylation of 66. To further investigate the potential activity of Sirt2 on Klip, a fluorogenic probe with O-NBD group that can detect delipoylation activity in a single step was next developed. The following kinetic data demonstrated that Sirt2 had a strong delipoylation capacity in vitro, even better than that of Sirt 4, previously the only identified mammalian delipidylase. These innovative findings thus suggest that sirtuins may regulate cellular Klip through different delipoylation mechanisms.
Methylation events in various organisms are essential for the regulation of DNA transcription, RNA stability, and protein activity.162,163 They are catalyzed by a host of methyltransferases in cells. Among them, the S-adenosine methionine (SAM)-dependent methyltransferases (MTs), capable of transferring a methyl group from SAM to their biological substrates, are the most prominent class in methylation.164 The resulting metabolite byproduct of this reaction, S-adenosyl-L-homoCys (SAH), may be further metabolized to methione (an amino acid) which is then reintegrated into SAM biosynthesis.165 Inspired from previous work of Dalhoff et al.,166 and crystallographic data of SAM- and SAH-binding enzymes, Cravatt's group functionalized the N-6 position of SAH with different linkers to generate a series of SAH-based AfBPs (67, Fig. 18A).167 Among them, 67b was used in the analysis and enrichment of a large number of endogenous MTs from the lysates of human cancer cells (Fig. 18B), with excellent selectivity over other classes of proteins.
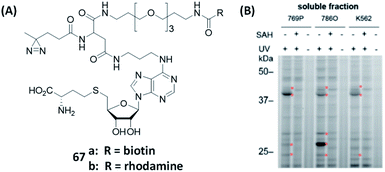 | ||
| Fig. 18 Gel-based analysis of MTs was performed using AfBPs. (A) Structures of reported AfBPs for MTs. (B) Gel-based profiling with 67b in human renal cancer cell (769P and 786O) and human leukemia cell (K562) lysates. 67 successfully labeled MTs in a variety of cancer cells. The addition of SAH (natural substrate) or the lack of UV irradiation resulted in the loss of MT labeling. Reproduced from ref. 167 with permission from American Chemical Society, copyright 2019. | ||
Forward genetics starts with a phenotype which subsequently leads to the identification of interesting genotypes, while reverse genetics starts with a known genotype and finally ends up with various phenotypes.168 The various aforementioned substrate/ligand screening strategies fit in the field of reverse genetics, but due to the limited size of available chemical libraries, their results are not always optimal. Cravatt et al. recently applied the concept of forward genetics in their screening of ABPs by incubating probes that contain covalent binding WHs and click handles with whole-cell proteome.31,169,170 Therefore, the pulled-down proteins were the target proteins of the probes. Furthermore, due to the ease of synthesis, it is relatively less complicated to form a small probe library. By using forward genetic screening of the probe library with the whole proteome, the authors confirmed that rapid discovery of ABPs that possess high selectivity against specific sets of enzymes could be realized.
Conclusions and outlook
As an emerging technological approach in the field of chemical proteomics, ABPP is a powerful tool that enables unbiased and quantitative detection of enzymatic activity by measuring cellular and tissue profiles, thereby allowing researchers to gain insights into the functional state of an enzyme rather than only its abundance.The use of ABPs and AfBPs is critical in ABPP. The main enzymes for which ABPs or AfBPs have been developed and are available including kinases, proteases, glycosidases, phosphatases and oxidoreductases. The successful application of ABPP in cellulo and animal models has identified a range of enzymatic activities associated with diseases that are closely related to each other. In addition, the development of these probes and ABPP has facilitated functional studies of enzymes involved in many biological processes, including neurotransmission, neurodegenerative diseases, signal transduction in tumors and immune diseases. All these events demonstrate the important role of ABPs and AfBPs in the discovery and study of enzyme function. In this review, we have summarized design strategies of different ABPs and AfBPs. Inhibitor-based probes have been widely used to identify off-targets, thereby aiding drug discovery and optimization. Substrate-based probes are favorable tools for enzyme functional studies and substrate discovery. The discovery of drug targets is one of the important tasks in the initial studies of clinical drugs. In addition, most of the current proteomics platforms are only able to capture enzymes in a small fraction of samples of cells and tissues. It is still challenging to apply ABPP to routine biological systems. In addition, non-specific labeling of probes and inefficient PAL remain a great challenge. The emergence of various new photo-crosslinkers and the development of bioorthogonal chemistry in recent years have provided some assistance for this purpose. How to obtain high PAL efficiency while reducing the impact of biological systems is also an issue to be addressed in the design of ABPs and AfBPs.
The visualization of important enzymatic activity could also be achieved by using highly selective fluorescent ABPs or AfBPs. For instance, Shrivastav et al. developed a near-infrared fluorescence (NIRF) ABP that was capable of imaging large polyps using a dual-laser NIRF endoscope.171 Recent years have seen a dramatic increase in the number of near-infrared region II (NIR-II) fluorescent molecules being used in a wide range of bioimaging and biosensing applications.172,173 The introduction of NIR-II dyes into the fabrication of ABPs and AfBPs could potentially expand their applications to disease diagnosis, surgical resection, and therapeutic response validation. Such novel ABPP strategies could offer new opportunities in interdisciplinary research collaborations.
However, due to the complexity and diversity of enzymes, ABPP still faces several issues such as poor specificity and sensitivity. For example, no ABP that can label inverting glycosidases has been reported, but AfBPs that were reported by Stubbs et al. successfully labeled them.174 This opens up the possibility of targeting other enzyme families that do not have nucleophilic residues and cannot be labeled by ABPs. In addition, most human enzymes lack selective chemical ligands and some classes of proteins are even considered undruggable. Therefore, it is difficult to develop probes to study these proteins.170 To overcome such problems, tremendous efforts have been put into the development of more advanced probe design strategies. Bump-and-hole strategy is one solution. By attaching one bulky group at the natural substrate, and at the same time mutating the “gatekeeper” residue of the substrate binding pocket of the enzyme, the orthogonal pair of the probe and the mutated enzyme is formed. Using this orthogonal “bump-hole” system, the specific target proteins of the corresponding enzyme can be identified. This method was successfully used in the ABPP of PARP and Ub.134,136,141 Bump-and-hole is a powerful method but this chemical genetic approach is limited to in cellulo studies, and is less utilized in the ABPP studies of other enzymes. Another approach, has isoTOP-ABPP, developed by Cravatt's group,29 which uses isotope-labeled probes to obtain more reliable results than other quantitative protein analysis methods. They have also developed fluoPol-ABPP30 that could be used in combination with HTS to discover new protein ligands. RP-ABPP,31 an unbiased method which employs nucleophilic hydrazine probes to capture active protein-bound electrophiles in cells,175 was also developed by the same group. High-throughput forward genetic screening of the whole proteome is another effective method that has been used in the discovery of highly selective chemical probes capable of targeting less-studied enzymes.169
In summary, ABPP is a multidisciplinary technology involving chemistry, biology, medicine and other disciplines. There is no single design strategy that is a universal remedy for all research needs. The design and development of better ABPs and AfBPs also require interdisciplinary efforts. We hope that this review will contribute to the design and development of more powerful ABPs and AfBPs to facilitate further advancement in the research of enzymes.
