Open Access Article
Open Access ArticleNonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search†
Mengyi
Yang
ab,
Ruirui
Sun
a,
Pujuan
Deng
c,
Yuzhuo
Yang
a,
Wenjuan
Wang
d,
Jun-Jie Gogo
Liu
 *c and
Chunlai
Chen
*c and
Chunlai
Chen
 *a
*a
aSchool of Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Tsinghua University, Beijing, China. E-mail: chunlai@mail.tsinghua.edu.cn
bLaboratory of Nutrition and Development, Key Laboratory of Major Diseases in Children, Ministry of Education, Beijing Pediatric Research Institutue, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing 100045, China
cSchool of Life Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, Tsinghua University, Beijing, China. E-mail: junjiegogoliu@tsinghua.edu.cn
dSchool of Life Sciences, Technology Center for Protein Sciences, Tsinghua University, Beijing, China
First published on 20th August 2021
Abstract
RNA-guided Streptococcus pyogenes Cas9 (SpCas9) is a sequence-specific DNA endonuclease that works as one of the most powerful genetic editing tools. However, how Cas9 locates its target among huge amounts of dsDNAs remains elusive. Here, combining biochemical and single-molecule fluorescence assays, we revealed that Cas9 uses both three-dimensional and one-dimensional diffusion to find its target with high efficiency. We further observed surprising apparent asymmetric target search regions flanking PAM sites on dsDNA under physiological salt conditions, which accelerates the target search efficiency of Cas9 by ∼10-fold. Illustrated by a cryo-EM structure of the Cas9/sgRNA/dsDNA dimer, non-specific interactions between DNA ∼8 bp downstream of the PAM site and lysines within residues 1151–1156 of Cas9, especially lys1153, are the key elements to mediate the one-dimensional diffusion of Cas9 and cause asymmetric target search regions flanking the PAM. Disrupting these non-specific interactions, such as mutating these lysines to alanines, diminishes the contribution of one-dimensional diffusion and reduces the target search rate by several times. In addition, low ionic concentrations or mutations on PAM recognition residues that modulate interactions between Cas9 and dsDNA alter apparent asymmetric target search behaviors. Together, our results reveal a unique searching mechanism of Cas9 under physiological salt conditions, and provide important guidance for both in vitro and in vivo applications of Cas9.
Introduction
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRIPSR-associated (Cas) protein function as an adaptive immune system to protect bacteria and archaea from invading bacteriophages and plasmids.1–5 In the class two type II system, the signature Cas9 endonuclease, which is guided by a CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA) or by a chimeric single-guide RNA (sgRNA), specifically locates the protospacer adjacent motif (PAM) sites and binds and cleaves a 20 bp-length complementary DNA flanked by a PAM site.6–10 CRISPR-Cas9 has been exploited as one of the most powerful tools in genomic engineering in living cells.11–15At the cellular level, Cas9 has to search among huge amounts of dsDNA to find its target with high accuracy and efficiency. After an initial interaction with dsDNA, the Cas9/RNA complex non-specifically binds at non-PAM sites and is quickly released, whereas interactions between Cas9/RNA and the PAM of a cognate target triggers local melting of dsDNA at the PAM-adjacent nucleation site to promote further R-loop formation and extension.8,9,16 A stable Cas9/RNA/dsDNA complex forms when a PAM-proximal 8–12 nt region (seed) is fully matched towards the guide RNA. Single-molecule fluorescence assays have previously been employed to understand how Cas9 searches for its target on dsDNA. DNA curtain experiments revealed a random collision (three-dimensional diffusion, 3D diffusion) model of Cas9 to search for its targets.16 With improved spatial resolution, single-molecule fluorescence resonance energy transfer (FRET) assays under low salt conditions capture one-dimensional (1D) diffusion of Cas9 among multiple PAM sites on the same dsDNA molecule.17 However, how Cas9 finds its target under physiological salt conditions remains largely unknown. In addition, there is no structural basis to provide mechanistic details regarding how Cas9 interacts with dsDNA during 1D diffusion.
Here, by combining ensemble biochemical and single-molecule fluorescence assays, we discovered that the Streptococcus pyogenes Cas9 (SpCas9) combines both 3D and 1D diffusion to find its targets, which increases the rate of target search by 10-fold or more than the one without 1D diffusion. Surprisingly, under physiological salt conditions, Cas9 displays an apparent asymmetric target search flanking the PAM, which is from ∼10 bp upstream of the PAM to ∼30 bp downstream of the PAM. To ensure rapid target search, the accessibility of target sites and ∼30 bp downstream of the PAM is essential for Cas9 to interact and diffuse. With a cryo-EM structure of the Cas9/sgRNA/dsDNA dimer, we clearly visualized a non-specific interaction site formed between lysine1153 of Cas9 and DNA ∼8 bp downstream of the PAM, which provides mechanistic insights into the molecular basis of the apparent asymmetric search behavior. Herein, we revealed a new kind of target search mechanism of proteins on dsDNA, in which the binding site to maintain 1D diffusion and the recognition site to locate the target are ∼8 bp away. Our discoveries provide new guidance to design and optimize Cas9 target sites for in vitro and in vivo applications and shed light on how two sites of distinctive functions coordinate with each other to achieve diffusional facilitated target searching.
