Highly sensitive and selective surface plasmon resonance biosensor for the detection of SARS-CoV-2 spike S1 protein†
Qiong
Wu
a,
Wen
Wu
a,
Fangfang
Chen
a and
Ping
Ren
 *b
*b
aNanomedicine Translational Research Center, China-Japan Union Hospital of Jilin University, 126 Sendai Street, Changchun 130033, Jilin, China
bDepartment of Thoracic Surgery, The First Hospital of Jilin University, 71 Xinmin Street, Changchun 130021, Jilin, China. E-mail: rpemail@jlu.edu.cn
First published on 4th May 2022
Abstract
The reality that the coronavirus disease 2019 (COVID-19) is still raging around the world and making a comeback with a strong presence has highlighted the need for rapid and sensitive quantitative detection methods of viral RNA, antibody and antigen for widespread tracking and screening applications. Surface plasmon resonance (SPR) detection technology has achieved rapid development and become a standard measurement method in the fields of biosensing, biomedicine, biochemistry and biopharmaceuticals due to its advantages of high sensitivity, fast response and no need for labelling. Here, we report a sandwiched structure-based SPR biosensor for detecting a specific viral antigen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike S1 protein. The sensor combines a Ti3C2-MXene nanosheet modified sensing platform and polydopamine (PDA)–Ag nanoparticle (AgNP)/anti-SARS-CoV-2 spike S1 protein nanoconjugate signal enhancers, exhibiting a wide linear range of 0.0001 to 1000 ng mL−1 with a low detection limit of 12 fg mL−1 (S/N = 3). In the analysis of artificial saliva and human serum samples, the proposed SPR biosensor exhibits good reproducibility and high specificity, which indicates its potential for application in complex bodily fluids. The exploitation of the MXene-based SPR biochip for recognizing the SARS-CoV-2 antigen provides an accessible and rapid way for COVID-19 diagnosis, and promotes the application of 2D nanomaterial-based sensing chips in clinical diagnosis and disease screening. Significantly, the proposed method possesses general applicability that can be reprogrammed to detect any protein antigen if a corresponding specific nanobody is available.
1. Introduction
The coronavirus disease 2019 (COVID-19) is still circulating globally and the number of cases continues to increase. Research on developing new detection technologies for achieving rapid, accurate, sensitive, and low-cost screening is of great significance for responding to the threat of COVID-19.1–4 Currently, there are three main approaches for diagnosing COVID-19: nucleic acid detection, antibody detection and antigen detection.5–7 The genes of the single-stranded RNA virus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are highly similar to SARS-CoV and other coronaviruses. Reverse transcription-polymerase chain reaction (RT-PCR) for measuring SARS-CoV-2 is the gold standard for detecting COVID-19. However, RT-PCR measurement is tedious and expensive, and RNA degrades easily. Antigen detection uses the principle of specific binding between antigen and antibody, and has long been used to quickly diagnose diseases such as influenza, Legionnaires’ disease, streptococcal pharyngitis, and acquired immunodeficiency syndrome (AIDS). The rapid epidemic of the new coronavirus has prompted new antigen-based or other rapid testing methods. In areas where the risk of new coronavirus infection is high, fast and simple, antigen testing can effectively share the pressure of RT-PCR testing. There are at least four main structural proteins in the SARS-CoV-2, including spike proteins (S protein), small envelope proteins (E protein), membrane proteins (M protein), and nucleocapsid proteins (N protein), which can be used as biomarkers for viral diagnostics.8–12 Moreover, virus copies present in saliva range from 9.9 × 102 to 1.2 × 1011 copies per mL throughout the duration of the infection.13 Because of these low numbers, developing sensitive detection methods and kits of SARS CoV-2 protein in bodily fluids will break through the key technical bottleneck of clinical sample inactivation and virus degradation that lead to decreased detection sensitivity.Surface plasmon resonance (SPR) technology possesses the advantages of fast response, not needing labeling of the analyte, real-time qualitative and quantitative analysis, and providing reactant molecular bonding information, thus exhibiting great potential in clinical applications.14–18 However, the inability of traditional SPR sensing technology to detect trace targets in complex matrices limits its practical application. The growing use of nanomaterials in fabricating SPR sensor chips and response signal enhancers is promising for SPR sensing.19–23 Among them, two-dimensional (2D) materials have become a hot spot in the biosensing field due to their unique dimensional characteristics.24–28 2D nanomaterials provide a large specific surface area for loading foreign molecules, and the interaction between 2D nanomaterials and the foreign molecules causes changes in electrical response. These 2D nanomaterial-based sensing platforms exhibit fast response speeds, high sensitivity, and good selectivity.
In 2011, Gogotsi and his colleagues proposed a new family of 2D nanomaterials called MXenes,29 attracting widespread interest among researchers. MXenes specifically refer to a kind of material system with the structural formula of Mn+1XnTx (n = 1–3), and can be produced by the selective etching of A layers from Mn+1AXn phases, where M is an early transition metal, A represents an A-group element, X is either carbon and/or nitrogen and T refers to the group/modifier on the surface of the 2D material.30–32 During the etching process, A atoms are replaced by OH or F atoms, and the massive MAX phase becomes a loose structure similar to an accordion. The interaction between Mn+1Xn layers becomes weaker and easier to separate. Afterwards, ultrasonic treatment of the MAX phase treated with the etchant can easily achieve the stratification of MXene.33–35 Unlike the challenge of modifying graphene, MXene materials allow for flexible chemical modification and show good compatibility with water, organic solvents and organisms. The unique properties of excellent electrical conductivity, hydrophilicity, large specific area, easy modification and high stability make MXene materials competitive among 2D materials in the field of biosensing.36–38 Very little research has investigated the potential of MXene-based SPR sensing chip in SARS-CoV-2 antigen detection.
