Electrochemical and spectroelectrochemical characterization of bacteria and bacterial systems
Vignesh
Sundaresan
 a,
Hyein
Do
a,
Hyein
Do
 b,
Joshua D.
Shrout
b,
Joshua D.
Shrout
 cde and
Paul W.
Bohn
cde and
Paul W.
Bohn
 *ab
*ab
aDepartment of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN 46556, USA. E-mail: pbohn@nd.edu
bDepartment of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, IN 46556, USA
cDepartment of Civil and Environmental Engineering and Earth Sciences, University of Notre Dame, Notre Dame, IN 46556, USA
dEck Institute for Global Health, University of Notre Dame, Notre Dame, IN 46556, USA
eDepartment of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA
First published on 7th December 2021
Abstract
Microbes, such as bacteria, can be described, at one level, as small, self-sustaining chemical factories. Based on the species, strain, and even the environment, bacteria can be useful, neutral or pathogenic to human life, so it is increasingly important that we be able to characterize them at the molecular level with chemical specificity and spatial and temporal resolution in order to understand their behavior. Bacterial metabolism involves a large number of internal and external electron transfer processes, so it is logical that electrochemical techniques have been employed to investigate these bacterial metabolites. In this mini-review, we focus on electrochemical and spectroelectrochemical methods that have been developed and used specifically to chemically characterize bacteria and their behavior. First, we discuss the latest mechanistic insights and current understanding of microbial electron transfer, including both direct and mediated electron transfer. Second, we summarize progress on approaches to spatiotemporal characterization of secreted factors, including both metabolites and signaling molecules, which can be used to discern how natural or external factors can alter metabolic states of bacterial cells and change either their individual or collective behavior. Finally, we address in situ methods of single-cell characterization, which can uncover how heterogeneity in cell behavior is reflected in the behavior and properties of collections of bacteria, e.g. bacterial communities. Recent advances in (spectro)electrochemical characterization of bacteria have yielded important new insights both at the ensemble and the single-entity levels, which are furthering our understanding of bacterial behavior. These insights, in turn, promise to benefit applications ranging from biosensors to the use of bacteria in bacteria-based bioenergy generation and storage.
Introduction
Microorganisms are tremendously important to human life, both because of our symbiotic relationships with them and for their utility in many areas of technology, such as food sciences, biomedicine and genetic engineering.1 On the other hand, microbial exposure can lead to deleterious outcomes ranging from the unpleasant, e.g. body odor, to the life threatening, e.g. systemic inflammatory response syndrome.2 As just one example, a broad range of microbes, including bacteria, molds, and yeasts, are used in food production, yet some bacteria contribute to contamination and food spoilage.3,4 In the context of human health, there are ca. 1400 known species of human pathogens, spanning bacteria, viruses, and fungi, and some of the bacterial pathogens responsible for high mortality and morbidity, e.g. Streptococcus pneumoniae and Pseudomonas aeruginosa.5–7 Therefore, the problem of microbial characterization has two main components: (1) the ability to detect and identify microbes at the species, and even strain, level in order to correlate them with pathogenic exposure, and (2) developing tools to better understand the molecular underpinnings of microbial behavior, including the structures, metabolites, electron transfer mechanisms, that collectively determine their functional characteristics.1,8 The detection and identification of microbial species, especially pathogens, is an area of intense investigation that has been the subject of a number of recent reviews.9–11 Therefore, this mini-review will concentrate on the second area – specifically on electrochemical and spectroelectrochemical means of elucidating microbial behavior at the molecular level.A wide range of analytical methods have been applied for detecting and analyzing microbes, depending on the purpose and the level of information needed.12 Some techniques have been developed for rapid and reliable bacterial identification, e.g. polymerase chain reaction-based methods,13–15 mass spectrometry,16–18 flow cytometry,19,20 and fluorescence immunoassay.21–23 In the last two decades, absorption, scattering, and vibrational techniques, e.g. uv-visible absorption, Raman, and Fourier-transform infrared (FTIR) spectroscopies, have demonstrated their great utility in microbial identification and have also been applied to obtain detailed information on the chemical composition of complex heterogeneous microbial systems.24–26 In addition to being non-destructive, label-free, and needing only minimal sample pretreatment, both IR and Raman spectra provide spectral fingerprints, thus delivering comprehensive chemical information about the main characteristics of biological systems at the molecular level.26,27 As just two examples, our group has used confocal Raman hyperspectral imaging to characterize how P. aeruginosa signaling molecules respond to different environmental conditions in both two- and three-dimensions,28,29 and Holman and coworkers used FTIR spectromicroscopy to monitor and characterize Escherichia coli biofilm activity at a molecular level over long times.30
Electrochemical methods, which have also been widely used to investigate microbial systems, are particularly powerful, because they provide information that is complementary to spectroscopy, especially those involving the redox properties of microbial analytes. Electrochemical approaches, in general, provide rapid response times, simple operation, good sensitivity, and are cost-efficient.31,32 Generally, bacteria can transport electrons across their cell membrane such that they electrically interact with their environment.33 Therefore, electrochemical methods have the advantage of being able to explore the interaction between an electrode surface and living microbial cells, which is especially useful, for example, in applications such as electricity production and bioremediation.34,35 In addition, electrochemical approaches can address both technological useful applications, such as microbial fuel cells, as well as potentially harmful processes such as bacterial-initiated metal corrosion.35–37 Recently, Simoska et al. demonstrated in vitro detection of three redox-active phenazine metabolites from the opportunistic human pathogen P. aeruginosa using carbon-based ultramicroelectrode (UME) sensing electrodes to monitor and characterize the production of the phenazines in real-time.38 Qiao et al. demonstrated that E. coli evolves under electrochemical tension in a microbial fuel cell in a such a way that it secretes hydroquinone derivatives through a highly permeable outer membrane, which then act as mediators for electron transport between cell and electrode.39
These examples, and those to be discussed below, illustrate the ability of electrochemical techniques to rapidly provide quantitative information about electroactive species. However, electrochemistry is limited in providing information about molecular structure. This provides powerful motivation for coupling electrochemistry with spectroscopy, since the two approaches generally offer complementary information. For example, changes in spectral line profiles, reflecting changes in the electronic structure of the molecule, are typically observed when the redox state is changed electrochemically.40,41 This feature has driven the application of spectroelectrochemical approaches in microbial sciences – targeting species as diverse as redox enzymes, electroactive bacteria, and microbial biofilms.40,42–44
In this review, we describe how electrochemical and spectroelectrochemical approaches are useful in understanding the characteristics of bacteria at both ensemble and single-entity levels. The review is meant to highlight the way in which advanced spectroelectrochemical measurements can be used to discern important operational characteristics of complex microbial electron transfer systems. It is specifically not intended to be an exhaustive or comprehensive examination of microbial external electron transfer (EET) for which other excellent recent reviews are available.45–48 The review is organized in three sections. In the first, we discuss recent mechanistic insights obtained on direct and mediated electron transfer processes occurring in bacteria. The discussion on direct electron transfer is exemplified by electron transport in Geobacter sulfurreducens whereas, the discussion of mediated electron transfer focuses on the necessary attributes of redox mediators. Next, we discuss how (spectro)electrochemical methods can be used to analyze the spatiotemporal distribution of secreted metabolites in order to understand the manner in which bacteria sense and react to their environment, by using P. aeruginosa as an example. In the final section, we describe how (spectro)electrochemical strategies have been exploited to study single bacterial cells. While earlier reports are highlighted to provide historical context, the primary emphasis is on papers published in the past five years.