Results and discussion
One-dimensional (1D) diffusion of Cas9 on dsDNA
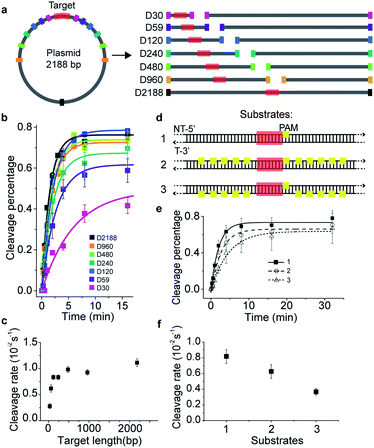
To investigate the target search mechanisms of SpCas9 under close to physiological conditions, 150 mM KCl was used, unless stated otherwise. Time-dependent in vitro cleavage assay was performed towards a series of dsDNAs with different lengths, which were linearized from the same plasmid containing a single cleavage site (Fig. 1a and Table S1†). The cleavage site was centered in the linearized dsDNA fragments, whose length varied from 30 bp to 2188 bp. When needed, a linearized fragment containing no cleavage site was supplied to maintain an identical total amount and base composition of dsDNAs. The apparent cleavage rates of Cas9/RNA towards target-containing dsDNAs of different lengths were resolved and quantified (Fig. 1b, c, S1a–g and Tables S1 and S2†), which are contributed by the overall target search rate and the enzymatic cleavage rate after Cas9 locating its target site. If Cas9 locates its cleavage target simply by 3D diffusion, the cleavage rates towards target-containing dsDNAs should be independent of the dsDNA length, because the concentrations of Cas9 and its target site remained constant in these experiments. Clearly, apparent cleavage rates increased significantly when the length of target-containing dsDNA increased from 30 bp to 120 bp and remained almost constant when they were 120 bp or longer. Therefore, we speculated that extended dsDNA fragments flanking cleavage sites accelerate the target searching process via a previously proposed facilitated diffusion model,18–21 which involves initial association of Cas9 at random dsDNA sites via random collisions (3D diffusion) and non-specific interactions, followed by 1D diffusion along dsDNA to locate the PAM sites of its specific targets. According to the facilitated diffusion model, when the length of dsDNA is shorter than the 1D diffusion length of Cas9 on dsDNA and the contribution of 1D diffusion is diminished, and the overall target search rate and the apparent cleavage rate would decrease, which agrees with our results (Fig. 1a–c).Cas9 is capable of transient binding to PAM sites in the absence of heteroduplex formation.22 In agreement with previous reports,8,16,22 multiple PAM sites surrounding the cleavage site were able to significantly reduce the apparent cleavage rate (Fig. 1d–f, S1h, Tables S1 and S2†). Notably, the cleavage site flanked by ten PAM sites on the non-target strand (substrate #2) was cleaved faster than that flanked by ten PAMs on the target strand (substrate #3, Fig. 1d–f and S1h†). Such phenomena contradicted the model that Cas9 uses only 3D random collision with dsDNA to search for its target. A more plausible search model is that, after initial random collision, Cas9 non-specifically binds to and one-dimensionally translocates on dsDNA to search for PAMs of cleavage sites on one single strand. To search for PAM sites on the other DNA single strand, Cas9 has to dissociate and re-associate with the dsDNA in an inverted orientation. A similar 1D diffusion model has been proposed and experimentally verified using the BbvCI restriction enzyme23 and DNA glycosylase hUNG and hOGG1.24,25 Consistent with this model, for substrate #2, Cas9 enriched by multiple PAM sites can diffuse one-dimensionally and locate their nearby PAM of the cleavage site in the same strand without the need of dissociation, whereas, for substrate #3, Cas9 enriched by PAM sites has to dissociate and re-associate with the dsDNA in the inverted orientation to find the PAM site of the cleavage site. This explained why the apparent cleavage rate of substrate #2 is faster than that of substrate #3. Therefore, our results support the model in which the Cas9/RNA complex can transiently interact with dsDNA in two inverted orientations via 3D diffusion and diffuse between nearby PAMs in the same strand via 1D diffusion.
