Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Towards identification of protein–protein interaction stabilizers via inhibitory peptide-fragment hybrids using templated fragment ligation†
Sonja
Srdanović
ab,
Zsofia
Hegedüs
 abc,
Stuart L.
Warriner
ab and
Andrew J.
Wilson
abc,
Stuart L.
Warriner
ab and
Andrew J.
Wilson
 *ab
*ab
aAstbury Centre for Structural Molecular Biology, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK. E-mail: a.j.wilson@leeds.ac.uk
bSchool of Chemistry, University of Leeds, Woodhouse Lane, Leeds LS2 9JT, UK
cDepartment of Medical Chemistry, University of Szeged, Dóm tér 8, H-6720 Szeged, Hungary
First published on 1st April 2022
Abstract
Using the hDMX/14-3-3 interaction, acylhydrazone-based ligand-directed fragment ligation was used to identify protein–protein interaction (PPI) inhibitory peptide-fragment hybrids. Separation of the peptide-fragment hybrids into the components yielded fragments that stabilized the hDMX/14-3-3 interaction.
Despite continued development of new ligand discovery methods,1,2 it remains challenging to identify hits for a significant palette of targets e.g. protein–protein interaction (PPI) inhibitors/stabilizers.3–6 Fragment-based-drug-discovery (FBDD)7,8 has proven powerful in furnishing starting points for PPI inhibitor candidates and led to development of clinically approved drugs.9 However, identification of fragments that can serve as starting points for the elaboration of PPI stabilizers10 presents specific challenges given the need to form a termolecular complex paired with typically weak fragment binding. Disulfide tethering11–14 has proven useful in developing PPI inhibitors,15 allosteric modulators,16 and tools to study aggregation.17 Recently the method was shown to be useful for site-directed fragment identification using the 14-3-3σ/ERα interface as a model. 40-Fold stabilization was achieved for binding of a C-terminal ERα peptide by 14-3-3 in the presence of the protein-fragment adduct,18 although stabilization in the absence of the disulfide tether remains to be reported. The approach has been inverted to exploit cysteines present within a peptide ligand to create “covalent-molecular glues”; fragment-peptide hybrids linked via a disulfide.19 Similarly, imine based tethering has subsequently been described whereby aldehydes react with surface exposed lysine residues on the surface of 14-3-3 to stabilize its interaction with peptides.20,21 A further complementary approach, whereby a scaffold compatible with dynamic combinatorial exchange by virtue of a central acylhydrazone bond, was used as a template to identify small-molecule 14-3-3ζ/synaptopodin stabilizers.22
We recently used ligand-directed fragment ligation as a tool to identify peptide-fragment hybrids that inhibit a β-strand mediated PPI.23 Reliant on hydrazone exchange,24,25 this approach is advantageous in that it unites the synthesis and assay steps within a single step and rapidly explores SAR using commercially available aldehyde fragments. Inspired also by the work of Ohkanda on the use of oxime ligation for in situ assembly of a bivalent diterpene-peptide conjugate as an intracellular 14-3-3ζ inhibitor,26 in this work we used our recently reported data on the hDMX/14-3-3 interaction27 as a model to further elaborate acylhydrazone-based ligand-directed fragment ligation to identify PPI inhibitory peptide-fragment hybrids which upon separation into the components can yield fragments as candidate PPI stabilizers (Fig. 1a). In contrast to the earlier work on covalent protein-18 and peptide-based19 molecular glues, this approach allowed us to reveal that non-covalent fragments which stabilize PPIs can be identified using site-directed ligation methods. Furthermore, the two methods are complementary in terms of the conditions under which the bonds are stable and under which exchange occurs.28 Finally, the asymmetry of the hydrazone linkage when compared to the disulfide, may be advantageous in terms of yielding only unsymmetrical peptide fragment hybrids.