Author contributions
H. F., B. P., L. L. and S. Q. Y. presented the outline of this review. H. F. wrote the draft. H. F. and B. P. performed the graphic design. O. S. Y., Q. W., L. L. and S. Q. Y. revised and edited the draft. B. P., L. L., and S. Q. Y. conceived and supervised this project. All authors participated in the discussion of the draft and carefully revised the draft before the final submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by GSK-EDB Trust Fund (R-143-000-88-592), the Synthetic Biology Research & Development Programme (SBP) of National Research Foundation (SBP-P4 and SBP-P8) of Singapore, the National Key R&D Program of China (2020YFA0709900), the National Natural Science Foundation of China (22077101, 22004099), the Joint Research Funds of Department of Science & Technology of Shaanxi Province and Northwestern Polytechnical University (2020GXLH-Z-008, 2020GXLH-Z-021 and 2020GXLH-Z-023), the Natural Science Foundation of Ningbo (202003N4049, 202003N4065), the Open Project Program of the Analytical & Testing Center of Northwestern Polytechnical University (2020T018), the Open Project Program of Wuhan National Laboratory for Optoelectronics (No. 2020WNLOKF023), Key Research and Development Program of Shaanxi (2020ZDLGY13-04, 2021KW-49), China-Sweden Joint Mobility Project (51811530018) and Fundamental Research Funds for the Central Universities.Notes and references
- U. Markel, K. D. Essani, V. Besirlioglu, J. Schiffels, W. R. Streit and U. Schwaneberg, Chem. Soc. Rev., 2020, 49, 233–262 RSC.
- R. H. Wijdeven, J. Neefjes and H. Ovaa, Trends Cell Biol., 2014, 24, 751–760 CrossRef CAS PubMed.
- M. Uttamchandani, C. H. Lu and S. Q. Yao, Acc. Chem. Res., 2009, 42, 1183–1192 CrossRef CAS PubMed.
- M. J. Evans and B. F. Cravatt, Chem. Rev., 2006, 106, 3279–3301 CrossRef CAS PubMed.
- L. E. Sanman and M. Bogyo, Annu. Rev. Biochem., 2014, 83, 249–273 CrossRef CAS PubMed.
- M. J. Niphakis and B. F. Cravatt, Annu. Rev. Biochem., 2014, 83, 341–377 CrossRef CAS PubMed.
- Y. Liu, M. P. Patricelli and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14694–14699 CrossRef CAS PubMed.
- D. Greenbaum, K. F. Medzihradszky, A. Burlingame and M. Bogyo, Chem. Biol., 2000, 7, 569–581 CrossRef CAS PubMed.
- Y. Su, J. Ge, B. Zhu, Y. G. Zheng, Q. Zhu and S. Q. Yao, Curr. Opin. Chem. Biol., 2013, 17, 768–775 CrossRef CAS PubMed.
- S. Pan, H. Zhang, C. Wang, S. C. Yao and S. Q. Yao, Nat. Prod. Rep., 2016, 33, 612–620 RSC.
- E. M. Sletten and C. R. Bertozzi, Acc. Chem. Res., 2011, 44, 666–676 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, Chem. Rev., 2014, 114, 4764–4806 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596 CrossRef CAS PubMed.
- J. C. Jewett, E. M. Sletten and C. R. Bertozzi, J. Am. Chem. Soc., 2010, 132, 3688–3690 CrossRef CAS PubMed.
- M. L. Blackman, M. Royzen and J. M. Fox, J. Am. Chem. Soc., 2008, 130, 13518–13519 CrossRef CAS PubMed.
- A. Singh, E. R. Thornton and F. H. Westheimer, J. Biol. Chem., 1962, 237, 3006–3008 CrossRef CAS.
- H. Shi, C. J. Zhang, G. Y. Chen and S. Q. Yao, J. Am. Chem. Soc., 2012, 134, 3001–3014 CrossRef CAS PubMed.
- F. Ragaini, A. Penoni, E. Gallo, S. Tollari, C. Li Gotti, M. Lapadula, E. Mangioni and S. Cenini, Chem.–Asian J., 2003, 9, 249–259 CAS.
- A. Saghatelian, N. Jessani, A. Joseph, M. Humphrey and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10000–10005 CrossRef CAS PubMed.
- G. L. Chee, J. C. Yalowich, A. Bodner, X. Wu and B. B. Hasinoff, Bioorg. Med. Chem., 2010, 18, 830–838 CrossRef CAS PubMed.
- Z. Li, L. Qian, L. Li, J. C. Bernhammer, H. V. Huynh, J. S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2016, 55, 3254 CrossRef CAS PubMed.
- J. Xu, X. Li, K. Ding and Z. Li, Chem.–Asian J., 2019, 15, 34–41 CrossRef PubMed.
- E. Smith and I. Collins, Future Med. Chem., 2015, 7, 159–183 CrossRef CAS PubMed.
- W. Lang, C. Yuan, B. Zhu, S. Pan, J. Liu, J. Luo, S. Nie, Q. Zhu, J. S. Lee and J. Ge, Org. Biomol. Chem., 2019, 17, 3010–3017 RSC.
- C. Guo, Y. Chang, X. Wang, C. Zhang, P. Hao, K. Ding and Z. Li, Chem. Commun., 2019, 55, 834–837 RSC.
- H. Zhang, R. Liu, J. Liu, L. Li, P. Wang, S. Q. Yao, Z. Xu and H. Sun, Chem. Sci., 2016, 7, 256–260 RSC.
- Z. Li, D. Wang, L. Li, S. Pan, Z. Na, C. Y. Tan and S. Q. Yao, J. Am. Chem. Soc., 2014, 136, 9990–9998 CrossRef CAS PubMed.