Asymmetric target search regions flanking the PAM under physiological buffer conditions
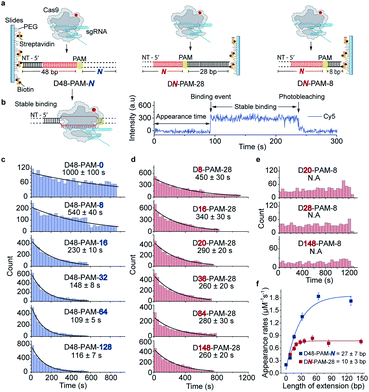
Single-molecule fluorescence assays were used to examine the 1D diffusion of Cas9 along dsDNA. Single Cy5 fluorophore-labeled Cas9/RNA complexes were allowed to flow into a reaction chamber containing immobilized biotinylated dsDNA (Fig. 2 and Table S3†). The time from injection of Cy5-Cas9 until the appearance of individual stable Cy5 fluorescence spots was recorded, which represents the total target search time (appearance time in Fig. 2b) spent by Cas9 to locate and recognize a PAM and to form a stable complex with its target site via both 3D diffusion and 1D diffusion (Fig. 2a and b). The distributions of the target search time on different dsDNAs were fitted by single exponential decay to quantify their time constants and apparent target search rates (Fig. 2c–f).For the target dsDNAs of the D48-PAM-N series (details of the nomenclature are shown in Fig. 2a), the distance between the PAM site and the 5′ end of the non-target strand remains the same (48 bp), whereas the distance between the PAM sites and the 3′ end of non-target strand varied from 0 bp to 128 bp (Fig. 2a and c, Table S3†). With the D48-PAM-N series, we were able to quantify how the extension of the dsDNA downstream of the PAM (towards 3′ end of the non-target strand) contributes to 1D diffusion facilitated target search of Cas9. We discovered that the downstream of the PAM greatly facilitated target search of Cas9 by ∼10-fold (Fig. 2c and Table S4†), which confirmed the model that Cas9 utilizes 1D diffusion along dsDNA to increase its target search efficiency. The maximal effective target search rate of Cas9 is ∼2 μM−1 s−1 under our experimental conditions (Table S4†), which is close to a previously reported value (∼4 μM−1 s−1).10
On the other hand, the DN-PAM-28 series (Fig. 2a, d and Table S3†), whose distance between the PAM site and the 5′ end of non-target strand varied from 8 bp to 148 bp (as the ∼8 bp seed region is necessary for Cas9/RNA to form a stable complex with dsDNA), were used to quantify how the extension of the dsDNA upstream of the PAM (towards 5′ end of the non-target strand) contributes to 1D diffusion facilitated target search of Cas9. For the DN-PAM-28 series, the apparent target search rates of Cas9 remained almost the same except for the one with D8-PAM-28, which was only ∼2-fold slower than the others (Fig. 2d and Table S4†). Similar behaviors were captured when three different sets of dsDNAs were used (Fig. S2, Tables S3 and S4†), which eliminated the possibility of sequence-dependent mechanisms. As a further support, when we used the DN-PAM-8 series, the extension of the dsDNA upstream of the PAM has no detectable effect on the apparent target search rates of Cas9 (Fig. 2a, e, Tables S3 and S4†). Together, these single-molecule fluorescence assays revealed an unusual asymmetric target search behavior. The extension of the dsDNA downstream of the PAM greatly facilitates target search of Cas9 via 1D diffusion, whereas the PAM upstream extension has only minor effects. Therefore, longer extension of the PAM downstream region causes the DN-PAM-28 series to exhibit a faster target search rate than the DN-PAM-8 series. We quantified the asymmetric region contributing to the facilitated 1D diffusion of Cas9 to be from 10 ± 3 bp upstream of the PAM to 27 ± 7 bp downstream of the PAM (Fig. 2f), which is close to the previously reported ∼20 bp diffusion length.17
Asymmetric target search regions verified by single-molecule FRET assay
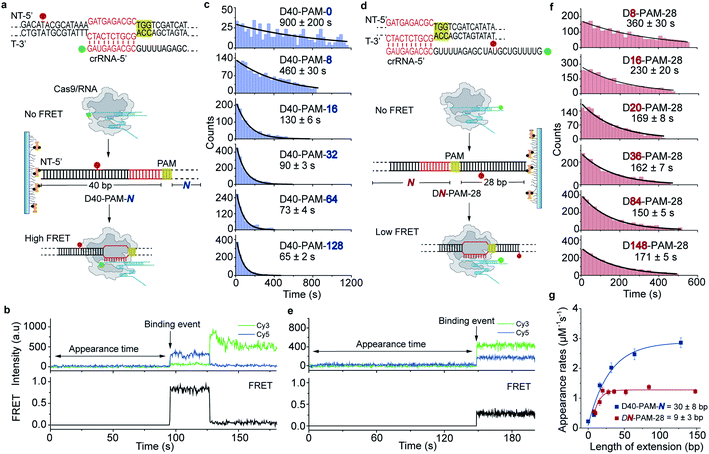
The single-molecule fluorescence assay described above cannot pinpoint the position where the Cas9 was stably bound on the dsDNA because only the Cas9/RNA complex was labeled with a fluorophore. Herein, we developed single-molecule FRET assays utilizing FRET signals between Cy3 labeled crRNA and Cy5 labeled dsDNA to probe stable interactions between Cas9 and dsDNA at the correct target site. To avoid potential influence of the dsDNA-attaching Cy5 fluorophore on the 1D diffusion of Cas9, two different FRET pairs were designed (Fig. 3a, d and Table S5†). In the design used to quantify the contribution of downstream extension of the PAM to the 1D diffusion facilitated target search of Cas9 (the D40-PAM-N series, Fig. 3a–c), Cy3 was labeled on the 5′ end of crRNA and