Microbial electron transfer
Metabolism in bacteria can be regulated by internal or external electron transfer events, or by a combination of the two. A great deal of attention has been given to external electron transfer (EET) because it's direct relevance in fields such as microbial fuel cells, corrosion, and sensors.49–51 EET can occur in microbes by one of two mechanisms: (1) direct electron transfer (Fig. 1, left), in which electron transfer occurs through membrane-associated redox proteins, and (2) mediated electron transfer (Fig. 1, right), in which electron transfer between the cell membrane and electrode occurs through the agency of a redox mediator.52 Since there are recent reviews focusing on EET as it relates to microbial fuel cells,49,50,53 corrosion51,54,55 and sensors,32,56 here we discuss only the recent advances in developing mechanistic understanding of EET using electrochemical and spectroelectrochemical methods.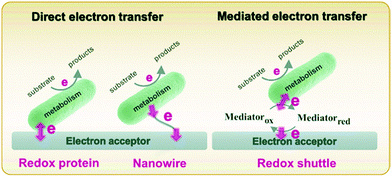 | ||
| Fig. 1 Schematic diagram describing direct (left) and mediated (right) electron transfer in the microbial system. Adapted with permission from ref. 57. Copyright 2020 Progress in Chemistry. | ||
Direct electron transfer
Direct electron transfer between the bacterial cell and electrode typically occurs via one of three mechanisms: through (1) redox proteins, such as C-type cytochromes and flavoproteins, (2) conductive pili, and (3) endogenously produced mediators that are bound to the cell membrane.49 Mechanistic understanding of the direct electron transfer process is important in order to improve understanding of bacterial metabolism and better design bioelectrochemical systems, and also as a starting point for the development of engineered redox proteins bound to the cell membrane.Owing to its ability to form thick biofilms and conduct long-distance electron transport, Geobacter sulfurreducens is commonly used in bioelectrochemical systems, specifically microbial fuel cells.58,59 EET occurs in G. sulfurreducens through membrane-resident c-type cytochromes and/or conductive pili.60–63 Electrochemical methods have been used to decipher the EET mechanism, track biofilm formation, and identify the charge state of the cells. For example, open circuit potentiometry was used to measure the charge stored in G. sulfurreducens.64 The results indicated three cytochrome proteins in the periplasm capable of storing charges. Rova and co-workers developed a dynamic model for EET in G. sulfurreducens using data obtained by a combination of in situ resonance Raman microscopy and chronoamperometry. Using this approach, they were able to calculate quantitative rate constants for electron transfer at different steps from the inner membrane to the electrode surface.65 Interestingly, G. sulfurreducens can be used for both anodic and cathodic bioelectrochemical systems. While the electron transfer mechanism of the anodic reaction, which occurs via cytochromes in the membrane and/or pili is reasonably well-understood, a deeper mechanistic understanding of the cathodic electron transfer process is needed.
Reisner and co-workers addressed this issue spectroelectrochemically by employing in situ resonance Raman spectroscopy and uv-visible absorption in both anodic and cathodic environments.66 First, anodically-grown G. sulfurreducens biofilms were interrogated for their activity towards oxidation of acetate to CO2. Then, the same biofilm containing electrode was operated in cathodic mode for the reduction of fumarate to succinate. The corresponding cyclic voltammograms of both anodic (Fig. 2A) and cathodic (Fig. 2B) modes show expected sigmoidal responses indicating reversible redox reactions. The uv-visible absorption spectrum (Fig. 2C) exhibits Soret bands at 409 and 419 nm in anodic mode, which are associated with the heme-type cytochromes. However, these bands disappear in cathodic mode, indicating the depletion of cytochromes at more negative potentials. Moreover, the electrode becomes red and increasingly darker over multiple cathodic cycles, and resonance Raman spectroscopy and electron microscopy suggest the presence of iron oxide nanoparticles on or near the cells. Taken together, the authors proposed that the EET in anodic mode occurs mainly via cytochromes, while EET in cathodic mode could be mediated by iron species and/or iron oxide nanoparticles produced by heme-containing cytochromes, as shown in Fig. 2D. Similarly, Yi et al. used electrochemical methods to study the mechanism of EET in Shewanella loihica, which is also capable of bidirectional electron transport.67 They postulated that riboflavin acts as a redox mediator in two different modes – freely diffusing for outward EET (electron transfer from bacteria to the electrode), or as a bound species for inward EET (electron transfer from the electrode to bacteria). Spectroelectrochemical studies have also been used to understand direct interspecies electron transfer (electron transfer directly between bacterial species) in Geobacter co-cultures involving Geobacter metallireducens and G. sulfurreducens.68In situ Raman scattering and FTIR revealed that interspecies electron transport is mediated by c-type cytochromes. Additionally, electrochemical studies and confocal laser scanning microscopy imaging with a pilR-deficient G. sulfurreducens mutant showed that this mutant strain forms thinner and less conductive biofilms, giving support to the importance of PilR in regulating PilA and overall type IV pilus (TFP) appendage production, as TFP are known to have important roles in biofilm formation and EETs.69
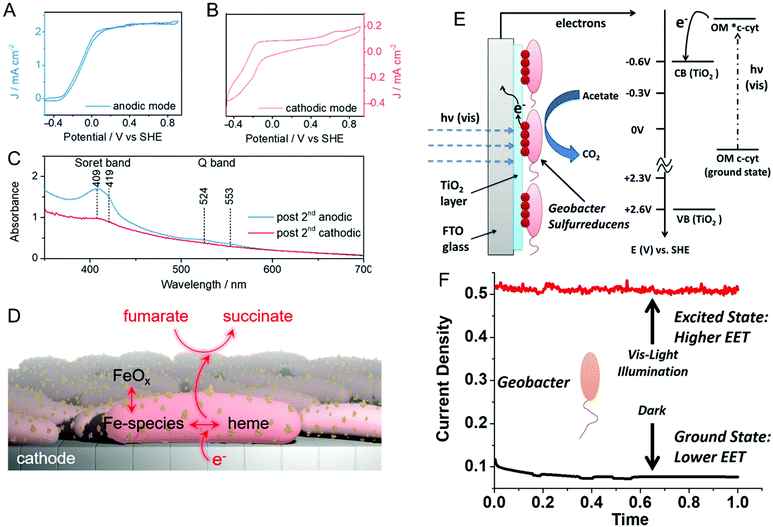 | ||
| Fig. 2 Cyclic voltammograms of a G. sulfurreducens biofilm on an indium-tin-oxide (ITO) electrode in (A) anodic (acetate to CO2) and (B) cathodic (fumarate to succinate) modes. (C) UV-visible spectra of biofilms after anodic and cathodic scans. (D) Schematic illustrating plausible EET mechanism in cathodic mode. (E) Schematic and energy diagram showing EET from G. sulfurreducens directly to TiO2 under visible illumination. OM = outer membrane. (F) Current density obtained from G. sulfurreducens under dark and illuminated conditions. Panels A–D are adapted with permission from ref. 66. Copyright 2020 American Chemical Society. Panel E and F are adapted with permission from ref. 70. Copyright 2020 Elsevier B.V. | ||
Recent reports suggest that visible light can play an important role in dictating the electron transfer through cell membrane proteins. For example, Zhang et al. show that visible illumination of G. sulfurreducens can excite c-type cytochromes (OM c-cyts) in the cell membrane to an energy level high enough to easily transfer electrons to TiO2 as shown in Fig. 2E.70 This visible-light driven approach produced an 8× improvement in EET as compared to the non-illuminated condition (Fig. 2F). Apart from enhancing the EET using light, Tefft and TerAvest showed that illuminating Shewanella oneidensis with green light can generate a proton pump or proton motive force within the cell, which can reverse the direction of electron transfer such that electrons can be transferred from the cathode to a proximal bacterial cell for reduction.71 These reports highlight how visible light can be used to improve and manipulate EET, enabling the design of bioelectrochemical systems with enhanced performance. Apart from G. sulfurreducens, electrochemical and spectroelectrochemical methods have been used to understand and characterize a wide variety of bacteria under a wide variety of other conditions,72–78 including recognizing EETs in mammalian gut microbiota,73 and long-distance electron transfer in a Gram-positive bacterium, Lysinibacillus isolate GY32.72
Mediated electron transfer
Most bacteria do not contain accessible redox-active species in their (outer) membrane and even in bacteria that do, transferring electrons directly via membrane-resident redox proteins is challenging because of factors like poorly electrically conducting cell membranes, and inaccessibility to redox proteins.79,80 To overcome these issues, redox mediators have been employed to shuttle electrons from the electrode to the bacterial redox site and vice versa.81–83 Redox mediators play an important role not only in accelerating EET processes but also in enhancing the efficiency of bioelectrochemical systems. Some of the commonly encountered redox mediators utilized by different bacteria include flavins, quinones, and phenazines.81,84,85 Both voltammetric and amperometric methods have been used to examine the redox mediator competency and its interaction with the bacterial cell.86–88 Recently, Minteer and co-workers performed a comprehensive electrochemical study to understand the phenazine-based redox mediators and their interaction with E. coli.88 They employed nine different phenazine-based redox mediators, out of which neutral red (NR) exhibited the highest current density, as shown in Fig. 3A. Additionally, cytotoxicity studies showed that NR did not affect cell growth in this system. Expanding to all mediators, the measured current densities can be correlated to cytotoxicity to an extent that dictates that redox potential is not the only criteria in choosing redox mediators. Analogously, Liu and co-workers studied the effect of redox mediators on S. oneidensis MR-1 biofilm formation.89 Even though previous reports showed that redox mediators promote biofilm formation, the mechanisms by which this occurs are poorly understood. The authors employed five different redox mediators, all of which promoted biofilm formation as evident by both the increase in current density (Fig. 3B) and the robust morphology shown in the SEM images (Fig. 3C). The improvement in EET efficiency was attributed to synergies between mediators promoting biofilm formation and upregulating gene expression for cell membrane constituents. In some cases, endogenously produced molecules can mediate electron transfer.90–92 For example, Mulla and co-workers showed that at elevated temperatures (55 °C), thermophilic Geobacillus sp. produce extracellular polysaccharide (EPS) which contains flavins that mediate EET.93 In addition, Zhuang and co-workers showed that the production and composition of EPS can be controlled by the application of oxidizing potentials in a mixed community biofilm.94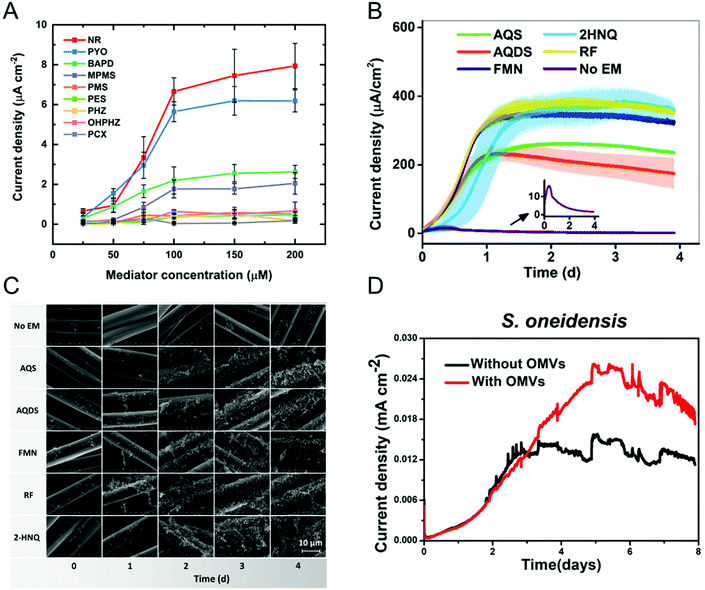 | ||