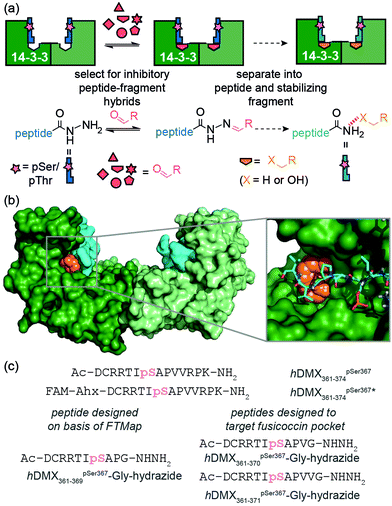
The central binding groove of 14-3-3 proteins accommodates phosphorylated peptides. Beside a phospho-binding groove on 14-3-3 proteins, a defined pocket has been shown to recognize Fusicoccin and other ligands resulting in stabilization of certain client/14-3-3 interactions.29hDMX361–374pSer367 was found to partially occupy the Fusicoccin binding pocket in the co-crystal structure with 14-3-3σ-ΔC. Thus, the challenge was to identify novel ligandable sites30 between 14-3-3 proteins and hDMX361–374pSer367. Computational ‘solvent mapping’ using FTMap31 was therefore used to identify suitable targetable pockets on the hDMX361–374pSer367/14-3-3σ interface. For the hDMX361–374pSer367/14-3-3σ-ΔC structure, FTMap identified several consensus sites on the 14-3-3 protein dimer interface but only one high quality consensus site was found to occupy a pocket on 14-3-3 abutting hDMX361–374pSer367 (Fig. 1b and Fig. S1, ESI†), To probe this pocket using a peptide based acylhydrazide, a truncated version of hDMX361–374pSer367; hDMX361–369pSer367Gly-hydrazide (Fig. 1c) was synthesized using hydrazone resin.32 The key residues (Arg at position −3 and Pro at position +2) needed to maximise binding affinity were kept, whilst a C-terminal Gly was used to maximize resin loading and conformational flexibility proximal to the acylhydrazide to support exploration of the 14-3-3 surface.
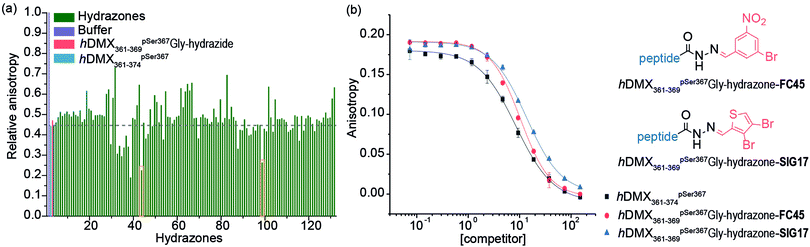
A fluorescence anisotropy (FA) competition assay was subsequently carried out using conditions previously found suitable for acylhydrazone formation (pH 6.5, 50 mM NH4OAc, 10 mM aniline) with added DTT (1 mM) to supress disulfide formation. hDMX361–369pSer367Gly-hydrazide was used to compete with a tracer peptide: hDMX361–374pSer367* for binding to 14-3-3η (Fig S2, ESI†). The resulting IC50 values were comparable over 24 hrs indicating good stability over the expected timeframe for hydrazone exchange. Initial attempts to screen in cocktails, established the aldehydes had no affect on their own (Fig. S3, ESI†). We then performed an assay where each aldehyde was screened individually with hDMX361–369pSer367Gly-hydrazide. A decrease in anisotropy can be attributed to a particular hydrazone product, avoiding the additional step of product identification e.g. by HRMS, and, combines the equilibration step for hydrazone formation with 14-3-3η screening (Fig. 2a). A number of hydrazones exhibiting lower anisotropy than hDMX361–374pSer367 were identified as hits, and we selected two bearing handles for further future functionalization to be assessed: hDMX361–369pSer367Gly-hydrazone-FC45 and hDMX361–369pSer367Gly-hydrazone-SIG17 (Fig. 2a, orange boxes). To confirm the single-concentration results, a competition assay was performed (Fig. 2b). hDMX361–369pSer367Gly-hydrazide was equilibrated with 5 equivalents of each aldehyde (product formation confirmed by LC-MS, see Fig. S4, ESI†), and serially diluted against the tracer peptide (hDMX361–374pSer367*) and 14-3-3η protein. The IC50 values for hDMX361–369pSer367Gly-hydrazone-FC45 (IC50 = 10.4 ± 0.3 μM) and hDMX361–369pSer367Gly-hydrazone-SIG17 (IC50 = 13.7 ± 0.5 μM) were found to be similar to hDMX361–374pSer367 (IC50 = 8.5 ± 0.2 μM) and hDMX361–369pSer367Gly-hydrazide (IC50 = 8.2 ± 1.3 μM). These data suggest the small-molecule fragments are tolerated adjacent to the peptide sequence, but that they likely contribute little to the binding energy possibly due to minimal contribution of the residues they replaced in the template peptide sequence.