Cy5 was labeled at the 5′ end of the non-target strand. In the other design used to quantify the contribution of upstream extension of the PAM to 1D diffusion facilitated target search of Cas9 (the DN-PAM-28 series, Fig. 3d–f), Cy3 was labeled on the 3′ end of crRNA and Cy5 was labeled close to the 3′ end of the non-target strand. The time from injection of Cy3-Cas9/RNA onto Cy5-dsDNA until the appearance of an individual stable FRET signal was extracted from time-dependent fluorescence trajectories and assigned as the total target search time spent by Cas9 via both 3D and 1D diffusion (appearance time in Fig. 3b and e).Time constants and appearance target search rates were quantified from the distributions of the target search time by single exponential fitting (Fig. 3c, f and Table S6†). The target search rates quantified via the single-molecule FRET assay (Fig. 3g) are slightly faster than the ones determined via the single-molecule fluorescence assay (Fig. 2f), because the wild-type Cas9 displays higher activity than the labeled one.26,27 Still, the results of both assays showed similar behaviors that only the dsDNA extension located PAM downstream facilitates target search of Cas9 by ∼10-fold, whereas the extension of the dsDNA upstream of the PAM has minor effects. The asymmetric region contributing to the facilitated 1D diffusion of Cas9 is from 9 ± 3 bp upstream of the PAM to 30 ± 8 bp downstream of the PAM (Fig. 3g), which also agrees with the values determined above (10 ± 3 bp and 27 ± 7 bp, Fig. 2f). Therefore, both assays confirmed that Cas9 displays unreported apparent asymmetric target search flanking PAM sites.
To rule out the possibility that the asymmetric target search of Cas9 is an artifact caused by surface immobilization, we switched the biotinylation site to the opposite end of dsDNA relative to the design in Fig. 3 (Fig. S3, Tables S5 and S6†). We discovered that the location of the dsDNA biotinylation site and the orientation of surface immobilization only caused <2-fold change in apparent target search rates, and thus the extension of dsDNA downstream of the PAM is still the dominant factor to accelerate apparent target search rates (>7-fold change, Fig. S3†). Taken together, we observed an asymmetric target search process of Cas9 flanking the PAM on dsDNA, whose target search rate is mainly accelerated by the facilitated 1D diffusion originating from the downstream region of the PAM.
Symmetric target search in low-salt buffer
Ionic strength is known to affect the kinetics and interactions between DNA and DNA-binding proteins.21,28–31 To evaluate how ionic strength affects the 1D diffusion of Cas9, we performed single-molecule fluorescence and FRET assays as shown in Fig. 2 and 3 in low-salt buffer containing 10 mM NaCl (Fig. 4, Tables S4 and S6†). Strikingly, extension of either end of dsDNA greatly increased the target search rates of Cas9, indicating a symmetric target search process, which agreed with previous single-molecule FRET measurements in the same low-salt buffer.17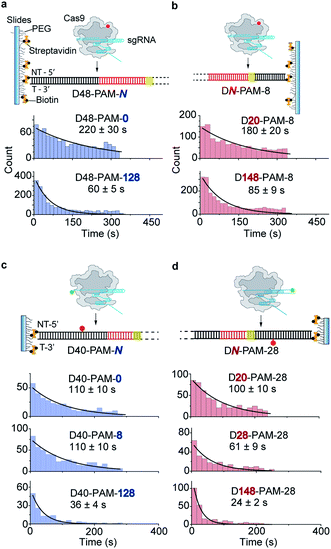 | ||
| Fig. 4 1D diffusion under low salt conditions. (a and b) Scheme of single-molecule fluorescence assay and distributions of the appearance time of labeled Cas9 on D48-PAM-N DNAs (a) and DN-PAM-8 DNAs (b) in low salt buffer (50 mM HEPES–NaOH pH 7.5, 10 mM NaCl, 2 mM MgCl2, 1 mM DTT). (c and d) Scheme of single-molecule FRET assay and distributions of the appearance time of Cas9 on D40-PAM-N DNAs (c) and DN-PAM-28 (d) in low salt buffer. The FRET pair is designed the same as indicated in Fig. 3a and d, respectively. Single exponential decay was used to extract the average appearance time from injection until stable binding of Cas9 on dsDNAs. | ||
Non-specific interactions between Cas9 and dsDNA governing 1D diffusion and target search
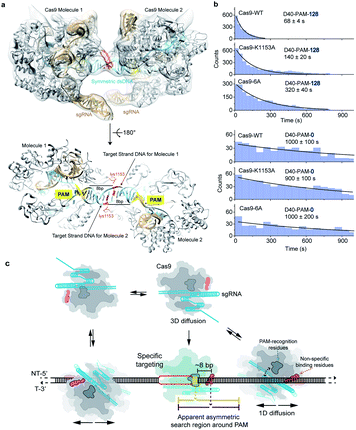