| Fig. 3 (A) Current densities obtained from E. coli immobilized on carbon paper electrode with nine different redox mediators at six different concentrations. NR: neutral red, PYO: pyocyanin, BAPD: benzo(A)phenazine-7,12-dioxide, MPMS: 1-methoxy-5-methylphenazinium methyl sulfate (MPMS), PMS: phenazine methosulfate, PES: phenazine ethosulfate, PHZ: phenazine, OHPHZ: 1-hydroxyphenazine, and PCX: phenazine-1-carboxamide. (B) Current densities as a function of time for S. oneidensis with five different redox mediators. AQS: 9,10-anthraquinone-2-sulfonic acid, AQDS: 9,10-anthraquinone-2,6-disulfonic acid, FMN: flavin mononucleotide, 2HNQ: 2-hydroxy-1,4-napthoquinone, and RF: riboflavin. (C) Scanning electron micrographs showing S. oneidensis biofilm formation as a function of redox mediator and time. (D) Current density obtained from S. oneidensis with and without outer membrane vesicles (OMVs). Panel A is adapted with permission from ref. 88. Copyright 2021 The Electrochemical Society. Panels B and C are adapted with permission from ref. 89. Copyright 2020 American Chemical Society. Panel D is adapted with permission from ref. 95. Copyright 2019 American Chemical Society. | ||
Augmenting traditional redox mediators, Liu et al. demonstrated that bacterial vesicles can also mediate EET.95 Several Gram-negative bacteria release outer membrane vesicles (OMVs) that contain c-type cytochromes which facilitate electron transfer. Electrochemical studies showed a ∼1.7× enhancement in current density when OMVs were used as mediators (Fig. 3D). Additionally, OMVs can enable electron transport in non-exoelectrogens including E. coli. It is important to note that a variety of bacteria secrete redox-active metabolites, which can also be used as mediators. Clearly, characterizing these metabolites would do much to enable the understanding of metabolic state, as discussed in the next section.
Spatiotemporal analysis of metabolites
Bacteria are social organisms, so in response to cell-population density and external stimuli, they commonly regulate gene expression and secrete signaling molecules as a means to communicate and coordinate with neighboring organisms, a phenomenon known as quorum sensing (QS) or quorum signaling.96,97 Many Gram-negative bacteria secrete N-acyl homoserine lactones (AHL) as the signaling molecule to effect this collective mode of communication. For example, the bioluminescence of Aliivibrio fischeri is related to the concentration and action of AHLs.98,99 In one species of opportunistic human pathogenic bacteria, P. aeruginosa, four interconnected QS systems, including las, rhl, pqs and iqs, with unique signaling molecules associated with each system that are used to coordinate collective activities such as biofilm formation and swarming motility.100–102 Secreted phenazines, for example, are known to act as virulence factors facilitating the survival of P. aeruginosa infections in the host organism.103,104 Apart from sensing and quantifying phenazine molecules to track P. aeruginosa pathogenesis, spatiotemporal quantification of phenazines can also aid in understanding collective behavior and biofilm formation. P. aeruginosa produces a multiplicity of phenazine derivatives, e.g. phenazine-1-carboxylic acid (PCA), 5-methyl-phenazine-1carboxylic acid (5-MCA), pyocyanin (PYO), and phenazine-1-carboxamide (PCN). Of these, PYO is highly virulent and responsible for both chronic and acute infections.105–107 Moreover, all of these phenazines are redox-active, undergoing reversible proton-coupled electron transfer oxidation/reduction reactions, thus positioning electrochemical methods to detect and quantify them.108–110Bard and co-workers used scanning electrochemical microscopy to map the concentration of PYO produced in a P. aeruginosa biofilm in three-dimensions.111 In their approach, a microelectrode biased at the oxidation potential of PYO was positioned above the biofilm (Fig. 4A), and reduced PYO produced by the biofilm diffused to the microelectrode where it could be oxidized, with the measured oxidation current being proportional to the PYO concentration. Then, the microelectrode was raster-scanned across the biofilm to obtain the electrochemical map shown in Fig. 4B, in which the redox current is proportional to PYO concentration. Moreover, raster scanning was done at a different microelectrode-biofilm separations to build a 3D profile of secreted PYO. Later, SECM technique was coupled with micro-3D printing (capable of fabricating protein-based walls around an individual or small population of bacteria) to understand the aggregate size and community-dependent behavior of P. aeruginosa,112 revealing that at least 500 cells per aggregate are required to initiate quorum sensing. By utilizing the wild-type and mutant strain aggregates at defined spatial locations, the authors observed at least 2000 cells were required to induce quorum sensing in neighboring aggregate positioned 8 μm away. In a complementary approach Bellin et al., developed an integrated circuit-based platform for spatial monitoring of phenazine produced by a P. aeruginosa biofilm.113 In this structure, an array of electrodes was placed under the bacterial colony separated by a thin agar layer. Because the electrodes were interrogated in spatially-dependent manner, PYO concentrations were obtained in different locations to obtain a spatial map of PYO concentration within the colony. With a spatial resolution of 750 μm, they detected higher concentrations of PYO at the colony edges than at the center. Interestingly, this result was opposite to the Bard and co-workers’ observation, where the redox current determined by the SECM was higher at the center of the biofilm than the edges. This difference could be explained by differences in mass transport mechanisms in the two experimental geometries freely diffusing PYO in the SECM map, as opposed to diffusion through an agar matrix in the integrated circuit-based platform. The integrated circuit-based platform was later extended to image the spatial distribution of multiple phenazine metabolites produced by the P. aeruginosa PA14 biofilms, finding that while PCA was distributed throughout the colony, 5-MCA and PYO were localized near the colony edges.114
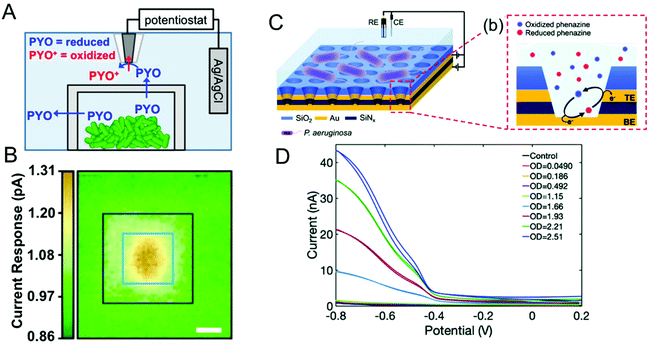 | ||
| Fig. 4 (A) Schematic diagram showing SECM in combination with 3D printed microtrap system for mapping PYO spatial distributions. (B) Electrochemical current maps of PYO secreted by P. aeruginosa using the SECM approach of panel A. Scale bar = 10 μm. (C) Left: Schematic of dual ring nanopore electrode array (NEA) in contact with P. aeruginosa. Right: Schematic showing redox cycling of phenazine in NEA. (D) Redox cycling induced cyclic voltammetry of P. aeruginosa as a function of optical density (OD). Panels A and B are adapted with permission from ref. 112. Copyright 2014 National Academy of Sciences. Panel C and D are adapted with permission from ref. 117. Copyright 2021 The Royal Society of Chemistry. | ||
Stevenson and co-workers have also carried out extensive studies of P. aeruginosa, based on electrochemical detection of phenazines using transparent carbon ultramicroelectrode arrays.38,109,115,116 For example, they demonstrated temporal tracking of phenazines, showing that PYO concentration increases over time in the first 21 h corresponding to the exponential growth of bacteria, after which it stabilizes.38 However, 5-MCA, the precursor to PYO, increases until intermediate times and decreases later, most likely reflecting the conversion of 5-MCA to PYO. In addition, both PYO and 5-MCA production vary slightly as a function of growth medium, providing a way to understand the environmental effects on bacterial growth and quorum sensing. Using the same approach, they have explored a range of environmental effects on P. aeruginosa To explore the effect of other bacterial pathogens, P. aeruginosa was cultured with other pathogens such as Staphyococcus aureus and E. coli in different growth media, and phenazine production was monitored to understand the effect of co-culture.109 In the presence of S. aureus, phenazine production was diminished in one growth medium, but not altered in the other. However, the presence of E. coli in co-culture substantially altered phenazine production independent of growth medium. To study the effect of the anti-bacterial agents, these authors targeted the antimicrobial properties of Ag+ with a specific focus on dynamic effects. Ag+ was introduced to bacterial solution grown for 6 h in two different media, and phenazine production decreased substantially within 30 min in both media,116 indicating either inhibition of metabolic process or cell death.