One of the SARS-CoV-2 structural proteins, the S protein coated at the outer shell of the virus, can be cleaved by the host cell furin-like proteases into two separate S1 and S2 subunits. The S1 subunit contains the RBD sequence and is surface exposed while S2 subunit is membrane bound (Fig. 1), and they are the major targets for SARS-CoV-2 therapy and detection.39–41,52 Antigen detection is a good choice for testing viral loading in early stages of the COVID-19 disease. This work explored 2D MXene-based SPR sensing strategy for detecting SARS-CoV-2 spike S1 protein abbreviated as S1 protein. In this study, we developed a Ti3C2-MXene-based biosensing chip functionalized with anti-S1 protein (CAP) as a sensing platform. After S1 protein was captured, PDA–AgNPs nanohybrids conjugated with anti-S1 protein (DET) as signal amplifiers were flowed over the sensing chip interface to further enhance detection sensitivity by forming a sandwiched structure. Our SPR biosensor targeting S1 protein exhibited an ultralow limit of detection (LOD) of 12 fg mL−1. More importantly, the selective detection of S1 protein in artificial saliva and human serum confirmed its potential for clinical applications.
2. Materials and methods
2.1 Materials and reagents
Mouse anti-SARS-COV-2 spike S1 protein (Catalog No. RM17568) abbreviated as Ab1, mouse anti-SARS-COV-2 spike S1 protein (Catalog No. RM17569) abbreviated as Ab2, and recombinant SARS-COV-2 spike S1 protein with its tag (Catalog No. RP01262) abbreviated as S1 protein were purchased from ABclonal. Bovine serum albumin (BSA) was purchased from Ding Guo Biotechnology Company. Ti3AlC2 MAX phase was purchased from Carbon-Ukraine Ltd. Dopamine (DA) and 3-mercaptopropinic acid (MPA) were purchased from Jk Chemical. Hydrogen tetrachloroauratehydrate (HAuCl4·3H2O) was purchased from Acros. Artificial saliva was purchased from Shenzhen Zhongwei Instrument Equipment Co., LTD. Human serum samples were provided by China–Japan Union Hospital of Jilin University. Sodium phosphate buffered saline (PBS, 0.01 mol L−1, pH 7.4) was prepared before use. All water used in this assay was prepared through a Millipore purification device. Antibody and antigen solutions were subpackaged and stored at −20 °C, and other biological reagents were kept at 4 °C.2.2 Synthesis of PDA–AgNPs nanohybrids
PDA nanospheres were synthesized according to the previous report with slight modifications.42 Briefly, 1.5 mL of ammonia solution (25–28%) was added to a mixture of ethanol (30 mL) and DI water (60 mL) under vigorous stirring at room temperature. Thirty minutes later, 6 mL of dopamine hydrochloride solution (0.05 g mL−1) was added to the above mixture and stirred vigorously for 40 h. The dark brown supernatant was centrifuged with ethanol and DI water in sequence at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The obtained PDA nanospheres were then dispersed in 25 mL of DI water.
000 rpm for 10 min. The obtained PDA nanospheres were then dispersed in 25 mL of DI water.
To prepare PDA–AgNPs nanohybrids, 60 mL of AgNO3 (25 mmol L−1) solution was mixed with 12 mL of PDA nanosphere solution and stirred continuously for 30 min to promote Ag+ adsorption on the surface of PDA nanospheres. Then, 20 μL of NaBH4 solution (0.15 mol L−1) was added to the mixture and stirred for 1 h. The resulting product was washed with DI water by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The obtained PDA–AgNPs nanohybrids were redispersed in 15 mL of PBS and stored at 4 °C for further use.
000 rpm for 10 min. The obtained PDA–AgNPs nanohybrids were redispersed in 15 mL of PBS and stored at 4 °C for further use.
2.3 Preparation of PDA–AgNPs–Ab2 nanoconjugates
PDA–AgNPs–Ab2 nanoconjugates were prepared by adding 150 μg of Ab2 into 1.5 mL of PDA–AgNPs nanohybrid solution prepared above under gentle stirring and kept at 4 °C for 12 h. Antibodies can be attached firmly to the surface of PDA nanospheres by Schiff-base reaction between amino residues of Ab2 and quinone groups formed in PDA. The redundant binding sites of PDA–AgNPs nanohybrids were blocked by 10 mg mL−1 BSA in PBS to eliminate nonspecific binding. Free BSA was removed by centrifugation, and the precipitate was redispersed in 1.5 mL of PBS containing 1% BSA. The obtained conjugates were stored at 4 °C prior to use.2.4 Fabrication of the sensing platform