- Z. Q. Li, P. L. Hao, L. Li, C. Y. J. Tan, X. M. Cheng, G. Y. J. Chen, S. K. Sze, H. M. Shen and S. Q. Yao, Angew. Chem., Int. Ed., 2013, 52, 8551–8556 CrossRef CAS PubMed.
- E. Weerapana, C. Wang, G. M. Simon, F. Richter, S. Khare, M. B. D. Dillon, D. A. Bachovchin, K. Mowen, D. Baker and B. F. Cravatt, Nature, 2010, 468, 790–795 CrossRef CAS PubMed.
- D. A. Bachovchin, S. J. Brown, H. Rosen and B. F. Cravatt, Nat. Biotechnol., 2009, 27, 387–394 CrossRef CAS PubMed.
- M. L. Matthews, L. He, B. D. Horning, E. J. Olson, B. E. Correia, J. R. Yates, P. E. Dawson and B. F. Cravatt, Nat. Chem., 2017, 9, 234–243 CrossRef CAS PubMed.
- G. Blum, G. von Degenfeld, M. J. Merchant, H. M. Blau and M. Bogyo, Nat. Chem. Biol., 2007, 3, 668–677 CrossRef CAS PubMed.
- L. Qian, L. Li and S. Q. Yao, Acc. Chem. Res., 2016, 49, 626–634 CrossRef CAS PubMed.
- F. V. Filipp, Canc. Metastasis Rev., 2017, 36, 91–108 CrossRef PubMed.
- B. R. Lannine, L. R. Whitby, M. M. Dix, J. Douhan, A. M. Gilbert, E. C. Hett, T. Johnson, C. Joslynl, J. C. Kath, S. Niessen, L. R. Roberts, M. E. Schnute, C. Wang, J. J. Hulce, B. X. Wei, L. O. Whiteley, M. M. Hayward and B. F. Cravatt, Nat. Chem. Biol., 2014, 10, 760–767 CrossRef PubMed.
- H. Fu, H. Fang, J. Sun, H. Wang, A. Liu, J. Sun and Z. Wu, Curr. Med. Chem., 2014, 21, 3271–3280 CrossRef CAS PubMed.
- N. Berndt, R. M. Karim and E. Schonbrunn, Curr. Opin. Chem. Biol., 2017, 39, 126–132 CrossRef CAS PubMed.
- J. A. Woyach, E. Bojnik, A. S. Ruppert, M. R. Stefanovski, V. M. Goettl, K. A. Smucker, L. L. Smith, J. A. Dubovsky, W. H. Towns, J. MacMurray, B. K. Harrington, M. E. Davis, S. Gobessi, L. Laurenti, B. Y. Chang, J. J. Buggy, D. G. Efremov, J. C. Byrd and A. J. Johnson, Blood, 2014, 123, 1207–1213 CrossRef CAS PubMed.
- J. C. Byrd, R. R. Furman, S. E. Coutre, I. W. Flinn, J. A. Burger, K. A. Blum, B. Grant, J. P. Sharman, M. Coleman, W. G. Wierda, J. A. Jones, W. Q. Zhao, N. A. Heerema, A. J. Johnson, J. Sukbuntherng, B. Y. Chang, F. Clow, E. Hedrick, J. J. Buggy, D. F. James and S. O'Brien, N. Engl. J. Med., 2013, 369, 32–42 CrossRef CAS PubMed.
- J. Chen, X. Wang, F. He and Z. Pan, Bioconjugate Chem., 2018, 29, 1640–1645 CrossRef CAS PubMed.
- X. Wang, N. Ma, R. Wu, K. Ding and Z. Li, Chem. Commun., 2019, 55, 3473–3476 RSC.
- S. Kobayashi, T. J. Boggon, T. Dayaram, P. A. Janne, O. Kocher, M. Meyerson, B. E. Johnson, M. J. Eck, D. G. Tenen and B. Halmos, N. Engl. J. Med., 2005, 352, 786–792 CrossRef CAS PubMed.
- S. Planken, D. C. Behenna, S. K. Nair, T. O. Johnson, A. Nagata, C. Almaden, S. Bailey, T. E. Ballard, L. Bernier, H. Cheng, S. Cho-Schultz, D. Dalvie, J. G. Deal, D. M. Dinh, M. P. Edwards, R. A. Ferre, K. S. Gajiwala, M. Hemkens, R. S. Kania, J. C. Kath, J. Matthews, B. W. Murray, S. Niessen, S. T. M. Orr, M. Pairish, N. W. Sach, H. Shen, M. Shi, J. Solowiej, K. Tran, E. Tseng, P. Vicini, Y. Wang, S. L. Weinrich, R. Zhou, M. Zientek, L. Liu, Y. Luo, S. Xin, C. Zhang and J. Lafontaine, J. Med. Chem., 2017, 60, 3002–3019 CrossRef CAS PubMed.
- S. Niessen, M. M. Dix, S. Barbas, Z. E. Potter, S. Y. Lu, O. Brodsky, S. Planken, D. Behenna, C. Almaden, K. S. Gajiwala, K. Ryan, R. Ferre, M. R. Lazear, M. M. Hayward, J. C. Kath and B. F. Cravatt, Cell Chem. Biol., 2017, 24, 1388–1400 CrossRef CAS PubMed.
- J. C. Byrd, B. Harrington, S. O'Brien, J. A. Jones, A. Schuh, S. Devereux, J. Chaves, W. G. Wierda, F. T. Awan, J. R. Brown, P. Hillmen, D. M. Stephens, P. Ghia, J. C. Barrientos, J. M. Pagel, J. Woyach, D. Johnson, J. Huang, X. Wang, A. Kaptein, B. J. Lannutti, T. Covey, M. Fardis, J. McGreivy, A. Hamdy, W. Rothbaum, R. Izumi, T. G. Diacovo, A. J. Johnson and R. R. Furman, N. Engl. J. Med., 2016, 374, 323–332 CrossRef CAS PubMed.
- N. Shindo, H. Fuchida, M. Sato, K. Watari, T. Shibata, K. Kuwata, C. Miura, K. Okamoto, Y. Hatsuyama, K. Tokunaga, S. Sakamoto, S. Morimoto, Y. Abe, M. Shiroishi, J. M. M. Caaveiro, T. Ueda, T. Tamura, N. Matsunaga, T. Nakao, S. Koyanagi, S. Ohdo, Y. Yamaguchi, I. Hamachi, M. Ono and A. Ojida, Nat. Chem. Biol., 2019, 15, 250–258 CrossRef CAS PubMed.