Since metabolite concentrations are low (<1 μM) during the initial bacterial growth stages, ultra-sensitive methodologies are needed. To address this challenge, ring-disk nanopore electrode arrays (NEAs), in which the disk electrode at the bottom and the ring electrode at the top of the nanopore is separated by an insulator, were developed in the authors’ laboratory.118 Biasing the two electrodes at differing potentials (one oxidizing and one reducing) enables redox cycling with accompanying current amplification thus making ultra-sensitive detection possible. When P. aeruginosa-containing solution is added to the top of the NEA devices, the bacteria are excluded from the pores since their diameter is much smaller than the cell size, admitting only metabolites into the nanopore where they undergo redox cycling, thus minimizing biofouling the electrode surface, as illustrated in Fig. 4C.117 Redox cycling enhanced voltammetry of PYO produced by the P. aeruginosa is shown in Fig. 4D. NEA voltammetry detected phenazines as low as 10 nM (PYO) in buffer, and PYO concentrations of 1.5 μM were recovered from the bacterial supernatant. Apart from ultra-sensitive detection, it is also possible to obtain semi-quantitative estimates of families secreted phenazines. Recently, our group extended this work by employing an NEA device with a block copolymer (BCP) membrane to selectively determine PCA concentration using both electrochemical and surface-enhanced Raman scattering (SERS). Since the BCP membrane is both pH- and charge-selective, by adjusting the pH of the bacterial medium above the pKa of PCA – but below the pKa of PYO and PCN – it is possible to selectively transport anionic PCA into the nanopores for both electrochemical and spectroscopic quantification.106 In addition, Zór and co-workers developed a centrifugal microfluidic lab-on-a-disk platform based on a supported liquid membrane (SLM) for extracting, enriching, and detecting hydroxycinnamic acid (pHCA), a metabolite produced by E. coli.119,120 The platform was constructed with donor and acceptor units separated by an SLM acting as a charge selective layer. By tuning the pH of the solution in the donor unit, neutral pHCA is able to diffuse through SLM and reach the acceptor unit, while the interferants are blocked. This platform detected pHCA concentration as low as 250 μM.
With the exception of SECM, most electrochemical methods discussed here can provide temporal analysis, but obtaining spatially-dependent information is more challenging.38,109,115–117,119,120 Spectroelectrochemistry offers one possible solution to this conundrum. Our group has coupled electrochemistry and surface-enhanced Raman spectroscopy (EC-SERS) to map phenazines produced by both wild type and mutant P. aeruginosa biofilms.121 Both pH- and potential-dependent changes were observed in PYO, as shown in Fig. 5. Raman band shifts and intensity changes were attributed to changes in the electronic structure, especially the central ring of PYO induced by proton-coupled electron transfer. In addition, EC-SERS mapping revealed localized PYO deposits approximately the size of P. aeruginosa cells, suggesting that PYO secretion remains localized near the cell of origin, at least initially. Thus, EC-SERS is as an elegant tool for unearthing effect of external conditions on P. aeruginosa while also providing spatial information about the distribution of secreted metabolites.
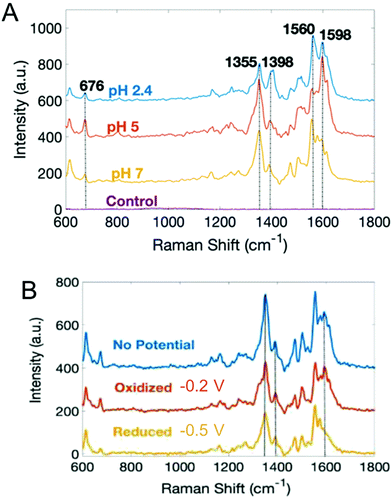 | ||
| Fig. 5 Surface-enhanced Raman spectra of PYO as a function of (A) pH, and (B) electrochemical potential. Adapted with permission from ref. 121. Copyright 2019 American Chemical Society. | ||
Single-cell characterization
In the last two sections, (spectro)electrochemical characterization of bacteria was considered at the ensemble level. Even though the measurements described are enormously powerful in, for example, deciphering EET mechanisms and characterizing the effect of environmental conditions on bacterial behavior, with these measurements alone it is not possible to isolate the contribution of individual cells to the behavior of the ensemble. On the other hand, characterizing bacteria at the single-cell level makes it possible to understand the relationship between cell-to-cell heterogeneity and bacterial behavior.122 One commonly used method for investigating individual cells electrochemically is the single-entity collision or nano-impact methodology,123 in which an ultramicroelectrode (UME) with a diameter typically less than 50 μm is used to detect the single entities impacting electrode–solution interface. Based on the redox-active nature of the entity these events can be classified into either physical blocking or catalytic amplification events. In physical blocking events, a small area of the electrode surface is occluded by the impact of a non-redox active entity, thereby causing the faradaic current to decrease. Catalytic amplification events, on the other hand, occurs when a redox-active catalytic entity contacts the UME under conditions where no electrochemical reaction would otherwise occur, thus producing an increase in current. Sepunaru et al. and Frkonja-Kuczin et al. detected E. coli by decorating them with Ag nanoparticles (NPs) and poising the UME at a potential to oxidize Ag, so the collision of single AgNP-decorated E. coli cells would be signaled by a transient increase in faradaic current resulting from the oxidation of the AgNPs.124,125 While AgNP labeling is effective, the anti-microbial property of Ag can potentially kill the cells.126 To overcome this issue, Lee et al. used the blocking approach to detect E. coli, using the collision of E. coli at a UME to impede the oxidation of ferrocyanide, as shown in Fig. 6A, top panel.127 A similar response was observed by Lebègue et al. for single S. oneidensis cells on a carbon UME.128 Ronspees and Thorgaard incorporated a fluorescence microscope in the blocking experiment to simultaneously track the attachment and movement of bacterial cells on the UME surface.129 In their experiment, E. coli showed a step current decrease as observed by other groups, however they observed that B. subtilis produced a transient current blockage. Fluorescence imaging confirmed that the step response is associated with the attachment of bacterial cells (Fig. 6B and C), while the transient response corresponds to short-lived cell attachment. Thus, spectroelectrochemical studies events can further elucidate the dynamics of single bacterial cell collision events at UMEs.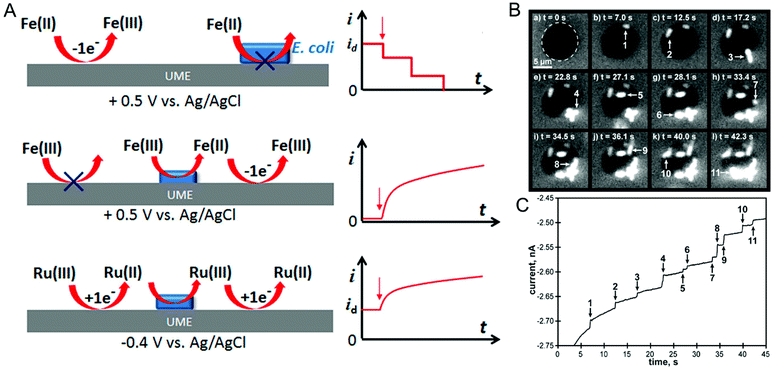 | ||
| Fig. 6 (A) Different ways of detecting bacteria-electrode collisions along with their current–time responses. Top panel: Bacterial cell blocking the UME electroactive area; Middle panel: bacterial cell catalyzing reduction; Bottom panel: regeneration of redox species by bacterial cell. (B) Time-lapse fluorescence images showing E. coli attaching to the UME surface. (C) Current–time trace corresponding to panel B. Panel A is adapted with permission from ref. 130. Copyright 2018 American Chemical Society. Panels B and C are adapted with permission from ref. 129. Copyright 2018 Elsevier Ltd. | ||