Ti3C2-MXene nanosheets were prepared by etching bulk Ti3AlC2 as we reported previously.43 The bare gold film was ultrasonically cleaned in ethanol and dried with N2 flow. The cleaned gold film was coated on a glass slide and then fixed on the bottom of a reactor to be used as a sensing chip. Ti3C2-MXene (500 μL, 0.8 mg mL−1) solution was injected into the reactor and incubated for 3 h at room temperature to complete the assembly on the bare gold film. Next, free Ti3C2-MXene nanosheets in the reactor were gently washed with DI water, and PBS was injected to balance the baseline. Afterwards, Ab1 solution (500 μL of 100 μg mL−1) was injected into the reactor and immobilized on Ti3C2-MXene nanosheets. After 3 h, free Ab1 in the reactor was removed by PBS, and then 10 mg mL−1 BSA solution was injected and incubated for 20 min to block the non-specific binding sites on the sensing platform. Finally, PBS was injected to wash off free BSA in the reactor, and the prepared sensing platform was ready to capture S1 protein in samples.2.5 SPR detection
In this work, S1 protein was used as the detection model to develop an SPR-based antigen detection method for COVID-19 diagnosis. Sample solutions were prepared by spiking PBS with S1 protein standard solutions at various concentration levels. SPR measurements were performed on an angle modulation SPR biosensor based on the conventional Kretschmann configuration, and the in situ SPR responses that originated from refractive index changes of the sensing interface were collected by a TR2005 spectrometer (RES-TEC Resonant Sensor Technology, Germany). For S1 protein detection, samples with different concentrations were flowed over the sensing platform and incubated for 30 min, respectively. The SPR chip surface was washed with PBS to remove S1 protein that was not captured, and then 8 mg mL−1 PDA–AgNPs–Ab2 nanoconjugates were injected into the reactor, forming the sandwiched immunocomplexes to enhance response signals. After incubation for 30 min, unreacted PDA–AgNPs–Ab2 nanoconjugates were removed from the reactor through PBS washing. The obtained SPR responses were recorded as shifts of resonant angle before injecting S1 protein samples and after PBS rinsing of PDA–AgNPs–Ab2 nanoconjugates in the sensing system. A schematic of the sensing procedure is illustrated in Fig. 1. The immunoassay was executed at normal pressure and room temperature, and all SPR measurements in this work were repeated three times to guarantee repeatability.3. Results and discussion
3.1 Characterization
Ti3C2-MXene nanosheets were synthesized from parent MAX-phase ceramics with a series of procedures of delamination, disintegration, and probe sonication breakage. X-ray powder diffraction (XRD) was employed to identify the crystalline structures of the prepared Ti3C2-MXene nanosheets, and the diffraction peaks located at ∼6.72°, 13.7°, 38.8° agree well with the previous report44 (Fig. 2a). X-ray photoelectron spectroscopy (XPS) measurement was used to evaluate the chemical status of the Ti3C2-MXene nanosheets. The survey spectrum demonstrates that the prepared Ti3C2-MXene is mainly composed of elements Ti, C, O and F (Fig. 2b), and O and F originated from the –OH and F terminal groups formed on the surface of theTi3C2-MXene nanosheets during etching.29 The prepared Ti3C2-MXene nanosheets were thin, highly transparent, and wrinkled from the scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images (Fig. 2c and d).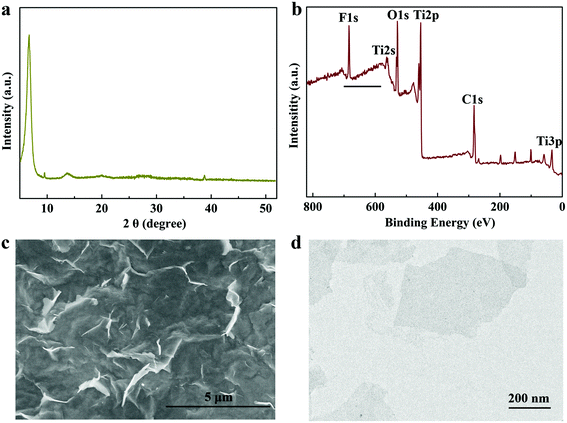 | ||
| Fig. 2 Structural characterization of Ti3C2-MXene nanosheets. XRD spectrum, XPS spectrum, SEM image and TEM image (a–d). | ||
The components of the PDA–AgNPs nanohybrids were measured through XPS analysis. Compared with the survey spectrum of PDA, the existence of an Ag peak demonstrated the successful introduction of Ag content, and the Ag 3d spectrum resolved into two spin–orbit components located at the binding energies of 374.25 eV and 368.15 eV is consistent with the Ag 3d3/2 and 3d5/2 peaks of metallic silver, respectively45 (Fig. 3a and b). TEM was used to characterize the morphology of the prepared PDA nanospheres and PDA–AgNPs nanohybrids. As shown in Fig. 3c, PDA nanospheres exhibited a uniform and regular spherical morphology, and the average diameter was approximately 70 nm. By in situ reduction of AgNO3 solution, PDA nanospheres were covered with dense AgNPs with an average diameter of 7 nm, and no isolated AgNPs were observed in the TEM image (Fig. 3d). Through Schiff-base reaction between the quinone groups in PDA and amino group in the antibody, Ab2 can be conjugated with PDA–AgNP nanohybrids to prepare PDA–AgNPs–Ab2 signal enhancers without using any further surface activation or treatment and maintain high reaction activity. The UV-Vis absorption spectrum of PDA–Ag nanohybrids showed the characteristic adsorption peak of AgNPs at 400 nm. After conjugating with Ab2, the appearance of a typical protein absorbance peak at 285 nm indicates the successful binding of Ab2, and peak position of AgNPs shifted from 400 nm to 410 nm due to a part of Ab2 adsorbing on the surfaces of AgNPs via the interaction of AgNPs and amino groups of Ab2 (Fig. S1†).