- A. M. Manning and R. J. Davis, Nat. Rev. Drug Discovery, 2003, 2, 554–565 CrossRef CAS PubMed.
- L. Qian, S. Pan, J.-S. Lee, J. Ge, L. Li and S. Q. Yao, Chem. Commun., 2019, 55, 1092–1095 RSC.
- G. F. Desrochers, C. Cornacchia, C. S. McKay and J. P. Pezacki, ACS Infect. Dis., 2018, 4, 752–757 CrossRef CAS PubMed.
- J. E. Burke, Mol. Cell, 2018, 71, 653–673 CrossRef CAS PubMed.
- S. C. Stolze, N. Liu, R. H. Wijdeven, A. W. Tuin, A. M. C. H. van den Nieuwendijk, B. I. Florea, M. van der Stelt, G. A. van der Marel, J. J. Neefjes and H. S. Overkleeft, Mol. BioSyst., 2016, 12, 1809–1817 RSC.
- E. K. Grant, D. J. Fallon, H. C. Eberl, K. G. M. Fantom, F. Zappacosta, C. Messenger, N. C. O. Tomkinson and J. T. Bush, Angew. Chem., Int. Ed., 2019, 131, 17483–17488 CrossRef.
- Q. Zhao, X. Ouyang, X. Wan, K. S. Gajiwala, J. C. Kath, L. H. Jones, A. L. Burlingame and J. Taunton, J. Am. Chem. Soc., 2017, 139, 680–685 CrossRef CAS PubMed.
- A. Cuesta, X. Wan, A. L. Burlingame and J. Taunton, J. Am. Chem. Soc., 2020, 142, 3392–3400 CrossRef CAS PubMed.
- X. Wan, T. Yang, A. Cuesta, X. Pang, T. E. Balius, J. J. Irwin, B. K. Shoichet and J. Taunton, J. Am. Chem. Soc., 2020, 142, 4960–4964 CrossRef CAS PubMed.
- M. Drag and G. S. Salvesen, Nat. Rev. Drug Discovery, 2010, 9, 690–701 CrossRef CAS PubMed.
- G. Blum, S. R. Mullins, K. Keren, M. Fonovic, C. Jedeszko, M. J. Rice, B. F. Sloane and M. Bogyo, Nat. Chem. Biol., 2005, 1, 203–209 CrossRef CAS PubMed.
- M. Hu, L. Li, H. Wu, Y. Su, P. Y. Yang, M. Uttamchandani, Q. H. Xu and S. Q. Yao, J. Am. Chem. Soc., 2011, 133, 12009–12020 CrossRef CAS PubMed.
- J. C. Widen, M. Tholen, J. J. Yim, A. Antaris, K. M. Casey, S. Rogalla, A. Klaassen, J. Sorger and M. Bogyo, Nat. Biomed. Eng., 2021, 5, 264–277 CrossRef CAS PubMed.
- J. B. Denault and G. S. Salvesen, Methods Mol. Biol., 2008, 414, 191–220 CAS.
- N. Aggarwal and B. F. Sloane, Proteonomics Clin. Appl., 2014, 8, 427–437 CrossRef CAS PubMed.
- L. E. Edgington, A. B. Berger, G. Blum, V. E. Albrow, M. G. Paulick, N. Lineberry and M. Bogyo, Nat. Med., 2009, 15, 967–973 CrossRef CAS PubMed.
- L. E. Edgington, M. Verdoes, A. Ortega, N. P. Withana, J. Lee, S. Syed, M. H. Bachmann, G. Blum and M. Bogyo, J. Am. Chem. Soc., 2013, 135, 174–182 CrossRef CAS PubMed.
- Y. Shaulov-Rotem, E. Merquiol, T. Weiss-Sadan, O. Moshel, S. Salpeter, D. Shabat, F. Kaschani, M. Kaiser and G. Blum, Chem. Sci., 2016, 7, 1322–1337 RSC.
- K. B. Sexton, M. D. Witte, G. Blum and M. Bogyo, Bioorg. Med. Chem. Lett., 2007, 17, 649–653 CrossRef CAS PubMed.
- M. Poreba, K. Groborz, M. Vizovisek, M. Maruggi, D. Turk, B. Turk, G. Powis, M. Drag and G. S. Salvesen, Chem. Sci., 2019, 10, 8461–8477 RSC.
- N. Amara, I. T. Foe, O. Onguka, M. Garland and M. Bogyo, Cell Chem. Biol., 2019, 26, 35–47 CrossRef CAS PubMed.
- M. Tholen, J. J. Yim, K. Groborz, E. Yoo, B. A. Martin, N. S. van den Berg, M. Drag and M. Bogyo, Angew. Chem., Int. Ed., 2020, 59, 19143–19152 CrossRef CAS PubMed.
- E. Walker, Y. Liu, I. Kim, M. Biro, S. R. Iyer, H. Ezaldein, J. Scott, M. Merati, R. Mistur, B. Zhou, B. Straight, J. J. Yim, M. Bogyo, M. Mann, D. L. Wilson, J. P. Basilion and D. L. Popkin, Cancer Res., 2020, 80, 2045–2055 CrossRef CAS PubMed.
- L. O. Ofori, N. P. Withana, T. R. Prestwood, M. Verdoes, J. J. Brady, M. M. Winslow, J. Sorger and M. Bogyo, ACS Chem. Biol., 2015, 10, 1977–1988 CrossRef CAS PubMed.
- F. V. Suurs, S. Q. Qiu, J. J. Yim, C. P. Schroder, H. Timmer-Bosscha, E. S. Bensen, J. T. Santini Jr, E. G. E. de Vries, M. Bogyo and G. M. van Dam, EJNMMI Res., 2020, 10, 111 CrossRef CAS.
- M. Verdoes, K. Oresic Bender, E. Segal, W. A. van der Linden, S. Syed, N. P. Withana, L. E. Sanman and M. Bogyo, J. Am. Chem. Soc., 2013, 135, 14726–14730 CrossRef CAS PubMed.
- A. Page-McCaw, A. J. Ewald and Z. Werb, Nat. Rev. Mol. Cell Biol., 2007, 8, 221–233 CrossRef CAS PubMed.
- M. Morell, T. Nguyen Duc, A. L. Willis, S. Syed, J. Lee, E. Deu, Y. Deng, J. Xiao, B. E. Turk, J. R. Jessen, S. J. Weiss and M. Bogyo, J. Am. Chem. Soc., 2013, 135, 9139–9148 CrossRef CAS PubMed.