A disadvantage of the blocking approach is that it is useful only for detecting the bacteria; it cannot be used to understand their redox activity. However, by taking advantage of the redox-active nature of E. coli, Gao et al., evaluated the redox activity of a single bacterial cell.130 As shown in Fig. 6A, middle panel, E. coli reduces ferricyanide to ferrocyanide, which in turn can be re-oxidized back to ferricyanide at the UME. Thus, the measured current is directly proportional to the reduction efficiency of individual E. coli cells. Alternatively, reduced (oxidized) species generated at the UME can be oxidized (reduced) by the bacterial cell attached to the UME (Fig. 6A, bottom panel). The regeneration of species by the bacterial cell increases the overall current providing an efficient way to understand microbial redox activity. Furthermore, this approach has been extended to assess the effect of antimicrobial agents such as cobalt ions and colistin. Compton and co-workers used N,N,N′,N′-tetramethyl-para-phenylene-diamine (TMPD) as a redox mediator to characterize the behavior of single E. coli cells, exploiting the fact that cytochrome c oxidase expressed in E. coli oxidizes TMPD to TMPD˙+ which gets regenerated (reduced) at the UME making it possible to assess both redox activity and cell viability.131
Spectroelectrochemical approaches have also been employed to reveal variation in EET at the single entity level. El-Naggar and co-workers used Thioflavin T, a fluorescent cationic dye, and Nernstian membrane potential indicator to study the dynamics of S. oneidensis MR-1 membrane potential during potential-induced EET.132 Application of a positive external potential (+0.3 V) was observed to result in a negative EET membrane potential, leading to accumulation of positive Thioflavin T in the membrane, thus increasing fluorescence, as shown in Fig. 7A, first panel. The reverse process occurred when a negative external potential (−0.5 V) was applied (Fig. 7A, second panel). The potential-dependent fluorescence was followed over three consecutive potential step cycles. Moreover, the potential- and time-dependent fluorescence intensity traces for three different cells shown in Fig. 7B indicate considerable cell-to-cell variation, revealing heterogeneity in EET. Recent work from our laboratory in collaboration with Willets used a coupled fluorescence and electrochemical approach to probe direct EET in Myxococcus xanthus, a soil-dwelling bacterium important in the degradation of woody plant materials.133 Instead of adding external flavins, the potential dependent fluorescence dynamics was tracked using intrinsic membrane-associated flavoproteins, which contain flavin molecules such flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), whose fluorescence changes based on redox state, i.e. fluorescent and non-fluorescent in the oxidized and reduced forms, respectively.134 The observation of a non-canonical potential response from bacteria led to a detailed investigation of the potential, concentration, and irradiance dependence of the model electrofluorogenic compound FMN molecule, showing that the carrier dynamics of the ITO substrate play an important role in determining the potential-dependent fluorescence response, thus emphasizing the importance of the electrode in spectroelectrochemical experiments, especially at the single-cell level.
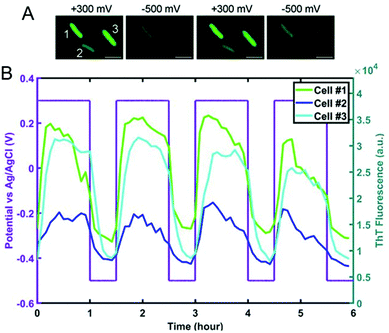 | ||
| Fig. 7 (A) Fluorescence images of S. oneidensis with thioflavin T obtained as a function of electrochemical potential. (B) Electrochemical potential- and fluorescence intensity-time traces for three individual cells marked in panel A. Adapted with permission from ref. 132. 2020 National Academy of Sciences. | ||
Conclusion and outlook
This review addressed recent advances in the use of electrochemical and spectroelectrochemical approaches to characterize bacterial systems in three broad areas. In microbial electron transfer, electrochemical and spectroelectrochemical techniques have been used to obtain mechanistic understanding of direct EET. The effect of redox mediator properties on EET and biofilm formation, which facilitate efficiency of bioelectrochemical systems, was also discussed. Apart from using spectroelectrochemical techniques to obtain more accurate mechanistic insight on EET, future research direction can productively focus on understanding the role of external factors, such as irradiation, on EET and gene expression. Further, in situ electrochemical inactivation and cell lysis could be used to analyze altered gene expression and correlate these with EET responses.135,136Second, approaches to reveal the spatiotemporal distributions of secreted factors, e.g. metabolites, were highlighted. SECM and integrated electrochemical chip-based detection provide three- and two-dimensional spatial distribution of metabolites, respectively. Carbon-based ultramicroelectrode and nanopore-electrode arrays have been used to achieve ultra-sensitive detection of metabolites enabling the development of electrochemical sensors for pathogenic bacteria. Moreover, EC-SERS is able to visualize metabolite spatial distributions as a function of changing environmental conditions. Given that chemical identity and concentration of secreted metabolites are a sensitive function of the environment, efforts to culture bacteria on miniaturized scales coupled to dynamic in situ strategies to alter the environment coupled to the metabolites measurement using spectroelectrochemistry with high spatial and temporal resolution would appear to hold much promise.
In the final section, we described electrochemical collision-based techniques for detecting single bacteria cells and measuring EET at the single-cell level. Further, fluorescence microscopy coupled with electrochemistry was found to be useful for high-throughput measurement of EETs in individual cells and to uncover cell-to-cell heterogeneity in EET kinetics. Future studies that can exploit the capability to isolate or compartmentalize individual bacteria in defined locations and study them using spectroelectrochemistry would enable us to understand not only heterogeneity between the cells but also to realize their intercellular behavior under well-defined conditions. Moreover, owing to the emergence of powerful new electrochemical imaging approaches, such as SECM, scanning ion conductance microscopy (SICM) scanning electrochemical cell microscopy (SECCM), and scanning photoelectrochemical microscopy (SPECM), it is possible to measure pH, surface charge, intracellular substances, membrane proteins, mechanical properties, and redox mediator transport by mapping them at the single-cell level.137–139 Additionally, advances in optical imaging, such as super-resolution,140 structured illumination,141 appear more ripe for combining with electrochemistry to achieve unprecedented advances in understanding behavior of bacteria and bacterial assemblies.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would like to acknowledge financial support from the following agencies for work described here from their laboratory: the Department of Energy Office of Science (Grant DE-SC0019312), the National Science Foundation (CHE-1904196), and the National Institutes of Health (R01 AI113219-06).References
- R. Franco-Duarte, L. Černáková, S. Kadam, K. S. Kaushik, B. Salehi, A. Bevilacqua, M. R. Corbo, H. Antolak, K. Dybka-Stępień, M. Leszczewicz, S. Relison Tintino, V. C. Alexandrino de Souza, J. Sharifi-Rad, H. D. Melo Coutinho, N. Martins and C. F. Rodrigues, Microorganisms, 2019, 7, 130 CrossRef CAS PubMed.
- Q. Zhang, M. Raoof, Y. Chen, Y. Sumi, T. Sursal, W. Junger, K. Brohi, K. Itagaki and C. J. Hauser, Nature, 2010, 464, 104–U115 CrossRef CAS PubMed.
- J. M. Lorenzo, P. E. Munekata, R. Dominguez, M. Pateiro, J. A. Saraiva and D. Franco, in Innovative Technologies for Food Preservation, ed. F. J. Barba, A. S. Sant'Ana, V. Orlien and M. Koubaa, Academic Press, 2018, pp. 53–107. DOI:10.1016/B978-0-12-811031-7.00003-0.