Previous reported structures have suggested the potential interactions between the stretching loop (Glu1150 to Leu1157) from the Cas9 PI domain and the DNA downstream of the PAM.32–35 This non-specific binding site located downstream of the PAM has been shown to affect the binding of Cas9.36 Due to the structural flexibility in this local region, it's hard to identify the key residues and the specific nucleotide involved in this interaction. To provide further structural details, we reconstituted a SpCas9 dimer complex on a symmetric dsDNA which helps to restrain the structural flexibility in this specific region. In this SpCas9 dimer complex, we are able to observe the clear interaction between lys1153 and the target strand DNA backbone ∼8 bp downstream of the PAM, which was not visible in previous structures (Fig. 5a and S4†).Next, our single-molecule FRET assays clearly revealed that disrupting these non-specific interactions significantly slows the 1D-diffusion driven target search of Cas9 (Fig. 5b). The search rate on DNA D40-PAM-128 was reduced by ∼2-fold when lys1153 was mutated to alanine (Cas9-K1153A). When adjacent residues 1151–1156 were mutated from KGKSKK to AAAAAA (Cas9-6A), the search rate was further reduced by additional ∼2-fold (total ∼5-fold decrease relative to Cas9-WT), suggesting that the nearby positively charged residues lys1551, lys1555 and lys1556 may also assist with the non-specific DNA binding through more flexible conformations during the 1D diffusion of Cas9 on dsDNA. In our control experiments, both Cas9-K1153A and Cas9-6A retained their DNA cleavage activities (Fig. S5†). In addition, when contribution of 1D diffusion on target search is diminished by truncating DNA downstream of the PAM, both Cas9-K1153A and Cas9-6A displayed similar search rates on DNA D40-PAM-0 to Cas9-WT (Fig. 5b and Table S7†), suggesting that target search attributed to 3D diffusion is not affected by these mutations. Together, our structural and single-molecule results demonstrated that the non-specific interactions between the DNA ∼8 bp downstream of the PAM site and residues 1151–1156 of Cas9 are the key elements to mediate the 1D diffusion of Cas9 and to facilitate target search.
Asymmetric target search of Cas9 affected by residues recognizing PAM sites
By modifying PAM-interacting residues, Cas9-VQR (D1135V/R1335Q/T1337R) and Cas9-EQR (D1135E/R1335Q/T1337R) have been engineered to recognize alternative 5′-TGAN-3′ PAM sequences.37,38 Their target search behaviors were examined by our single-molecule assays. Cas9-VQR displays similar asymmetric behaviors to the wild-type Cas9 (Table S8†). For the Cas9-EQR variant, extension of either end of dsDNA significantly increased rates of Cas9 target search, indicating a symmetric target search behavior (Fig. S6 and Table S8†). Such phenomena suggested that the Cas9 residues interacting with dsDNA to recognize PAM sites are also able to affect 1D diffusion and the asymmetric target search behaviors of Cas9.Modeling target search of Cas9
Our results support a model in which Cas9 combines both 3D and 1D diffusion to find its target with high efficiency and accuracy (Fig. 5c). A freely diffusing Cas9/RNA complex interacts with dsDNA through 3D random collision. After non-specific interaction with dsDNA through lysines within residues 1151–1156, Cas9 transiently binds and laterally diffuses along dsDNA (1D diffusion) to search for PAM sites. On average, Cas9 diffuses for ∼20 bp before dissociation, unless it encounters a PAM of a cognate target, recognizes the PAM and forms a stable binding complex through RNA strand invasion and R-loop expansion.35,39 Otherwise, Cas9 continues its 1D diffusion on dsDNA after transient binding with PAM sites flanking the non-target sequence.17Our results suggested that diffusion-facilitated target search of Cas9 is fulfilled by two Cas9–DNA interaction sites of distinctive functions, which are the non-specific binding site to maintain transient binding during 1D diffusion and the PAM-recognition site to locate PAMs. Under physiological buffer conditions, the interactions between the PAM-recognition site and dsDNA are weak and cannot mediate long-distance 1D diffusion of Cas9. However, the interactions between the non-specific binding site and dsDNA are strong enough to maintain transient binding of Cas9 on dsDNA to perform 1D-diffusion driven PAM search, which increases the target search efficiency by ∼10-fold. Because the non-specific binding site is located ∼8 bp downstream of the PAM, the symmetric random diffusion (∼20 bp) around the non-specific binding site leads to an apparent asymmetric search region from ∼10 bp upstream of the PAM to ∼30 bp downstream of the PAM (Fig. 5c). Under physiological salt conditions, the target search rate of Cas9 is mainly accelerated by the facilitated 1D diffusion from the downstream region of the PAM, providing important guidance for designing target sites of Cas9.