- N. Amara, M. Tholen and M. Bogyo, ACS Chem. Biol., 2018, 13, 2645–2654 CrossRef CAS PubMed.
- L. Wu, Z. Armstrong, S. P. Schroder, C. de Boer, M. Artola, J. M. Aerts, H. S. Overkleeft and G. J. Davies, Curr. Opin. Chem. Biol., 2019, 53, 25–36 CrossRef CAS PubMed.
- V. Cullen, S. P. Sardi, J. Ng, Y. H. Xu, Y. Sun, J. J. Tomlinson, P. Kolodziej, I. Kahn, P. Saftig, J. Woulfe, J. C. Rochet, M. A. Glicksman, S. H. Cheng, G. A. Grabowski, L. S. Shihabuddin and M. G. Schlossmacher, Ann. Neurol., 2011, 69, 940–953 CrossRef CAS PubMed.
- M. Czjzek, M. Cicek, V. Zamboni, D. R. Bevan, B. Henrissat and A. Esen, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 13555–13560 CrossRef CAS PubMed.
- B. Bissaro, P. Monsan, R. Faure and M. J. O'Donohue, Biochem. J., 2015, 467, 17–35 CrossRef CAS PubMed.
- W. W. Kallemeijn, K.-Y. Li, M. D. Witte, A. R. A. Marques, J. Aten, S. Scheij, J. Jiang, L. I. Willems, T. M. Voorn-Brouwer, C. P. A. A. van Roomen, R. Ottenhoff, R. G. Boot, H. van den Elst, M. T. C. Walvoort, B. I. Florea, J. D. C. Codee, G. A. van der Marel, J. M. F. G. Aerts and H. S. Overkleeft, Angew. Chem., Int. Ed., 2012, 51, 12529–12533 CrossRef CAS PubMed.
- L. I. Willems, T. J. M. Beenakker, B. Murray, S. Scheij, W. W. Kallemeijn, R. G. Boot, M. Verhoek, W. E. Donker-Koopman, M. J. Ferraz, E. R. van Rijssel, B. I. Florea, J. D. C. Codee, G. A. van der Marel, J. M. F. G. Aerts and H. S. Overkleeft, J. Am. Chem. Soc., 2014, 136, 11622–11625 CrossRef CAS PubMed.
- B. T. Adams, S. Niccoli, M. A. Chowdhury, A. N. K. Esarik, S. J. Lees, B. P. Rempel and C. P. Phenix, Chem. Commun., 2015, 51, 11390–11393 RSC.
- A. Alcaide, A. Trapero, Y. Perez and A. Llebaria, Org. Biomol. Chem., 2015, 13, 5690–5697 RSC.
- J. Jiang, T. J. M. Beenakker, W. W. Kallemeijn, G. A. van der Marel, H. van den Elst, J. D. C. Codee, J. M. F. G. Aerts and H. S. Overkleeft, Chem. - Eur. J., 2015, 21, 10861–10869 CrossRef CAS PubMed.
- T. M. Gloster, R. Madsen and G. J. Davies, Org. Biomol. Chem., 2007, 5, 444–446 RSC.
- M. D. Witte, W. W. Kallemeijn, J. Aten, K. Y. Li, A. Strijland, W. E. Donker-Koopman, A. M. van den Nieuwendijk, B. Bleijlevens, G. Kramer, B. I. Florea, B. Hooibrink, C. E. Hollak, R. Ottenhoff, R. G. Boot, G. A. van der Marel, H. S. Overkleeft and J. M. Aerts, Nat. Chem. Biol., 2010, 6, 907–913 CrossRef CAS PubMed.
- D. J. Vocadlo, H. C. Hang, E. J. Kim, J. A. Hanover and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9116–9121 CrossRef CAS PubMed.
- S. J. Williams, V. Notenboom, J. Wicki, D. R. Rose and S. G. Withers, J. Am. Chem. Soc., 2000, 122, 4229–4230 CrossRef CAS.
- R. O. Brady, J. N. Kanfer, R. M. Bradley and D. Shapiro, J. Clin. Invest., 1966, 45, 1112–1115 CrossRef CAS PubMed.
- E. van Meel, E. Bos, M. J. C. van der Lienden, H. S. Overkleeft, S. I. van Kasteren, A. J. Koster and J. M. F. G. Aerts, Traffic, 2019, 20, 346–356 CrossRef CAS PubMed.
- J. E. Deane, S. C. Graham, N. N. Kim, P. E. Stein, R. McNair, M. B. Cachon-Gonzalez, T. M. Cox and R. J. Read, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15169–15173 CrossRef CAS PubMed.
- A. R. Marques, L. I. Willems, M. D. Herrera, B. I. Florea, S. Scheij, R. Ottenhoff, C. P. van Roomen, M. Verhoek, J. K. Nelson, W. W. Kallemeijn, A. Biela-Banas, O. R. Martin, M. B. Cachon-Gonzalez, N. N. Kim, T. M. Cox, R. G. Boot, H. S. Overkleeft and J. M. Aerts, Chembiochem, 2017, 18, 402–412 CrossRef CAS PubMed.
- M. Artola, C.-L. Kuo, L. T. Lelieveld, R. J. Rowland, G. A. van der Marel, J. D. C. Codée, R. G. Boot, G. J. Davies, J. M. F. G. Aerts and H. S. Overkleeft, J. Am. Chem. Soc., 2019, 141, 4214–4218 CrossRef CAS PubMed.
- L. Wu, J. Jiang, Y. Jin, W. W. Kallemeijn, C.-L. Kuo, M. Artola, W. Dai, C. van Elk, M. van Eijk, G. A. van der Marel, J. D. C. Codée, B. I. Florea, J. M. F. G. Aerts, H. S. Overkleeft and G. J. Davies, Nat. Chem. Biol., 2017, 13, 867–873 CrossRef CAS PubMed.
- H. Van den Hout, A. J. Reuser, A. G. Vulto, M. C. Loonen, A. Cromme-Dijkhuis and A. T. Van der Ploeg, Lancet, 2000, 356, 397–398 CrossRef CAS.
- J. Jiang, C. L. Kuo, L. Wu, C. Franke, W. W. Kallemeijn, B. I. Florea, E. van Meel, G. A. van der Marel, J. D. Codee, R. G. Boot, G. J. Davies, H. S. Overkleeft and J. M. Aerts, ACS Cent. Sci., 2016, 2, 351–358 CrossRef CAS PubMed.