 | ||
| Fig. 3 XPS survey spectra of PDA and PDA–AgNPs nanohybrids (a), the high-resolution XPS spectrum of Ag 3d (b), and TEM image of PDA (c) and PDA–AgNPs nanohybrids (d). | ||
3.2 Fabrication of sensing platform
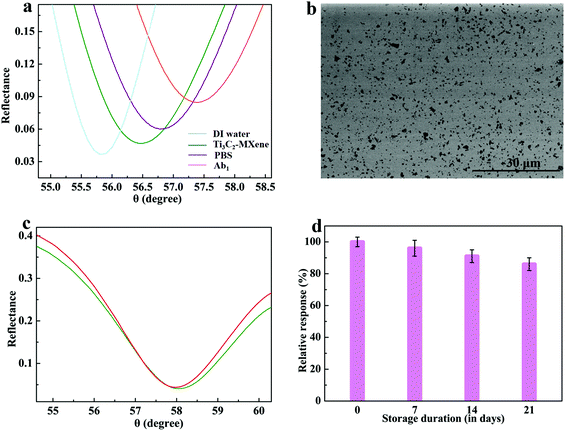
Ti3C2-MXene nanosheets were assembled on a flat gold film via the strong metal–carbon coupling between MXene and the gold film for immobilizing antibody,46 and the whole process of preparing the sensing platform was monitored by the change of the SPR spectrum. The assembly of Ti3C2-MXene nanosheets could be completed within 12 h, leading to a resonant angle shift of 0.64° (Fig. 4a). The SEM image of Ti3C2-MXene modified sensing chip showed a thin layer of Ti3C2-MXene nanosheets well distributed on the surface of gold film with satisfactory surface coverage (Fig. 4b). The immobilization of Ab1 on the sensing chip could be completed within 60 min, and the resulting resonant angle shift was 0.6°. The stability test was performed by washing the prepared sensing chip with PBS, and the obtained blueshift of the resonant angle with 10 washing cycles was less than 8% of the initial resonant angle shift (Fig. 4c), which ensures the stability of the proposed Ti3C2-MXene-based sensing platform. Moreover, we evaluated the shelf life of this sensing chip which was kept at 4 °C in a dark area by measuring its detection activity at regular intervals. The prepared Ti3C2-MXene-based sensing chip still showed 86% of the initial activity after storing for 21 days, further confirming its good stability (Fig. 4d).3.3 S1 protein detection
For the proposed strategy, the SPR response signals were effectively amplified by introducing PDA–AgNPs–Ab2 nanoconjugates into the sensing system after S1 protein was captured on the sensing platform. Hence, the optimal dosage of PDA–AgNPs–Ab2 nanoconjugates in this assay was investigated. PDA–AgNPs–Ab2 nanoconjugates were prepared with different concentrations and injected into the reactor that had incubated 10−6 g mL−1 S1 protein. The resonant angle exhibited a continuous redshift with the increase of the PDA–AgNPs–Ab2 nanoconjugate concentration up to 8 mg mL−1, and then the curve was level, which indicates that PDA–AgNPs–Ab2 nanoconjugates captured by the sensing platform had reached saturation (Fig. 5a). Hence, 8 mg mL−1 was selected to be the optimal concentration of PDA–AgNPs–Ab2 solution in this assay. Furthermore, blank human serum samples were determined by the present method to estimate the nonspecific adsorption of the sensing platform towards PDA–AgNPs–Ab2 nanoconjugates. The obtained resonant angle exhibited no obvious shift, confirming negligible nonspecific interaction between the prepared sensing chip and the signal amplifiers.3.4 Sensitivity of the immunoassay
Under the optimal experimental conditions, the obtained SPR response signals exhibited a linear increase with the logarithm of S1 protein concentration ranging from 10−13 to 10−6 g mL−1 (Fig. 5b), and the linear relationship of the relative resonant angle change (Δθ) versus concentration of S1 protein (C) was described as Δθ = 0.6098 + 0.0459![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9983). The obtained LOD was as low as 12 fg mL−1, which was defined as a signal-to-noise ratio of 3 s (where s is the standard deviation of the blank solution, n = 10). Fig. 5c displays the SPR spectra of detecting S1 protein with a concentration of 10−13 to 10−6 g mL−1 by the proposed biosensor. The high sensitivity achieved by the signal amplification strategy should be attributed to the following three aspects: (1) owing to hydrophilicity, a large specific surface area, high stability and good biocompatibility, the Ti3C2-MXene-modified sensing chip shows higher capacity of immobilizing antibodies and maintaining their biological activity compared with the conventional organic molecular layer, which effectively improves the detection sensitivity; (2) the formation of Ab1/S1 protein/Ab2–AgNPs–PDA sandwiched structure greatly increases the mass captured by the SPR sensing chip, amplifying the response signal by increasing the real part of the refractive index of the chip surface; (3) the intense electromagnetic coupling between the dense AgNPs located on PDA nanospheres and the underlying Au film leads to the increase of the imaginary part of the refractive index, enhancing the detection sensitivity. Our biosensor shows advantages of a better detection limit and detection range compared with those reported in the literatures (Table 1). Reproducibility of the proposed SPR biosensor was evaluated with the relative standard deviation (RSD) which was achieved by conducting independent testing of each sample in triplicate with different sensors. The obtained RSDs for detecting S1 protein were less than 5% (Fig. 5b), suggesting that the preparation procedure is highly reproducible.
C (R2 = 0.9983). The obtained LOD was as low as 12 fg mL−1, which was defined as a signal-to-noise ratio of 3 s (where s is the standard deviation of the blank solution, n = 10). Fig. 5c displays the SPR spectra of detecting S1 protein with a concentration of 10−13 to 10−6 g mL−1 by the proposed biosensor. The high sensitivity achieved by the signal amplification strategy should be attributed to the following three aspects: (1) owing to hydrophilicity, a large specific surface area, high stability and good biocompatibility, the Ti3C2-MXene-modified sensing chip shows higher capacity of immobilizing antibodies and maintaining their biological activity compared with the conventional organic molecular layer, which effectively improves the detection sensitivity; (2) the formation of Ab1/S1 protein/Ab2–AgNPs–PDA sandwiched structure greatly increases the mass captured by the SPR sensing chip, amplifying the response signal by increasing the real part of the refractive index of the chip surface; (3) the intense electromagnetic coupling between the dense AgNPs located on PDA nanospheres and the underlying Au film leads to the increase of the imaginary part of the refractive index, enhancing the detection sensitivity. Our biosensor shows advantages of a better detection limit and detection range compared with those reported in the literatures (Table 1). Reproducibility of the proposed SPR biosensor was evaluated with the relative standard deviation (RSD) which was achieved by conducting independent testing of each sample in triplicate with different sensors. The obtained RSDs for detecting S1 protein were less than 5% (Fig. 5b), suggesting that the preparation procedure is highly reproducible.