Any factor altering one or both of Cas9–DNA interaction sites would affect the 1D diffusion mediated target search. Mutating lysines in residues 1151–1156 greatly suppresses 1D diffusion without affecting 3D diffusion, which slows the overall target search rate as shown in Fig. 5b. The low-salt condition, which is known to strengthen interactions between DNA binding proteins and dsDNA21,28–30,40 and to increase the 1D sliding length of several endonucleases,41 is likely to significantly strengthen interactions between dsDNA and the PAM-recognition site of Cas9. Thus, the PAM-recognition site might contribute to mediating 1D diffusion in low-salt buffer, which causes Cas9 to display a symmetric search region around the PAM. In addition, Cas9-VQR with altered PAM preference still exhibits an asymmetric search region while Cas9-EQR shows close-to-symmetric search behaviors under physiological salt conditions (Fig. S6 and Table S8†). The D1135V and D1135E mutations, which are the only difference between Cas9-VQR and Cas9-EQR, are proposed to play auxiliary roles to accommodate the PAM duplex in a displaced conformation.37,38 The fourth G of the PAM sequence in the Cas9-EQR is fixed more tightly than that in the Cas9-VQR, because the aliphatic portion of the D1135E side chain forms a van der Waals contact with its ribose moiety. Therefore, tight interactions between the PAM-recognition site and dsDNA caused by Cas9 mutations might also increase the contributions of the PAM-recognition site in mediating 1D diffusion, which leads to symmetric search around the PAM as we discussed above.
A mathematical model has been developed by Halford and Marko to estimate the total time required for a DNA binding protein finding its target by combining 3D and 1D diffusion,18 in which the total search time is estimated via
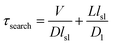 | (1) |
Our measurements showed that lsl = ∼6.8 nm (∼20 bp). With an improved time resolution of 2.5 ms per frame, we captured that the average dwell time of Cas9 from nonspecific binding until dissociation after 1D diffusion on a PAM-free dsDNA (τsl) is 11.2 ± 0.2 ms (Fig. S7a and b†). Introducing 1 or 3 PAMs into the PAM-free dsDNA has almost no effect on the nonspecific binding rate of Cas9 on dsDNA (Fig. S7a right column and Fig. S7e†), indicating that the initial binding between Cas9 and dsDNA is mediated by non-specific binding without PAM recognition. On the other hand, the presence of 1 or 3 PAMs slightly increased the dwell time of Cas9 on dsDNA after initial binding (Fig. S7a† left column). Based on the dwell time of Cas9 on the PAM-free dsDNA, we have D1 = lsl2/τsl = 4.1 × 103 nm2 s−1 (3.3 × 104 bp2 s−1), which is significantly slower than reported diffusion constants of Cas12a and many DNA glycosylases.31,42,43 For Escherichia coli (E. coli), V ≈ 109 nm3 and L ≈ 106 nm3.18 For the Cas9/RNA complex, D ≈ 5 × 107 nm2 s−1. Together, we have τsearch = 1.7 × 103 s, which suggests that a Cas9/RNA molecule needs 0.47 hour to search through the whole E. coli nucleoid to find its target site when there is no other PAM site at all. However, in reality, there are ∼106 PAM sites in the E. coli nucleoid,44 many of which contain partial complementary target sequences towards the guide sequence of the Cas9/RNA complex. These PAM sites and off-target sites would significantly lengthen their binding times with Cas9/RNA10 and greatly increase the total search time. Consistent with our speculation, a recent study revealed that a single Cas9 protein needs to spend 6 hours to find the correct target site in live E. coli.44 Therefore, our results suggested that the vast majority of time consumed by Cas9 in the target search process is to interact with endogenous PAM sites and off-target sites, whereas the diffusion process including both 3D and 1D motions to search for target sites is less time-consuming.
Our work provided important mechanistic insights into the target search of Cas9. The actual target search process in live cells is further complicated by inter-segmental transfer between nearby DNAs and hopping of Cas9 over other DNA binding proteins. Interestingly, the average dwell time of Cas9 on dsDNA during 1D-diffusion driven target search is 11–16 ms (Fig. S7a and b†), which agrees well with the value (17 ± 4 ms) measured in live Lactococcus lactis cells.45 Therefore, the mechanistic model and the kinetics determined by our work provided a foundation to understand the target search of Cas9 in vivo.