- Y. Chen, Z. Armstrong, M. Artola, B. I. Florea, C. L. Kuo, C. de Boer, M. S. Rasmussen, M. Abou Hachem, G. A. van der Marel, J. D. C. Codee, J. Aerts, G. J. Davies and H. S. Overkleeft, J. Am. Chem. Soc., 2021, 143, 2423–2432 CrossRef CAS.
- S. P. Schroder, C. de Boer, N. G. S. McGregor, R. J. Rowland, O. Moroz, E. Blagova, J. Reijngoud, M. Arentshorst, D. Osborn, M. D. Morant, E. Abbate, M. A. Stringer, K. Krogh, L. Raich, C. Rovira, J. G. Berrin, G. P. van Wezel, A. F. J. Ram, B. I. Florea, G. A. van der Marel, J. D. C. Codee, K. S. Wilson, L. Wu, G. J. Davies and H. S. Overkleeft, ACS Cent. Sci., 2019, 5, 1067–1078 CrossRef.
- C. Whidbey, N. C. Sadler, R. N. Nair, R. F. Volk, A. J. DeLeon, L. M. Bramer, S. J. Fansler, J. R. Hansen, A. K. Shukla, J. K. Jansson, B. D. Thrall and A. T. Wright, J. Am. Chem. Soc., 2018, 141, 42–47 CrossRef PubMed.
- A. Hershko and A. Ciechanover, Annu. Rev. Biochem., 1998, 67, 425–479 CrossRef CAS PubMed.
- M. Bogyo, J. S. McMaster, M. Gaczynska, D. Tortorella, A. L. Goldberg and H. Ploegh, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 6629–6634 CrossRef CAS PubMed.
- B. H. B. K. Lihao Meng, N. Sin and C. M. Crews, Cancer Res., 1999, 59, 2798–2801 Search PubMed.
- H. Ovaa, P. F. van Swieten, B. M. Kessler, M. A. Leeuwenburgh, E. Fiebiger, A. M. van den Nieuwendijk, P. J. Galardy, G. A. van der Marel, H. L. Ploegh and H. S. Overkleeft, Angew. Chem., Int. Ed., 2003, 42, 3626–3629 CrossRef CAS PubMed.
- G. de Bruin, B. T. Xin, M. Kraus, M. van der Stelt, G. A. van der Marel, A. F. Kisselev, C. Driessen, B. I. Florea and H. S. Overkleeft, Angew. Chem., Int. Ed., 2016, 55, 4199–4203 CrossRef CAS.
- J. A. Ward, L. McLellan, M. Stockley, K. R. Gibson, G. A. Whitlock, C. Knights, J. A. Harrigan, X. Jacq and E. W. Tate, ACS Chem. Biol., 2016, 11, 3268–3272 CrossRef CAS.
- N. Panyain, A. Godinat, T. Lanyon-Hogg, S. Lachiondo-Ortega, E. J. Will, C. Soudy, M. Mondal, K. Mason, S. Elkhalifa, L. M. Smith, J. A. Harrigan and E. W. Tate, J. Am. Chem. Soc., 2020, 142, 12020–12026 CrossRef CAS PubMed.
- B. A. Gibson and W. L. Kraus, Nat. Rev. Mol. Cell Biol., 2012, 13, 411–424 CrossRef CAS.
- R. T. Howard, P. Hemsley, P. Petteruti, C. N. Saunders, J. A. Molina Bermejo, J. S. Scott, J. W. Johannes and E. W. Tate, ACS Chem. Biol., 2020, 15, 325–333 CrossRef CAS.
- C. S. Lentz, J. R. Sheldon, L. A. Crawford, R. Cooper, M. Garland, M. R. Amieva, E. Weerapana, E. P. Skaar and M. Bogyo, Nat. Chem. Biol., 2018, 14, 609–617 CrossRef CAS PubMed.
- A. C. M. van Esbroeck, A. P. A. Janssen, A. B. R. Cognetta, D. Ogasawara, G. Shpak, M. van der Kroeg, V. Kantae, M. P. Baggelaar, F. M. S. de Vrij, H. Deng, M. Allara, F. Fezza, Z. Lin, T. van der Wel, M. Soethoudt, E. D. Mock, H. den Dulk, I. L. Baak, B. I. Florea, G. Hendriks, L. De Petrocellis, H. S. Overkleeft, T. Hankemeier, C. I. De Zeeuw, V. Di Marzo, M. Maccarrone, B. F. Cravatt, S. A. Kushner and M. van der Stelt, Science, 2017, 356, 1084–1087 CrossRef CAS PubMed.
- Z. Huang, D. Ogasawara, U. I. Seneviratne, A. B. Cognetta, C. W. Am Ende, D. M. Nason, K. Lapham, J. Litchfield, D. S. Johnson and B. F. Cravatt, ACS Chem. Biol., 2019, 14, 192–197 CrossRef CAS PubMed.
- R. E. Rau, B. A. Rodriguez, M. Luo, M. Jeong, A. Rosen, J. H. Rogers, C. T. Campbell, S. R. Daigle, L. S. Deng, Y. C. Song, S. Sweet, T. Chevassut, M. Andreeff, S. M. Kornblau, W. Li and M. A. Goodell, Blood, 2016, 128, 971–981 CrossRef CAS PubMed.
- B. Zhu, H. Zhang, S. Pan, C. Wang, J. Ge, J.-S. Lee and S. Q. Yao, Chem.–Asian J., 2016, 22, 7824–7836 CAS.
- M. E. Pacold, K. R. Brimacombe, S. H. Chan, J. M. Rohde, C. A. Lewis, L. J. Swier, R. Possemato, W. W. Chen, L. B. Sullivan, B. P. Fiske, S. Cho, E. Freinkman, K. Birsoy, M. Abu-Remaileh, Y. D. Shaul, C. M. Liu, M. Zhou, M. J. Koh, H. Chung, S. M. Davidson, A. Luengo, A. Q. Wang, X. Xu, A. Yasgar, L. Liu, G. Rai, K. D. Westover, M. G. Vander Heiden, M. Shen, N. S. Gray, M. B. Boxer and D. M. Sabatini, Nat. Chem. Biol., 2016, 12, 452–458 CrossRef CAS.
- S. Pan, S.-Y. Jang, S. S. Liew, J. Fu, D. Wang, J.-S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2018, 57, 579–583 CrossRef CAS PubMed.
- L. L. Lairson, B. Henrissat, G. J. Davies and S. G. Withers, Annu. Rev. Biochem., 2008, 77, 521–555 CrossRef CAS PubMed.
- A. Richman, A. Swanson, T. Humphrey, R. Chapman, B. McGarvey, R. Pocs and J. Brandle, Plant J., 2005, 41, 56–67 CrossRef CAS PubMed.
- Y. Zhou, W. Li, W. You, Z. Di, M. Wang, H. Zhou, S. Yuan, N.-K. Wong and Y. Xiao, Chem. Commun., 2018, 54, 7179–7182 RSC.
- N.-K. Wong, S. Zhong, W. Li, F. Zhou, Z. Deng and Y. Zhou, Chem. Commun., 2020, 56, 12387–12390 RSC.
- L. Mouchiroud, R. H. Houtkooper, N. Moullan, E. Katsyuba, D. Ryu, C. Canto, A. Mottis, Y. S. Jo, M. Viswanathan, K. Schoonjans, L. Guarente and J. Auwerx, Cell, 2013, 154, 430–441 CrossRef CAS PubMed.
- A. Ray Chaudhuri and A. Nussenzweig, Nat. Rev. Mol. Cell Biol., 2017, 18, 610–621 CrossRef CAS.
- H. Jiang, J. H. Kim, K. M. Frizzell, W. L. Kraus and H. Lin, J. Am. Chem. Soc., 2010, 132, 9363–9372 CrossRef CAS PubMed.
- A. Buntz, S. Wallrodt, E. Gwosch, M. Schmalz, S. Beneke, E. Ferrando-May, A. Marx and A. Zumbusch, Angew. Chem., Int. Ed., 2016, 55, 11256–11260 CrossRef CAS.
- M. C. Morris, J. Depollier, J. Mery, F. Heitz and G. Divita, Nat. Biotechnol., 2001, 19, 1173–1176 CrossRef CAS PubMed.
- S. Wallrodt, A. Buntz, Y. Wang, A. Zumbusch and A. Marx, Angew. Chem., Int. Ed., 2016, 55, 7660–7664 CrossRef CAS PubMed.
- S. Wallrodt, E. L. Simpson and A. Marx, Beilstein J. Org. Chem., 2017, 13, 495–501 CrossRef CAS PubMed.
- C. Canto, R. H. Houtkooper, E. Pirinen, D. Y. Youn, M. H. Oosterveer, Y. Cen, P. J. Fernandez-Marcos, H. Yamamoto, P. A. Andreux, P. Cettour-Rose, K. Gademann, C. Rinsch, K. Schoonjans, A. A. Sauve and J. Auwerx, Cell Metab., 2012, 15, 838–847 CrossRef CAS PubMed.
- M. Grammel, H. Hang and N. K. Conrad, Chembiochem, 2012, 13, 1112–1115 CrossRef CAS PubMed.
- N. P. Westcott, J. P. Fernandez, H. Molina and H. C. Hang, Nat. Chem. Biol., 2017, 13, 302–308 CrossRef CAS PubMed.
- K. Kalesh, S. Lukauskas, A. J. Borg, A. P. Snijders, V. Ayyappan, A. K. L. Leung, D. O. Haskard and P. A. DiMaggio, Sci. Rep., 2019, 9, 6655 CrossRef.
- X.-N. Zhang, Q. Cheng, J. Chen, A. T. Lam, Y. Lu, Z. Dai, H. Pei, N. M. Evdokimov, S. G. Louie and Y. Zhang, Nat. Commun., 2019, 10, 4196 CrossRef PubMed.
- D. Koley and A. J. Bard, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16783–16787 CrossRef CAS.
- I. Carter-O'Connell, H. Jin, R. K. Morgan, L. L. David and M. S. Cohen, J. Am. Chem. Soc., 2014, 136, 5201–5204 CrossRef PubMed.
- B. A. Gibson, Y. J. Zhang, H. Jiang, K. M. Hussey, J. H. Shrimp, H. N. Lin, F. Schwede, Y. H. Yu and W. L. Kraus, Science, 2016, 353, 45–50 CrossRef CAS PubMed.
- A. T. Lam, X. N. Zhang, V. V. Courouble, T. S. Strutzenberg, H. Pei, B. L. Stiles, S. G. Louie, P. R. Griffin and Y. Zhang, ACS Chem. Biol., 2021, 16, 389–396 CrossRef CAS PubMed.
- J. Sileikyte, S. Sundalam, L. L. David and M. S. Cohen, J. Am. Chem. Soc., 2021, 143, 6787–6791 CrossRef CAS.
- F. Ikeda, N. Crosetto and I. Dikic, Cell, 2010, 143, 677–681 CrossRef CAS PubMed.
- J. A. Harrigan, X. Jacq, N. M. Martin and S. P. Jackson, Nat. Rev. Drug Discovery, 2018, 17, 57–78 CrossRef CAS.
- D. Flierman, G. J. van der Heden Van Noort, R. Ekkebus, P. P. Geurink, T. E. T. Mevissen, M. K. Hospenthal, D. Komander and H. Ovaa, Cell Chem. Biol., 2016, 23, 472–482 CrossRef CAS.
- S. D. Whedon, N. Markandeya, A. S. J. B. Rana, N. A. Senger, C. E. Weller, F. Tureček, E. R. Strieter and C. Chatterjee, J. Am. Chem. Soc., 2016, 138, 13774–13777 CrossRef CAS PubMed.
- J. Liang, L. Zhang, X.-L. Tan, Y.-K. Qi, S. Feng, H. Deng, Y. Yan, J.-S. Zheng, L. Liu and C.-L. Tian, Angew. Chem., Int. Ed., 2017, 56, 2744–2748 CrossRef CAS.
- P. Xu, D. M. Duong, N. T. Seyfried, D. Cheng, Y. Xie, J. Robert, J. Rush, M. Hochstrasser, D. Finley and J. Peng, Cell, 2009, 137, 133–145 CrossRef CAS PubMed.
- R. Meledin, S. M. Mali, O. Kleifeld and A. Brik, Angew. Chem., Int. Ed., 2018, 57, 5645–5649 CrossRef CAS PubMed.
- J. R. Shaeffer, J. Biol. Chem., 1994, 269, 22205–22210 CrossRef CAS.
- J. Liu, C. F. Han, B. Xie, Y. Wu, S. X. Liu, K. Chen, M. Xia, Y. Zhang, L. J. Song, Z. Q. Li, T. Zhang, F. Ma, Q. Q. Wang, J. L. Wang, K. J. Deng, Y. Zhuang, X. H. Wu, Y. Z. Yu, T. Xu and X. T. Cao, Nat. Immunol., 2014, 15, 612–622 CrossRef CAS PubMed.