| Method | Analyte | Sample | LOD | Detection range | Ref. |
|---|---|---|---|---|---|
| Magnetic particle spectroscopy | Spike protein | PBS | 1.56 nM | 3.13–400 nM | 47 |
| Photoelectrochemical | S protein | Pharynx swabs | 72 ng mL−1 | 0.5–8 μg mL−1 | 48 |
| Colorimetric | S protein | Influenza A virus | 4.98 ng mL−1 | 4.98–139 ng mL−1 | 49 |
| ELISA | S-RBD | Blood | 20.6 fg mL−1 | 0.34–1065 pg mL−1 | 50 |
| Electrochemical | Spike protein | Saliva | 1 pg mL−1 | 1–1000 pg mL−1 | 51 |
| LSPR | S1 subunit | Saliva | 0.26 nM | 1–100 nM | 52 |
| Electrochemical | S-RBD | Saliva | 1.68 ng mL−1 | 1–100 ng mL−1 | 53 |
| Colorimetric | S1 subunit | PBS | 11 ng mL−1 | 10–100 ng mL−1 | 54 |
| Chemiluminescence | S-RBD | Pseudovirus | 0.1 ng mL−1 | 0.2–100 ng mL−1 | 55 |
| SPR | S-RBD | Serum | 37 nM | 25–1000 nM | 56 |
| SPR | S1 subunit | Serum | 12 fg mL−1 | 0.0001–1000 ng mL−1 | This work |
We further performed control experiments of a traditional mercaptopropionic acid (MPA)-based sensing platform, Ti3C2-MXene-based sensing platform, Ti3C2-MXene-based sensing platform combined with Ab2 signal amplifier, Ti3C2-MXene-based sensing platform combined with PDA–Ab2 signal amplifier, and Ti3C2-MXene-based sensing platform combined with PDA–AgNPs–Ab2 signal amplifiers for determining S1 protein analyte with a concentration of 10−6 g mL−1 to reveal their roles in enhancing the detection sensitivity with the present sensing strategy (Fig. 5d). The Ti3C2-MXene-based sensing platform was demonstrated to be more sensitive than a traditional MPA-modified sensing chip because of the increased number of available binding sites for loading antibodies. The classic sandwich method was used to further improve the detection sensitivity, and we evaluated Ab2, PDA–Ab2 and PDA–AgNPs–Ab2 as response signal amplifiers, respectively. PDA nanospheres offer a large surface area for loading Ab2 and enlarge the mass arriving on the interface of the sensing chip, resulting in more effective signal amplification compared with Ab2 signal amplifiers. AgNPs have been reported to be significant in enhancing SPR detection sensitivity and widely applied in SPR biosensing by virtue of the electromagnetic coupling between AgNPs and the gold chip to amplify SPR response signals. Electromagnetic coupling effect and considerable increase of mass captured on the sensing chip derived from the formation of dense AgNPs on PDA nanospheres results in the obvious signal amplification. The present dual signal amplification strategy achieves a ∼8.5-fold increase in response signal over that of an MPA-based sensing strategy for detecting 10−6 g mL−1 S1 protein. The prepared sensing platform can be reused by impulse injection of massive PBS with a large amount of ions into the reactor to dissociate antigen from antibodies immobilized on the sensing chip. The reusability of the fabricated biosensor was investigated by measuring the SPR responses to 10−8 g mL−1 S1 repeatedly, and 17% of its initial activity lost after six reuse cycles (Fig. S2†).
3.5 S1 protein detection in artificial saliva and serum samples
Evaluating the detection selectivity is critical due to the actual requirement of real sample analysis. The practicability of the S1 protein biosensor was tested in a complex matrix including artificial saliva and human serum to mimic the biological conditions. The utilized artificial saliva and human serum samples were diluted 10 times by PBS prior to the addition of S1 protein for reducing non-specific interference of other components. The resulting recoveries of S1 protein in artificial saliva ranged from 91% to 114% with RSDs of 3.8% to 6.2% (Table 2), indicating that the proposed biosensor could potentially be used to determine S1 protein content for clinical diagnosis and screening of COVID-19. In addition, the spiked serum samples also exhibited a satisfactory recovery of 92% to 120% with RSDs of 3.5% to 6.8% (Table S1†), further demonstrating good performance of the present sensing strategy in analysis of real biological samples.| Content of S1 protein (ng mL−1) | Spiked (ng mL−1) | Detected (ng mL−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| None | 100 | 98 | 98 | 5.3 |
| None | 10 | 9.1 | 91 | 6.0 |
| None | 1 | 0.96 | 96 | 4.5 |
| None | 0.1 | 0.105 | 105 | 5.7 |
| None | 0.01 | 0.0093 | 93 | 3.8 |
| None | 0.001 | 0.00114 | 114 | 6.2 |
4. Conclusion
In summary, we present an SPR-based antigen detection method capable of detecting SARS-CoV-2 via spike S1 protein recognition. Our biosensor provides a sensitive, and selective means for determining S1 protein using a sandwiched amplification strategy based on a Ti3C2-MXene modified sensing platform and PDA–AgNPs–Ab2 response signal amplifiers. The proposed SPR biosensor provides a wide detection range with a low detection limit of 12 fg mL−1. Moreover, this method exhibited satisfactory precision in evaluating S1 protein levels in artificial saliva, indicating its potential application of early diagnosis and screening of COVID-19. We believe that our work provides a rapid and real-time response, and sensitive and selective sensing strategy which could be easily applied in diagnosing other viral variants and diseases just by changing the antibodies on the sensing platform and signal enhancers. In addition, this work might offer rewarding profits and act as a reference for extending applications of MXene-based biochips in clinical diagnosis.Author contributions
Conceptualization, Q. W., P. R.; methodology, Q. W.; validation, Q. W., W. W.; formal analysis, Q. W., P. R.; investigation, Q. W.; data curation, Q. W., W. W.; writing – original draft preparation, Q. W.; writing – review and editing, Q. W., F. C. and P. R.; visualization, P. R.; supervision, P. R.; project administration, P. R.; funding acquisition, Q. W., F. C. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We greatly acknowledge the Natural Science Foundation of Jilin Province (YDZJ202101ZYTS024), the National Natural Science Foundation of China (31771093), and the Program for JLU Science and Technology Innovative Research Team (JLUSTIRT, 2019TD-36). Jim Hesson copyedited the manuscript (https://www.academicenglishsolutions.com/editing-service).References
- S. Suleman, S. K. Shukla, N. Malhotra, S. D. Bukkitgar, N. P. Shetti, R. Pilloton, J. Narang, Y. N. Tan and T. M. Aminabhavi, Point of care detection of COVID-19: Advancement in biosensing and diagnostic methods, Chem. Eng. J., 2021, 414, 128759 CrossRef CAS PubMed.