- L. Gram, L. Ravn, M. Rasch, J. B. Bruhn, A. B. Christensen and M. Givskov, Int. J. Food Microbiol., 2002, 78, 79–97 CrossRef PubMed.
- M. E. J. Woolhouse and S. Gowtage-Sequeria, Emerg. Infect. Dis. J., 2005, 11, 1842 CrossRef PubMed.
- K. W. McConnell, J. E. McDunn, A. T. Clark, W. M. Dunne, D. J. Dixon, I. R. Turnbull, P. J. Dipasco, W. F. Osberghaus, B. Sherman, J. R. Martin, M. J. Walter, J. P. Cobb, T. G. Buchman, R. S. Hotchkiss and C. M. Coopersmith, Crit. Care Med., 2010, 38, 223–241 CrossRef PubMed.
- P. Martens, S. W. Worm, B. Lundgren, H. B. Konradsen and T. Benfield, BMC Infect. Dis., 2004, 4, 21 CrossRef PubMed.
- D. Emerson, L. Agulto, H. Liu and L. Liu, BioScience, 2008, 58, 925–936 CrossRef.
- A. Ahmed, J. V. Rushworth, N. A. Hirst and P. A. Millner, Clin. Microbiol. Rev., 2014, 27, 631–646 CrossRef CAS PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Munoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- J. Hu and P. W. Bohn, J. Anal. Test., 2017, 1, 1–17 CrossRef.
- R. Karlsson, L. Gonzales-Siles, M. Gomila, A. Busquets, F. Salvà-Serra, D. Jaén-Luchoro, H. E. Jakobsson, A. Karlsson, F. Boulund, E. Kristiansson and E. R. B. Moore, PLoS One, 2018, 13, e0208804–e0208804 CrossRef CAS PubMed.
- S. A. Barghouthi, Indian J. Microbiol., 2011, 51, 430–444 CrossRef CAS PubMed.
- P. Athamanolap, K. Hsieh, L. Chen, S. Yang and T.-H. Wang, Anal. Chem., 2017, 89, 11529–11536 CrossRef CAS PubMed.
- L. Zhu, Y. Zhang, P. He, Y. Zhang and Q. Wang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1093–1094, 141–146 CrossRef CAS PubMed.
- F. Nomura, S. Tsuchida, S. Murata, M. Satoh and K. Matsushita, Clin. Proteomics, 2020, 17, 14 CrossRef CAS PubMed.
- B. Rodríguez-Sánchez and M. Oviaño, in Application and Integration of Omics-powered Diagnostics in Clinical and Public Health Microbiology, ed. J. Moran-Gilad and Y. Yagel, Springer International Publishing, Cham, 2021, pp. 175–189. DOI:10.1007/978-3-030-62155-1_10.
- C. R. Cox and K. J. Voorhees, Bacterial Identification by Mass Spectrometry. In: Banoub J. (eds) Detection of Chemical, Biological, Radiological and Nuclear Agents for the Prevention of Terrorism, NATO Science for Peace and Security Series A: Chemistry and Biology, Springer, Dordrecht, 2014, DOI:10.1007/978-94-017-9238-7_8.
- P. Rubbens, R. Props, N. Boon and W. Waegeman, PLoS One, 2017, 12, e0169754 CrossRef PubMed.
- L. X. Coggins, I. Larma, A. Hinchliffe, R. Props and A. Ghadouani, Water Res., 2020, 169, 115243 CrossRef CAS PubMed.
- Y. Shen, Y. Zhang, Z. F. Gao, Y. Ye, Q. Wu, H.-Y. Chen and J.-J. Xu, Nano Today, 2021, 38, 101121 CrossRef CAS.
- A. Qureshi and J. H. Niazi, Analyst, 2020, 145, 7825–7848 RSC.
- Y. Xu, M. M. Hassan, A. S. Sharma, H. Li and Q. Chen, Crit. Rev. Food Sci. Nutr., 2021, 1–19, DOI:10.1080/10408398.2021.1950117.
- S. A. Strola, J.-C. Baritaux, E. Schultz, A. C. Simon, C. Allier, I. Espagnon, D. Jary and J.-M. Dinten, J. Biomed. Opt., 2014, 19, 111610 CrossRef PubMed.
- J. P. Harrison and D. Berry, Front. Microbiol., 2017, 8, 675–675 Search PubMed.
- M. Harz, P. Rösch and J. Popp, Cytometry, Part A, 2009, 75, 104–113 CrossRef CAS PubMed.
- Y. Chen, C. Zou, M. Mastalerz, S. Hu, C. Gasaway and X. Tao, Int. J. Mol. Sci., 2015, 16, 30223–30250 CrossRef CAS PubMed.
- T. Cao, J. V. Sweedler, P. W. Bohn, J. D. Shrout and C. D. Ellermeier, mSphere, 2020, 5, e00426–e00420 CrossRef CAS PubMed.
- T. Cao, A. A. Weaver, S. Baek, J. Jia, J. D. Shrout and P. W. Bohn, J. Chem. Phys., 2021, 154, 204201 CrossRef CAS PubMed.
- H.-Y. N. Holman, R. Miles, Z. Hao, E. Wozei, L. M. Anderson and H. Yang, Anal. Chem., 2009, 81, 8564–8570 CrossRef CAS PubMed.
- S. Kuss, H. M. A. Amin and R. G. Compton, Chem. – Asian J., 2018, 13, 2758–2769 CrossRef CAS PubMed.
- O. Simoska and K. J. Stevenson, Analyst, 2019, 144, 6461–6478 RSC.
- M. C. Potter and A. D. Waller, Proc. R. Soc. London, Ser. B, 1911, 84, 260–276 Search PubMed.
- R. Y. A. Hassan, F. Febbraio and S. Andreescu, Sensors, 2021, 21, 1279 CrossRef CAS PubMed.
- F. Kracke, I. Vassilev and J. O. Krömer, Front. Microbiol., 2015, 6, 575–575 CrossRef PubMed.
- K.-M. Moon, H.-R. Cho, M.-H. Lee, S.-K. Shin and S.-C. Koh, Met. Mater. Int., 2007, 13, 211 CrossRef CAS.
- K.-Y. Chan, L.-C. Xu and H. H. P. Fang, Environ. Sci. Technol., 2002, 36, 1720–1727 CrossRef CAS PubMed.
- O. Simoska, M. Sans, M. D. Fitzpatrick, C. M. Crittenden, L. S. Eberlin, J. B. Shear and K. J. Stevenson, ACS Sens., 2019, 4, 170–179 CrossRef CAS PubMed.
- Y. Qiao, C. M. Li, S.-J. Bao, Z. Lu and Y. Hong, Chem. Commun., 2008, 1290–1292, 10.1039/B719955D.
- D. Millo, Biochem. Soc. Trans., 2012, 40, 1284–1290 CrossRef CAS PubMed.
- Y. Zhai, Z. Zhu, S. Zhou, C. Zhu and S. Dong, Nanoscale, 2018, 10, 3089–3111 RSC.
- M. Sezer, D. Millo, I. M. Weidinger, I. Zebger and P. Hildebrandt, IUBMB Life, 2012, 64, 455–464 CrossRef CAS PubMed.
- D. Millo, A. Bonifacio, M. R. Moncelli, V. Sergo, C. Gooijer and G. van der Zwan, Colloids Surf., B, 2010, 81, 212–216 CrossRef CAS PubMed.
- J. P. Busalmen, A. Esteve-Núñez, A. Berná and J. M. Feliu, Angew. Chem., Int. Ed., 2008, 47, 4874–4877 CrossRef CAS PubMed.
- X. B. Liu, L. Shi and J. D. Gu, Biotechnol. Adv., 2018, 36, 1815–1827 CrossRef CAS PubMed.
- B. E. Logan, R. Rossi, A. Ragab and P. E. Saikaly, Nat. Rev. Microbiol., 2019, 17, 307–319 CrossRef CAS PubMed.
- A. J. Slate, K. A. Whitehead, D. A. C. Brownson and C. E. Banks, Renewable Sustainable Energy Rev., 2019, 101, 60–81 CrossRef CAS.
- X. X. Xiao, H. Q. Xia, R. R. Wu, L. Bai, L. Yan, E. Magner, S. Cosnier, E. Lojou, Z. G. Zhu and A. H. Liu, Chem. Rev., 2019, 119, 9509–9558 CrossRef CAS PubMed.