- T. P. Yang, X. M. Li, X. C. Bao, Y. M. E. Fung and X. D. Li, Nat. Chem. Biol., 2016, 12, 70–72 CrossRef CAS PubMed.
- X.-D. Tan, M. Pan, S. Gao, Y. Zheng, J. Shi and Y.-M. Li, Chem. Commun., 2017, 53, 10208–10211 RSC.
- W. Gui, C. A. Ott, K. Yang, J. S. Chung, S. Shen and Z. Zhuang, J. Am. Chem. Soc., 2018, 140, 12424–12433 CrossRef CAS PubMed.
- G. Saito, J. A. Swanson and K. D. Lee, Adv. Drug Delivery Rev., 2003, 55, 199–215 CrossRef CAS.
- Z. Q. Li, L. H. Qian, L. Li, J. C. Bernhammer, H. V. Huynh, J. S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2016, 55, 2002–2006 CrossRef CAS.
- W. Gui, S. Shen and Z. Zhuang, J. Am. Chem. Soc., 2020, 142, 19493–19501 CrossRef CAS PubMed.
- Y. J. Machida, Y. Machida, A. A. Vashisht, J. A. Wohlschlegel and A. Dutta, J. Biol. Chem., 2009, 284, 34179–34188 CrossRef CAS PubMed.
- X. L. Zhuo, X. Guo, X. Y. Zhang, G. H. Jing, Y. Wang, Q. Chen, Q. Jiang, J. J. Liu and C. M. Zhang, J. Cell Biol., 2015, 210, 727–735 CrossRef CAS PubMed.
- M. P. C. Mulder, K. Witting, I. Berlin, J. N. Pruneda, K.-P. Wu, J.-G. Chang, R. Merkx, J. Bialas, M. Groettrup, A. C. O. Vertegaal, B. A. Schulman, D. Komander, J. Neefjes, F. El Oualid and H. Ovaa, Nat. Chem. Biol., 2016, 12, 523–530 CrossRef CAS PubMed.
- C. M. Pickart, E. M. Kasperek, R. Beal and A. Kim, J. Biol. Chem., 1994, 269, 7115–7123 CrossRef CAS.
- K.-C. Pao, N. T. Wood, A. Knebel, K. Rafie, M. Stanley, P. D. Mabbitt, R. Sundaramoorthy, K. Hofmann, D. M. F. van Aalten and S. Virdee, Nature, 2018, 556, 381–385 CrossRef CAS.
- S. Minucci and P. G. Pelicci, Nat. Rev. Cancer, 2006, 6, 38–51 CrossRef CAS PubMed.
- Y. Xie, J. Ge, H. Lei, B. Peng, H. Zhang, D. Wang, S. Pan, G. Chen, L. Chen, Y. Wang, Q. Hao, S. Q. Yao and H. Sun, J. Am. Chem. Soc., 2016, 138, 15596–15604 CrossRef CAS PubMed.
- P. Bheda, H. Jing, C. Wolberger and H. N. Lin, Annu. Rev. Biochem., 2016, 85, 405–429 CrossRef CAS PubMed.
- M. D. Spalding and S. T. Prigge, Microbiol. Mol. Biol. Rev., 2010, 74, 200–228 CrossRef CAS PubMed.
- Y. Xie, L. Chen, R. Wang, J. Wang, J. Li, W. Xu, Y. Li, S. Q. Yao, L. Zhang, Q. Hao and H. Sun, J. Am. Chem. Soc., 2019, 141, 18428–18436 CrossRef CAS PubMed.
- Y. Fu, D. Dominissini, G. Rechavi and C. He, Nat. Rev. Genet., 2014, 15, 293–306 CrossRef CAS PubMed.
- Y. N. Xing, Z. Li, Y. Chen, J. B. Stock, P. D. Jeffrey and Y. G. Shi, Cell, 2008, 133, 154–163 CrossRef CAS PubMed.
- T. C. Petrossian and S. G. Clarke, Mol. Cell. Proteomics, 2011, 10, M110.000976 CrossRef PubMed.
- S. C. Lu, Int. J. Biochem. Cell Biol., 2000, 32, 391–395 CrossRef CAS PubMed.
- C. Dalhoff, M. Huben, T. Lenz, P. Poot, E. Nordhoff, H. Koster and E. Weinhold, Chembiochem, 2010, 11, 256–265 CrossRef CAS PubMed.
- B. D. Horning, R. M. Suciu, D. A. Ghadiri, O. A. Ulanovskaya, M. L. Matthews, K. M. Lum, K. M. Backus, S. J. Brown, H. Rosen and B. F. Cravatt, J. Am. Chem. Soc., 2016, 138, 13335–13343 CrossRef CAS PubMed.
- J. S. Takahashi, L. H. Pinto and M. H. Vitaterna, Science, 1994, 264, 1724–1733 CrossRef CAS PubMed.
- C. G. Parker, A. Galmozzi, Y. Wang, B. E. Correia, K. Sasaki, C. M. Joslyn, A. S. Kim, C. L. Cavallaro, R. M. Lawrence, S. R. Johnson, I. Narvaiza, E. Saez and B. F. Cravatt, Cell, 2017, 168, 527–541 CrossRef CAS PubMed.
- S. M. Hacker, K. M. Backus, M. R. Lazear, S. Forli, B. E. Correia and B. F. Cravatt, Nat. Chem., 2017, 9, 1181–1190 CrossRef CAS PubMed.
- M. Shrivastav, E. Gounaris, M. W. Khan, J. Ko, S. H. Ryu, M. Bogyo, A. Larson, T. A. Barret and D. J. Bentrem, PloS One, 2018, 13, e0206568 CrossRef PubMed.
- Z. Shi, X. Han, W. Hu, H. Bai, B. Peng, L. Ji, Q. Fan, L. Li and W. Huang, Chem. Soc. Rev., 2020, 49, 7533–7567 RSC.
- F. Zhang and Z. Lei, Angew. Chem., Int. Ed., 2020 DOI:10.1002/anie.202007040.
- M. N. Gandy, A. W. Debowski and K. A. Stubbs, Chem. Commun., 2011, 47, 5037–5039 RSC.
- S. E. Dettling, M. Ahmadi, Z. Lin, L. He and M. L. Matthews, Curr. Protoc. Chem. Biol., 2020, 12, e86 CAS.
| This journal is © The Royal Society of Chemistry 2021 |