- H. E. Kim, A. Schuck, S. H. Lee, Y. Lee, M. Kang and Y. S. Kim, Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests, Biosens. Bioelectron., 2021, 182, 113168 CrossRef CAS PubMed.
- E. Valera, A. Jankelow, J. Lim, V. Kindratenko, A. Ganguli, K. White, J. Kumar and R. Bashir, COVID-19 Point-of-care diagnostics: present and future, ACS Nano, 2021, 15, 7899–7906 CrossRef CAS PubMed.
- A. Gowri, N. A. Kumar and B. S. S. Anand, Recent advances in nanomaterials based biosensors for point of care (POC) diagnosis of Covid-19-A minireview, TrAC, Trends Anal. Chem., 2021, 137, 116205 CrossRef CAS PubMed.
- G. Balkourani, A. Brouzgou, M. Archonti, N. Papandrianos, S. Song and P. Tsiakaras, Emerging materials for the electrochemical detection of COVID-19, J. Electroanal. Chem., 2021, 893, 115289 CrossRef CAS PubMed.
- X. Ding, K. Yin, Z. Li, M. M. Sfeir and C. Liu, Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay, Biosens. Bioelectron., 2021, 184, 113218 CrossRef CAS PubMed.
- Q. Bayin, L. Huang, C. Ren, Y. Fu, X. Ma and J. Guo, Anti-SARS-CoV-2 IgG and IgM detection with a GMR based LFIA system, Talanta, 2021, 227, 122207 CrossRef CAS PubMed.
- T. Ji, Z. Liu, G. Wang, X. Guo, S. A. Khan, C. Lai, H. Chen, S. Huang, S. Xia, B. Chen, H. Jia, Y. Chen and Q. Zhou, Detection of COVID-19: A review of the current literature and future perspectives, Biosens. Bioelectron., 2020, 166, 112455 CrossRef CAS PubMed.
- J. Sitjar, J. D. Liao, H. Lee, H. P. Tsai, J. R. Wang and P. Y. Liu, Challenges of SERS technology as a non-nucleic acid or -antigen detection method for SARS-CoV-2 virus and its variants, Biosens. Bioelectron., 2021, 181, 113153 CrossRef CAS PubMed.
- C. Yi, Y. Yi and J. Li, mRNA Vaccines: Possible tools to combat SARS-CoV-2, Virol. Sin., 2020, 35, 259–262 CrossRef CAS PubMed.
- A. Wu, Y. Peng, B. Huang, X. Ding, X. Wang, P. Niu, J. Meng, Z. Zhu, Z. Zhang, J. Wang, J. Sheng, L. Quan, Z. Xia, W. Tan, G. Cheng and T. Jiang, Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in china, Cell Host Microbe, 2020, 27, 325–328 CrossRef CAS PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, 2020, 180, 281–292 CrossRef PubMed.
- K. K. W. To, O. T. Y. Tsang, C. C. Y. Yip, K. H. Chan, T. C. Wu, J. M. C. Chan, W. S. Leung, T. S. H. Chik, C. Y. C. Choi, D. H. Kandamby, D. C. Lung, A. R. Tam, R. W. S. Poon, A. Y. F. Fung, I. F. N. Hung, V. C. C. Cheng, J. F. W. Chan and K. Y. Yuen, Consistent detection of 2019 novel coronavirus in saliva, Clin. Infect. Dis., 2020, 71, 841–843 CrossRef CAS PubMed.
- Y. F. Chang, Y. T. Chou, C. Y. Cheng, J. F. Hsu, L. C. Su and J. A. Ho, Amplification-free detection of cytomegalovirus miRNA using a modification-free surface plasmon resonance biosensor, Anal. Chem., 2021, 93, 8002–8009 CrossRef CAS PubMed.
- M. Garrido-Jareno, L. Puchades-Carrasco, L. Orti-Perez, J. M. Sahuquillo-Arce, M. d. C. Meyer-Garcia, J. Mollar-Maseres, C. Lloret-Sos, A. Gil-Brusola, J. L. López-Hontangas, J. M. Beltrán-Garrido, J. Pemán-García and A. Pineda-Lucena, A surface plasmon resonance based approach for measuring response to pneumococcal vaccine, Sci. Rep., 2021, 11, 6502 CrossRef CAS PubMed.
- E. Mauriz, P. Dey and L. M. Lechuga, Advances in nanoplasmonic biosensors for clinical applications, Analyst, 2019, 144, 7105 RSC.
- M. Drozd, S. Karon and E. Malinowska, Recent advancements in receptor layer engineering for applications in SPR-based immunodiagnostics, Sensors, 2021, 21, 3781 CrossRef CAS PubMed.
- A. Jebelli, F. Oroojalian, F. Fathi, A. Mokhtarzadeh and M. d. Guardia, Recent advances in surface plasmon resonance biosensors for microRNAs detection, Biosens. Bioelectron., 2020, 169, 112599 CrossRef CAS PubMed.
- J. Hu, J. Zhao, H. Zhu, Q. Chen, X. Hu, K. Koh and H. Chen, AuNPs network structures as a plasmonic matrix for ultrasensitive immunoassay based on surface plasmon resonance spectroscopy, Sens. Actuators, B, 2021, 340, 129948 CrossRef CAS.