- H. Chen, O. Simoska, K. Lim, M. Grattieri, M. Yuan, F. Dong, Y. S. Lee, K. Beaver, S. Weliwatte, E. M. Gaffney and S. D. Minteer, Chem. Rev., 2020, 120, 12903–12993 CrossRef CAS PubMed.
- E. Zhou, Y. Lekbach, T. Gu and D. Xu, Curr. Opin. Electrochem., 2021, 100830, DOI:10.1016/j.coelec.2021.100830.
- S. N. Victoria, A. Sharma and R. Manivannan, J. Indian Chem. Soc., 2021, 98, 100083 CrossRef.
- G. Pankratova, L. Hederstedt and L. Gorton, Anal. Chim. Acta, 2019, 1076, 32–47 CrossRef CAS PubMed.
- R. L. Heydorn, C. Engel, R. Krull and K. Dohnt, ChemBioEng Rev., 2020, 7, 4–17 CrossRef CAS.
- B. J. Little, D. J. Blackwood, J. Hinks, F. M. Lauro, E. Marsili, A. Okamoto, S. A. Rice, S. A. Wade and H. C. Flemming, Corros. Sci., 2020, 170, 108641 CrossRef CAS.
- Y. Ma, Y. Zhang, R. Zhang, F. Guan, B. Hou and J. Duan, Appl. Microbiol. Biotechnol., 2020, 104, 515–525 CrossRef CAS PubMed.
- F. McEachern, E. Harvey and G. Merle, Biotechnol. J., 2020, 15, 2000140 CrossRef CAS PubMed.
- Y. L. L. Chen, X. Tian and F. Zhao, Prog. Chem., 2020, 32, 1557–1563 Search PubMed.
- B. E. Logan, R. Rossi, A. A. Ragab and P. E. Saikaly, Nat. Rev. Microbiol., 2019, 17, 307–319 CrossRef CAS PubMed.
- K. P. Nevin, H. Richter, S. F. Covalla, J. P. Johnson, T. L. Woodard, A. L. Orloff, H. Jia, M. Zhang and D. R. Lovley, Environ. Microbiol., 2008, 10, 2505–2514 CrossRef CAS PubMed.
- N. S. Malvankar, M. T. Tuominen and D. R. Lovley, Energy Environ. Sci., 2012, 5, 8651–8659 RSC.
- D. R. Bond, S. M. Strycharz-Glaven, L. M. Tender and C. I. Torres, ChemSusChem, 2012, 5, 1099–1105 CrossRef CAS PubMed.
- A. Jain, G. Gazzola, A. Panzera, M. Zanoni and E. Marsili, Electrochim. Acta, 2011, 56, 10776–10785 CrossRef CAS.
- R. M. Snider, S. M. Strycharz-Glaven, S. D. Tsoi, J. S. Erickson and L. M. Tender, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15467–15472 CrossRef CAS PubMed.
- G. D. Schrott, P. S. Bonanni and J. P. Busalmen, Electrochim. Acta, 2019, 303, 176–182 CrossRef CAS.
- A. Krige, K. Ramser, M. Sjöblom, P. Christakopoulos, U. Rova and R. M. Kelly, Appl. Environ. Microbiol., 2020, 86, e01535–e01520 CrossRef CAS PubMed.
- N. Heidary, N. Kornienko, S. Kalathil, X. Fang, K. H. Ly, H. F. Greer and E. Reisner, J. Am. Chem. Soc., 2020, 142, 5194–5203 CrossRef CAS PubMed.
- Y. Yi, T. Zhao, Y. Zang, B. Xie and H. Liu, Electrochem. Commun., 2021, 124, 106966 CrossRef CAS.
- X. Liu, J. Zhan, L. Liu, F. Gan, J. Ye, K. H. Nealson, C. Rensing and S. Zhou, Environ. Sci. Technol., 2021, 55, 10142–10151 CrossRef CAS PubMed.
- G. A. Huerta-Miranda, A. I. Arroyo-Escoto, X. Burgos, K. Juárez and M. Miranda-Hernández, Bioelectrochemistry, 2019, 127, 145–153 CrossRef CAS PubMed.
- B. Zhang, H.-Y. Cheng and A. Wang, Bioelectrochemistry, 2021, 138, 107683 CrossRef CAS PubMed.
- N. M. Tefft and M. A. TerAvest, ACS Synth. Biol., 2019, 8, 1590–1600 CrossRef CAS PubMed.
- Y. Yang, Z. Wang, C. Gan, L. H. Klausen, R. Bonné, G. Kong, D. Luo, M. Meert, C. Zhu, G. Sun, J. Guo, Y. Ma, J. T. Bjerg, J. Manca, M. Xu, L. P. Nielsen and M. Dong, Nat. Commun., 2021, 12, 1709 CrossRef CAS PubMed.
- W. Wang, Y. Du, S. Yang, X. Du, M. Li, B. Lin, J. Zhou, L. Lin, Y. Song, J. Li, X. Zuo and C. Yang, Anal. Chem., 2019, 91, 12138–12141 CrossRef CAS PubMed.
- D. Naradasu, A. Guionet, T. Okinaga, T. Nishihara and A. Okamoto, ChemElectroChem, 2020, 7, 2012–2019 CrossRef CAS.
- S. Werwinski, J. A. Wharton, M. Nie and K. R. Stokes, ACS Appl. Mater. Interfaces, 2021, 13, 31393–31405 CrossRef CAS PubMed.
- A. A. Karbelkar, A. R. Rowe and M. Y. El-Naggar, Electrochim. Acta, 2019, 324, 134838 CrossRef CAS.
- I. Brand and B. Khairalla, Faraday Discuss., 2020 10.1039/D0FD00039F.
- B. Abada, S. Boumerfeg, A. Haddad and M. Etienne, J. Electrochem. Soc., 2020, 167, 135502 CrossRef CAS.
- H. P. Bennetto, J. L. Stirling, K. Tanaka and C. A. Vega, Biotechnol. Bioeng., 1983, 25, 559–568 CrossRef CAS PubMed.
- T. W. Seviour and J. Hinks, Crit. Rev. Biotechnol., 2018, 38, 634–646 CrossRef CAS PubMed.
- X. Liu, L. Shi and J.-D. Gu, Biotechnol. Adv., 2018, 36, 1815–1827 CrossRef CAS PubMed.
- D. H. Park and J. G. Zeikus, Appl. Environ. Microbiol., 2000, 66, 1292–1297 CrossRef CAS PubMed.
- S.-L. Li, Y.-J. Wang, Y.-C. Chen, S.-M. Liu and C.-P. Yu, Front. Microbiol., 2019, 10, 399 CrossRef PubMed.
- M. Grattieri, Z. Rhodes, D. P. Hickey, K. Beaver and S. D. Minteer, ACS Catal., 2019, 9, 867–873 CrossRef CAS.
- K. Rabaey, N. Boon, M. Höfte and W. Verstraete, Environ. Sci. Technol., 2005, 39, 3401–3408 CrossRef CAS PubMed.
- E. VanArsdale, J. Pitzer, G. F. Payne and W. E. Bentley, iScience, 2020, 23, 101545 CrossRef CAS PubMed.
- T. Tschirhart, E. Kim, R. McKay, H. Ueda, H.-C. Wu, A. E. Pottash, A. Zargar, A. Negrete, J. Shiloach, G. F. Payne and W. E. Bentley, Nat. Commun., 2017, 8, 14030 CrossRef CAS PubMed.
- O. Simoska, E. M. Gaffney, K. Lim, K. Beaver and S. D. Minteer, J. Electrochem. Soc., 2021, 168, 025503 CrossRef CAS.
- Y. Wu, X. Luo, B. Qin, F. Li, M. M. Häggblom and T. Liu, Environ. Sci. Technol., 2020, 54, 7217–7225 CrossRef CAS PubMed.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- R. Starwalt-Lee, M. Y. El-Naggar, D. R. Bond and J. A. Gralnick, Mol. Microbiol., 2021, 115, 1069–1079 CrossRef CAS PubMed.
- T. Tian, X. Fan, M. Feng, L. Su, W. Zhang, H. Chi and D. Fu, RSC Adv., 2019, 9, 40903–40909 RSC.
- D. M. Gurumurthy, R. N. Bharagava, A. Kumar, B. Singh, M. Ashfaq, G. D. Saratale and S. I. Mulla, Microbiol. Res., 2019, 229, 126324 CrossRef CAS PubMed.
- J. Guo, G. Yang, Z. Zhuang, Q. Mai and L. Zhuang, Sci. Total Environ., 2021, 797, 149207 CrossRef CAS PubMed.