Conclusions
For most DNA binding proteins, the site to bind DNA and the site to recognize target sequence are the same one or located adjacent to each other.31,42,46 Herein, we revealed that Cas9 utilizes two separated sites to fulfill these distinctive functions, DNA binding and PAM recognition, separately, which underlines the mechanistic diversity in diffusional facilitated target searching. We speculated that, the non-specific interactions between dsDNA and lysines within residues 1151–1156 of Cas9 not only promote the 1D diffusion of Cas9 on dsDNA to facilitate RNA-guided target search by ∼10-fold but also play important roles in mediating RNA-independent DNA cleavage activities.47,48 Due to ∼8 bp offset between this non-specific binding site and the PAM-recognition site, the facilitated diffusion of Cas9 is mainly contributed by the region from ∼10 bp upstream of the PAM to ∼30 bp downstream of the PAM, which causes apparent asymmetric search behavior. Our scheme clearly shows that the accessibility of ∼30 base pairs downstream of a PAM site dominates the efficiency of Cas9 to locate this PAM, which provides important guidance to design target sites for both in vivo and in vitro applications of Cas9.Data availability
The EM reconstruction of the Cas9 dimer and it's corresponding atomic coordinate have been deposited in the Electron Microscopy Data Bank, under accession code EMD-31721, and in the Protein Data Bank, under accession code 7V59.Author contributions
MY, JJGL and CC designed the experiments; MY, RS, YY and WW prepared materials and reagents and performed biochemical and single-molecule experiments; PD and JJGL solved the cryo-EM structure, and MY and CC analyzed the data and wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by grants from the National Natural Science Foundation of China (21877069, 21922704 and 22061160466 to CC and 22007054 to WW), grants from the Tsinghua-Peking Joint Center for Life Sciences and Beijing Advanced Innovation Center for Structural Biology to CC and JJGL, and grants from the Beijing Frontier Research Center for Biological Structure to CC and Lab Innovation Funding from Lab and Instrument Department, Tsinghua University to WW. The authors would like to acknowledge Lijingyao Zhang and Weixiong Zhong for the assistance in sample preparation.Notes and references
- S. J. Brouns, M. M. Jore, M. Lundgren, E. R. Westra, R. J. Slijkhuis, A. P. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin and J. van der Oost, Science, 2008, 321, 960–964 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- L. A. Marraffini, Nature, 2015, 526, 55–61 CrossRef CAS PubMed.
- A. V. Wright, J. K. Nunez and J. A. Doudna, Cell, 2016, 164, 29–44 CrossRef CAS PubMed.
- B. Wiedenheft, S. H. Sternberg and J. A. Doudna, Nature, 2012, 482, 331–338 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- M. Jinek, F. Jiang, D. W. Taylor, S. H. Sternberg, E. Kaya, E. Ma, C. Anders, M. Hauer, K. Zhou, S. Lin, M. Kaplan, A. T. Iavarone, E. Charpentier, E. Nogales and J. A. Doudna, Science, 2014, 343, 1247997 CrossRef PubMed.
- C. Anders, O. Niewoehner, A. Duerst and M. Jinek, Nature, 2014, 513, 569–573 CrossRef CAS PubMed.
- M. D. Szczelkun, M. S. Tikhomirova, T. Sinkunas, G. Gasiunas, T. Karvelis, P. Pschera, V. Siksnys and R. Seidel, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 9798–9803 CrossRef CAS PubMed.
- D. Singh, S. H. Sternberg, J. Fei, J. A. Doudna and T. Ha, Nat. Commun., 2016, 7, 12778 CrossRef CAS PubMed.
- G. J. Knott and J. A. Doudna, Science, 2018, 361, 866–869 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262–1278 CrossRef CAS PubMed.
- R. Barrangou and J. A. Doudna, Nat. Biotechnol., 2016, 34, 933–941 CrossRef CAS PubMed.
- J. A. Doudna and E. Charpentier, Science, 2014, 346, 1258096 CrossRef PubMed.
- S. H. Sternberg, S. Redding, M. Jinek, E. C. Greene and J. A. Doudna, Nature, 2014, 507, 62–67 CrossRef CAS PubMed.
- V. Globyte, S. H. Lee, T. Bae, J. S. Kim and C. Joo, EMBO J., 2019, 38(4), e99466 CrossRef PubMed.
- S. E. Halford and J. F. Marko, Nucleic Acids Res., 2004, 32, 3040–3052 CrossRef CAS PubMed.
- U. Gerland, J. D. Moroz and T. Hwa, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12015–12020 CrossRef CAS PubMed.
- P. H. von Hippel and O. G. Berg, J. Biol. Chem., 1989, 264, 675–678 CrossRef CAS.
- O. G. Berg, R. B. Winter and P. H. von Hippel, Biochemistry, 1981, 20, 6929–6948 CrossRef CAS PubMed.
- V. Mekler, L. Minakhin and K. Severinov, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5443–5448 CrossRef CAS PubMed.
- D. M. Gowers, G. G. Wilson and S. E. Halford, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 15883–15888 CrossRef CAS PubMed.
- J. D. Schonhoft and J. T. Stivers, Nat. Chem. Biol., 2012, 8, 205–210 CrossRef CAS PubMed.
- M. M. Rowland, J. D. Schonhoft, P. L. McKibbin, S. S. David and J. T. Stivers, Nucleic Acids Res., 2014, 42, 9295–9303 CrossRef CAS PubMed.
- M. Yang, S. Peng, R. Sun, J. Lin, N. Wang and C. Chen, Cell Rep., 2018, 22, 372–382 CrossRef CAS PubMed.
- S. H. Sternberg, B. LaFrance, M. Kaplan and J. A. Doudna, Nature, 2015, 527, 110–113 CrossRef CAS PubMed.
- T. M. Lohman, CRC Crit. Rev. Biochem., 1986, 19, 191–245 CrossRef CAS PubMed.
- I. Bonnet, A. Biebricher, P. L. Porte, C. Loverdo, O. Benichou, R. Voituriez, C. Escude, W. Wende, A. Pingoud and P. Desbiolles, Nucleic Acids Res., 2008, 36, 4118–4127 CrossRef CAS PubMed.
- A. Tempestini, C. Monico, L. Gardini, F. Vanzi, F. S. Pavone and M. Capitanio, Nucleic Acids Res., 2018, 46, 5001–5011 CrossRef CAS PubMed.
- S. Peng, X. Wang, L. Zhang, S. He, X. S. Zhao, X. Huang and C. Chen, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 21889–21895 CrossRef CAS PubMed.
- C. Huai, G. Li, R. J. Yao, Y. Zhang, M. Cao, L. L. Kong, C. Q. Jia, H. Yuan, H. Y. Chen, D. R. Lu and Q. Huang, Nat. Commun., 2017, 8(1), 1375 CrossRef PubMed.
- H. Nishimasu, F. A. Ran, P. D. Hsu, S. Konermann, S. I. Shehata, N. Dohmae, R. Ishitani, F. Zhang and O. Nureki, Cell, 2014, 156, 935–949 CrossRef CAS PubMed.
- F. Jiang, D. W. Taylor, J. S. Chen, J. E. Kornfeld, K. Zhou, A. J. Thompson, E. Nogales and J. A. Doudna, Science, 2016, 351, 867–871 CrossRef CAS PubMed.
- F. Jiang and J. A. Doudna, Annu. Rev. Biophys., 2017, 46, 505–529 CrossRef CAS PubMed.
- Q. Zhang, F. Wen, S. Zhang, J. Jin, L. Bi, Y. Lu, M. Li, X. G. Xi, X. Huang, B. Shen and B. Sun, Sci. Adv., 2019, 5, eaaw9807 CrossRef CAS PubMed.
- S. Hirano, H. Nishimasu, R. Ishitani and O. Nureki, Mol. Cell, 2016, 61, 886–894 CrossRef CAS PubMed.
- C. Anders, K. Bargsten and M. Jinek, Mol. Cell, 2016, 61, 895–902 CrossRef CAS PubMed.
- S. A. Gorski, J. Vogel and J. A. Doudna, Nat. Rev. Mol. Cell Biol., 2017, 18, 215–228 CrossRef CAS PubMed.
- A. Mondal and A. Bhattacherjee, Nucleic Acids Res., 2015, 43, 9176–9186 CrossRef CAS PubMed.
- R. M. Ferreira, A. D. Ware, E. Matozel and A. C. Price, Biochem. Biophys. Res. Commun., 2021, 534, 1059–1063 CrossRef CAS PubMed.
- P. C. Blainey, G. Luo, S. C. Kou, W. F. Mangel, G. L. Verdine, B. Bagchi and X. S. Xie, Nat. Struct. Mol. Biol., 2009, 16, 1224–1229 CrossRef CAS PubMed.
- Y. Jeon, Y. H. Choi, Y. Jang, J. Yu, J. Goo, G. Lee, Y. K. Jeong, S. H. Lee, I. S. Kim, J. S. Kim, C. Jeong, S. Lee and S. Bae, Nat. Commun., 2018, 9, 2777 CrossRef PubMed.
- D. L. Jones, P. Leroy, C. Unoson, D. Fange, V. Curic, M. J. Lawson and J. Elf, Science, 2017, 357, 1420–1424 CrossRef CAS PubMed.
- K. J. A. Martens, S. P. B. van Beljouw, S. van der Els, J. N. A. Vink, S. Baas, G. A. Vogelaar, S. J. J. Brouns, P. van Baarlen, M. Kleerebezem and J. Hohlbein, Nat. Commun., 2019, 10(1), 3552 CrossRef PubMed.
- A. Tafvizi, F. Huang, A. R. Fersht, L. A. Mirny and A. M. van Oijen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 563–568 CrossRef CAS PubMed.
- R. Sundaresan, H. P. Parameshwaran, S. D. Yogesha, M. W. Keilbarth and R. Rajan, Cell Rep., 2017, 21, 3728–3739 CrossRef CAS PubMed.
- C. Saha, P. Mohanraju, A. Stubbs, G. Dugar, Y. Hoogstrate, G. J. Kremers, W. A. van Cappellen, D. Horst-Kreft, C. Laffeber, J. H. G. Lebbink, S. Bruens, D. Gaskin, D. Beerens, M. Klunder, R. Joosten, J. A. A. Demmers, D. van Gent, J. W. Mouton, P. J. van der Spek, J. van der Oost, P. van Baarlen and R. Louwen, Sci. Adv., 2020, 6, eaaz4849 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc02633j |
| This journal is © The Royal Society of Chemistry 2021 |