- T. B. A. Akib, S. F. Mou, M. M. Rahman, M. M. Rana, M. R. Islam, I. M. Mehedi, M. A. P. Mahmud and A. Z. Kouzani, Design and numerical analysis of a graphene-coated SPR biosensor for rapid detection of the novel coronavirus, Sensors, 2021, 21, 3491 CrossRef PubMed.
- W. Wu, X. Yu, J. Wu, T. Wu, Y. Fan, W. Chen, M. Zhao, H. Wu, X. Li and S. Ding, Surface plasmon resonance imaging-based biosensor for multiplex and ultrasensitive detection of NSCLC-associated exosomal miRNAs using DNA programmed heterostructure of Au-on-Ag, Biosens. Bioelectron., 2021, 175, 112835 CrossRef CAS PubMed.
- D. E. P. Souto, J. Volpe, C. d. C. Goncalves, C. H. I. Ramos and L. T. Kubota, A brief review on the strategy of developing SPR-based biosensors for application to the diagnosis of neglected tropical diseases, Talanta, 2019, 205, 120122 CrossRef CAS PubMed.
- Vikas, M. K. Yadav, P. Kumar and R. K. Verma, Detection of adulteration in pure honey utilizing Ag-graphene oxide coated fiber optic SPR probes, Food Chem., 2020, 332, 127346 CrossRef CAS PubMed.
- B. Han, S. Jin, Q. Chu, Y. Jin, X. Xue, S. Guo, Y. Park, L. Chen and Y. M. Jung, New insight into SPR modulating by two-dimensional correlation spectroscopy: the case for an Ag/ITO system, Nanoscale, 2020, 12, 24357–24361 RSC.
- D. T. Nurrohman, Y. H. Wang and N. F. Chiu, Exploring graphene and MoS2 chips based surface plasmon resonance biosensors for diagnostic applications, Front. Chem., 2020, 8, 728 CrossRef CAS PubMed.
- T. Xue, W. Liang, Y. Li, Y. Sun, Y. Xiang, Y. Zhang, Z. Dai, Y. Duo, L. Wu, K. Qi, B. N. Shivananju, L. Zhang, X. Cui, H. Zhang and Q. Bao, Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor, Nat. Commun., 2019, 10, 28 CrossRef CAS PubMed.
- Y. Ke, X. Wen, D. Zhao, R. Che, Q. Xiong and Y. Long, Controllable fabrication of two-dimensional patterned VO2 nanoparticle, nanodome, and nanonet arrays with tunable temperature-dependent localized surface plasmon resonance, ACS Nano, 2017, 11, 7542–7551 CrossRef CAS PubMed.
- P. Zhao, Y. Chen, Y. Chen, S. Hu, H. Chen, W. Xiao, G. Liu, Y. Tang, J. Shi, Z. He, Y. Luo and Z. Chen, A MoS2 nanoflower and gold nanoparticle modified surface plasmon resonance biosensor for a sensitivity-improved immunoassay, J. Mater. Chem. C, 2020, 8, 6861–6868 RSC.
- M. Naguib, M. Kurtoglu, V. Presser, J. Lu, J. Niu, M. Heon, L. Hultman, Y. Gogotsi and M. W. Barsoum, Two-dimensional nanocrystals produced by exfoliation of Ti3AlC2, Adv. Mater., 2011, 23, 4248–4253 CrossRef CAS PubMed.
- P. Ma, D. Fang, Y. Liu, Y. Shang, Y. Shi and H. Yang, MXene-based materials for electrochemical sodium-ion storage, Adv. Sci., 2021, 8, 2003185 CrossRef CAS PubMed.
- X. Xu, Y. Zhang, H. Sun, J. Zhou, F. Yang, H. Li, H. Chen, Y. Chen, Z. Liu, Z. Qiu, D. Wang, L. Ma, J. Wang, Q. Zeng and Z. Peng, Progress and perspective: MXene and MXene-based nanomaterials for high-performance energy storage devices, Adv. Electron. Mater., 2021, 7, 2000967 CrossRef CAS.
- A. Ahmed, M. M. Hossain, B. Adak and S. Mukhopadhyay, Recent advances in 2D MXene integrated smart-textile interfaces for multifunctional applications, Chem. Mater., 2020, 32, 10296–10320 CrossRef CAS.
- J. Xu, T. Peng, X. Qin, Q. Zhang, T. Liu, W. Dai, B. Chen, H. Yu and S. Shi, Recent advances in 2D MXenes: preparation, intercalation and applications in flexible devices, J. Mater. Chem. A, 2021, 9, 14147 RSC.
- E. Saatçi and S. Natarajan, State-of-the-art colloidal particles and unique interfaces-based SARS-CoV-2 detection methods and COVID-19 diagnosis, Curr. Opin. Colloid Interface Sci., 2021, 55, 101469 CrossRef PubMed.
- R. Antiochia, Paper-based biosensors: Frontiers in point-of-care detection of COVID-19 disease, Biosensors, 2021, 11, 110 CrossRef CAS PubMed.
- T. Xia, G. Liu, J. Wang, S. Hou and S. Hou, MXene-based enzymatic sensor for highly sensitive and selective detection of cholesterol, Biosens. Bioelectron., 2021, 183, 113243 CrossRef CAS PubMed.
- Y. Xu, X. Wang, C. Ding and X. Luo, Ratiometric antifouling electrochemical biosensors based on multifunctional peptides and MXene loaded with Au nanoparticles and methylene blue, ACS Appl. Mater. Interfaces, 2021, 13, 20388–20396 CrossRef CAS PubMed.
- M. Li, L. Wang, R. Liu, J. Li, Q. Zhang, G. Shi, Y. Li, C. Hou and H. Wang, A highly integrated sensing paper for wearable electrochemical sweat analysis, Biosens. Bioelectron., 2021, 174, 112828 CrossRef CAS PubMed.
- X. Gao, K. Liang, S. Mei, S. Peng, E. G. Vong and J. Zhan, An efficient system to generate truncated human angiotensin converting enzyme 2 (hACE2) capable of binding RBD and spike protein of SARS-CoV2, Protein Expression Purif., 2021, 184, 105889 CrossRef CAS PubMed.
- M. Targosz-Korecka, A. Kubisiak, D. Kloska, A. Kopacz, A. G. Przeczek and M. Szymonski, Endothelial glycocalyx shields the interaction of SARS-CoV-2 spike protein with ACE2 receptors, Sci. Rep., 2021, 11, 12157 CrossRef CAS PubMed.
- J. C. Harbour, Z. L. Lyski, J. B. Schell, A. Thomas, W. B. Messer, M. K. Slifka and J. C. Nolz, Cellular and humoral immune responses in mice immunized with vaccinia virus expressing the SARS-CoV-2 spike protein, J. Immunol., 2021, 206, 2596–2604 CrossRef CAS PubMed.
- J. Xia, Y. Liu, M. Ran, W. Lu, L. Bi, Q. Wang, D. Lu and X. Cao, The simultaneous detection of the squamous cell carcinoma antigen and cancer antigen 125 in the cervical cancer serum using nano-Ag polydopamine nanospheres in an SERS-based lateral flow immunoassay, RSC Adv., 2020, 10, 29156 RSC.
- Q. Wu, N. Li, Y. Wang, Y. Liu, Y. Xu, S. Wei, J. Wu, G. Jia, X. Fang, F. Chen and X. Cui, A 2D transition metal carbide MXene-based SPR biosensor for ultrasensitive carcinoembryonic antigen detection, Biosens. Bioelectron., 2019, 144, 111697 CrossRef CAS PubMed.
- L. Wu, X. Lu, Dhanjai, Z. S. Wu, Y. Dong, X. Wang, S. Zheng and J. Chen, 2D transition metal carbide MXene as a robust biosensing platform for enzyme immobilization and ultrasensitive detection of phenol, Biosens. Bioelectron., 2018, 107, 69–75 CrossRef CAS PubMed.
- G. Yang, Q. Zou, P. Wang, H. Lai, T. Lai, X. Zeng, Z. Li, J. Luo, Y. Zhang and C. Cui, Towards understanding the facile synthesis of well-covered Cu-Ag core-shell nanoparticles from a complexing model, J. Alloys Compd., 2021, 874, 159900 CrossRef CAS.
- B. Song, D. Li, W. Qi, M. Elstner, C. Fan and H. Fang, Graphene on Au(111): a highly conductive material with excellent adsorption properties for high-resolution bio/nanodetection and identification, ChemPhysChem, 2010, 11, 585–589 CrossRef CAS PubMed.
- K. Wu, V. K. Chugh, V. D. Krishna, A. di Girolamo, Y. A. Wang, R. Saha, S. Liang, M. C. J. Cheeran and J. P. Wang, One-step, wash-free, nanoparticle clustering-based magnetic particle spectroscopy bioassay method for detection of SARS-CoV-2 spike and nucleocapsid proteins in the liquid phase, ACS Appl. Mater. Interfaces, 2021, 13, 44136–44146 CrossRef CAS PubMed.
- Z. W. Jiang, T. T. Zhao, C. M. Li, Y. F. Li and C. Z. Huang, 2D MOF-based photoelectrochemical aptasensor for SARS-CoV-2 spike glycoprotein detection, ACS Appl. Mater. Interfaces, 2021, 13, 49754–49761 CrossRef CAS PubMed.
- Y. T. Büyüksünetçi, B. E. Çitil, U. Tapan and Ü. Anik, Development and application of a SARS-CoV-2 colorimetric biosensor based on the peroxidase-mimic activity of γ-Fe2O3 nanoparticles, Microchim. Acta, 2021, 188, 335 CrossRef PubMed.
- Q. Cai, J. Mu, Y. Lei, J. Ge, A. A. Aryee, X. Zhang and Z. Li, Simultaneous detection of the spike and nucleocapsid proteins from SARS-CoV-2 based on ultrasensitive single molecule assays, Anal. Bioanal. Chem., 2021, 413, 4645–4654 CrossRef CAS PubMed.
- E. Karakus, E. Erdemir, N. Demirbilek and L. Liv, Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef CAS PubMed.
- T. Lewis, E. Giroux, M. Jovicb and S. Martic-Milne, Localized surface plasmon resonance aptasensor for selective detection of SARS-CoV-2 S1 protein, Analyst, 2021, 146, 7207–7217 RSC.
- V. J. Vezza, A. Butterworth, P. Lasserre, E. O. Blair, A. MacDonald, S. Hannah, C. Rinaldi, P. A. Hoskisson, A. C. Ward, A. Longmuir, S. Setford, E. C. W. Farmer, M. E. Murphy and D. K. Corrigan, An electrochemical SARS-CoV-2 biosensor inspired by glucose test strip manufacturing processes, Chem. Commun., 2021, 57, 3704 RSC.
- Z. Fu, W. Zeng, S. Cai, H. Li, J. Ding, C. Wang, Y. Chen, N. Han and R. Yang, Porous Au@Pt nanoparticles with superior peroxidase-like activity for colorimetric detection of spike protein of SARS-CoV-2, J. Colloid Interface Sci., 2021, 604, 113–121 CrossRef CAS PubMed.
- D. Liu, C. Ju, C. Han, R. Shi, X. Chen, D. Duan, J. Yan and X. Yan, Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen, Biosens. Bioelectron., 2021, 173, 112817 CrossRef CAS PubMed.
- N. Cennamo, L. Pasquardini, F. Arcadio, L. Lunelli, L. Vanzetti, V. Carafa, L. Altucci and L. Zeni, SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor, Talanta, 2021, 233, 122532 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an00426g |
| This journal is © The Royal Society of Chemistry 2022 |