- X. Liu, X. Jing, Y. Ye, J. Zhan, J. Ye and S. Zhou, Environ. Sci. Technol. Lett., 2020, 7, 27–34 CrossRef CAS.
- A. Sharma, P. Singh, B. K. Sarmah and S. P. Nandi, Res. Microbiol., 2020, 171, 159–164 CrossRef CAS PubMed.
- O. P. Duddy and B. L. Bassler, PLoS Pathog., 2021, 17, e1009074 CrossRef CAS PubMed.
- S. T. Rutherford and B. L. Bassler, Cold Spring Harbor Perspect. Med., 2012, 2, a012427 Search PubMed.
- M. Molnár, É. Fenyvesi, Z. Berkl, I. Németh, I. Fekete-Kertész, R. Márton, E. Vaszita, E. Varga, D. Ujj and L. Szente, Int. J. Pharm., 2021, 594, 120150 CrossRef PubMed.
- S. Mukherjee and B. Bassler, Nat. Rev. Microbiol., 2019, 17, 371–382 CrossRef CAS PubMed.
- J. Shrout, T. Tolker-Nielsen, M. Givskov and M. Parsek, MRS Bull., 2011, 36, 367–373 CrossRef CAS PubMed.
- K. Brindhadevi, F. LewisOscar, E. Mylonakis, S. Shanmugam, T. N. Verma and A. Pugazhendhi, Process Biochem., 2020, 96, 49–57 CrossRef CAS.
- A. J. Sabat, D. Pantano, V. Akkerboom, E. Bathoorn and A. W. Friedrich, Biol. Chem., 2021, 402(12), 1565–1573 CrossRef CAS PubMed.
- L. Vilaplana and M. P. Marco, Anal. Bioanal. Chem., 2020, 412, 5897–5912 CrossRef CAS PubMed.
- M. Z. El-Fouly, A. M. Sharaf, A. A. M. Shahin, H. A. El-Bialy and A. M. A. Omara, J. Radiat. Res. Appl. Sci., 2015, 8, 36–48 CrossRef CAS.
- J. Jia, S.-R. Kwon, S. Baek, V. Sundaresan, T. Cao, A. R. Cutri, K. Fu, B. Roberts, J. D. Shrout and P. W. Bohn, Anal. Chem., 2021, 93, 14481–14488 CrossRef CAS PubMed.
- J. Oziat, T. Cohu, S. Elsen, M. Gougis, G. G. Malliaras and P. Mailley, Bioelectrochemistry, 2021, 140, 107747 CrossRef CAS PubMed.
- A. Buzid, F. Shang, F. J. Reen, E. Ó. Muimhneacháin, S. L. Clarke, L. Zhou, J. H. T. Luong, F. O'Gara, G. P. McGlacken and J. D. Glennon, Sci. Rep., 2016, 6, 30001 CrossRef CAS PubMed.
- O. Simoska, M. Sans, L. S. Eberlin, J. B. Shear and K. J. Stevenson, Biosens. Bioelectron., 2019, 142, 111538 CrossRef CAS PubMed.
- F. A. a. Alatraktchi, W. E. Svendsen and S. Molin, Sensors, 2020, 20, 5218 CrossRef CAS PubMed.
- D. Koley, M. M. Ramsey, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 19996 CrossRef CAS PubMed.
- J. L. Connell, J. Kim, J. B. Shear, A. J. Bard and M. Whiteley, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 18255 CrossRef CAS PubMed.
- D. L. Bellin, H. Sakhtah, J. K. Rosenstein, P. M. Levine, J. Thimot, K. Emmett, L. E. P. Dietrich and K. L. Shepard, Nat. Commun., 2014, 5, 3256 CrossRef PubMed.
- D. L. Bellin, H. Sakhtah, Y. Zhang, A. Price-Whelan, L. E. P. Dietrich and K. L. Shepard, Nat. Commun., 2016, 7, 10535 CrossRef CAS PubMed.
- J. Elliott, O. Simoska, S. Karasik, J. B. Shear and K. J. Stevenson, Anal. Chem., 2017, 89, 6285–6289 CrossRef CAS PubMed.
- O. Simoska, J. Duay and K. J. Stevenson, ACS Sens., 2020, 5, 3547–3557 CrossRef CAS PubMed.
- H. Do, S.-R. Kwon, S. Baek, C. S. Madukoma, M. K. Smiley, L. E. Dietrich, J. D. Shrout and P. W. Bohn, Analyst, 2021, 146, 1346–1354 RSC.
- K. Fu, S.-R. Kwon, D. Han and P. W. Bohn, Acc. Chem. Res., 2020, 53, 719–728 CrossRef CAS PubMed.
- K. Sanger, K. Zór, C. Bille Jendresen, A. Heiskanen, L. Amato, A. Toftgaard Nielsen and A. Boisen, Sens. Actuators, B, 2017, 253, 999–1005 CrossRef CAS.
- S. Z. Andreasen, K. Sanger, C. B. Jendresen, A. T. Nielsen, J. Emnéus, A. Boisen and K. Zór, ACS Sens., 2019, 4, 398–405 CrossRef CAS PubMed.
- H. Do, S.-R. Kwon, K. Fu, N. Morales-Soto, J. D. Shrout and P. W. Bohn, Langmuir, 2019, 35, 7043–7049 CrossRef CAS PubMed.
- L. A. Baker, J. Am. Chem. Soc., 2018, 140, 15549–15559 CrossRef CAS PubMed.
- Y.-Y. Peng, R.-C. Qian, M. E. Hafez and Y.-T. Long, ChemElectroChem, 2017, 4, 977–985 CrossRef CAS.
- L. Sepunaru, K. Tschulik, C. Batchelor-McAuley, R. Gavish and R. G. Compton, Biomater. Sci., 2015, 3, 816–820 RSC.
- A. Frkonja-Kuczin, L. Ray, Z. Zhao, M. C. Konopka and A. Boika, Electrochim. Acta, 2018, 280, 191–196 CrossRef CAS.
- L. M. Stabryla, K. A. Johnston, J. E. Millstone and L. M. Gilbertson, Environ. Sci.: Nano, 2018, 5, 2047–2068 RSC.
- J. Y. Lee, B.-K. Kim, M. Kang and J. H. Park, Sci. Rep., 2016, 6, 30022 CrossRef CAS PubMed.
- E. Lebègue, N. L. Costa, R. O. Louro and F. Barrière, J. Electrochem. Soc., 2020, 167, 105501 CrossRef.
- A. T. Ronspees and S. N. Thorgaard, Electrochim. Acta, 2018, 278, 412–420 CrossRef CAS.
- G. Gao, D. Wang, R. Brocenschi, J. Zhi and M. V. Mirkin, Anal. Chem., 2018, 90, 12123–12130 CrossRef CAS PubMed.
- R. A. S. Couto, L. Chen, S. Kuss and R. G. Compton, Analyst, 2018, 143, 4840–4843 RSC.
- S. Pirbadian, M. S. Chavez and M. Y. El-Naggar, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 20171 CrossRef CAS PubMed.
- V. Sundaresan, A. R. Cutri, J. Metro, C. S. Madukoma, J. D. Shrout, A. J. Hoffman, K. A. Willets and P. W. Bohn, Electrochem. Sci. Adv., 2021, e2100094, DOI:10.1002/elsa.202100094.
- L. P. Zaino, D. A. Grismer, D. Han, G. M. Crouch and P. W. Bohn, Faraday Discuss., 2015, 184, 101–115 RSC.
- S. Wang, Y. Zhu, Y. Yang, J. Li and M. R. Hoffmann, Electrochim. Acta, 2020, 338, 135864 CrossRef CAS PubMed.
- A. K. Singh, J. Jaiswal and M. Dhayal, J. Electroanal. Chem., 2020, 868, 114119 CrossRef CAS.
- S. Bergner, J. Wegener and F.-M. Matysik, Anal. Methods, 2012, 4, 623–629 RSC.
- F. Zhao, F. Conzuelo, V. Hartmann, H. Li, S. Stapf, M. M. Nowaczyk, M. Rögner, N. Plumeré, W. Lubitz and W. Schuhmann, Biosens. Bioelectron., 2017, 94, 433–437 CrossRef CAS PubMed.
- J. Zhang, T. Zhu, J. Lang, W. Fu and F. Li, Curr. Opin. Electrochem., 2020, 22, 178–185 CrossRef.
- B. Huang, W. Q. Wang, M. Bates and X. W. Zhuang, Science, 2008, 319, 810–813 CrossRef CAS PubMed.
- M. G. L. Gustafsson, J. Microsc., 2000, 198, 82–87 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |