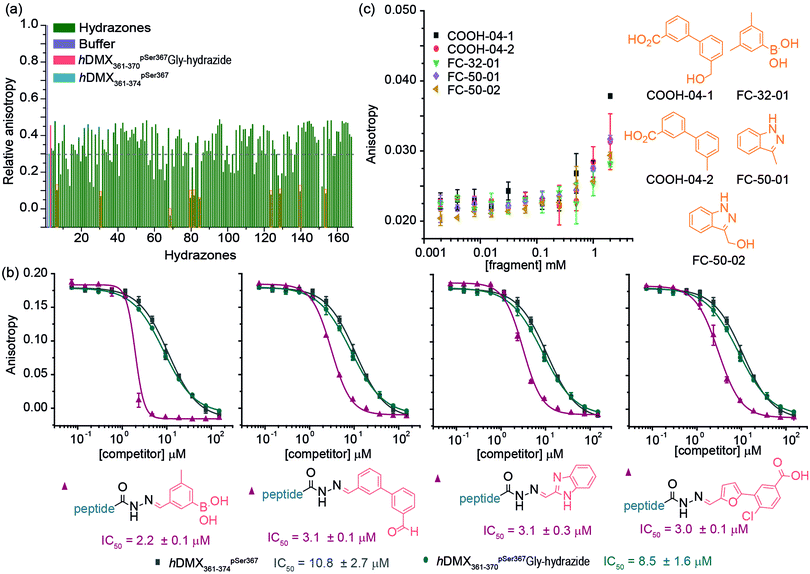
To demonstrate the approach could be used to target specific pockets on a protein, the Fusicoccin pocket on 14-3-3 was then targeted with an extended library of aldehydes (165 in total). Two longer hydrazide peptides were synthesized (Fig. 1c) extended by one (hDMX361–370pSer367Gly-hydrazide) and two (hDMX361–371pSer367Gly-hydrazide) residues so as to reach the Fusicoccin pocket. Similar results (in terms of potency) were obtained for each peptide in the dynamic ligation screen; thus for clarity, only data for hDMX361–370pSer367Gly-hydrazide are described here (Fig. 3a; screening and validation, including for hDMX361–371pSer367Gly-hydrazide is shown in the ESI† Fig. S5 and S6). More hits with a hDMX361–374pSer367*/14-3-3η response greater than the control were identified in this second round. Different fragments were identified as hits for each sequence pointing to the specificity of the selection process. For validation, a competition assay for ten hits was carried out (highlighted in the orange box in Fig. 3a, ESI† for additional results, Fig. S5). Fig. 3b illustrates IC50 data for four hydrazone hits that exceeded affinities of hDMX361–374pSer367* and the control hydrazide peptide (note: the steep Hill slope for the first derivative may suggest the boronic acid plays a role in covalent modification).
To explore the effect of non-linked fragments, 5 commercially available fragments with similarity to the reduced aldehyde form of COOH4, FC50 and FC32; analogues bearing alcohol or methyl groups in place of the aldehyde (Fig. 3c) were assessed. For consistency a modified tracer peptide was used (hDMX361–370pSer367Gly* i.e. analogous to the screening peptide but bearing FAM-Ahx). The 14-3-3 affinity of this peptide was determined by FA (Kd = 172.2 ± 9.3 nM, Fig. S7, ESI†), and activity of fragments tested in serial dilution. Stabilization of the peptide/14-3-3η interaction was observed as the anisotropy increased at the highest fragment concentration, consistent with millimolar affinities (Fig. 3c).
Conclusions
We used a ligand-directed dynamic-ligation approach to identify weak binding fragments that can augment the inhibitory potency of a peptide ligand and simultaneously serve as starting points for subsequent development of PPI stabilizers using the hDMX361–371pSer367/14-3-3η interaction as a model. We used acylhydrazone exchange, involving acyl hydrazide peptides and readily available aldehydes together with the surface of 14-3-3η protein to template peptide-fragment hybrids. Screening of representative fragments validated the method as a conceptually distinct approach for identification of PPI stabilizers. Although here, the screen was directed towards a known stabilizer pocket on 14-3-3, we note that inhibitory peptide-fragment hybrids could also reveal fragments that inhibit when separated from the peptide anchor. The acylhydrazide peptides described in this work varied in length and were designed to target distinct pockets on the 14-3-3 amphipathic groove. In obtaining different fragments from these screens, we demonstrated the site-selective specificity of the method. Future studies will centre on elaborating the identified fragments as small molecule stabilizers and broadening the approach to other targets.Author contributions
S. S., S. L. W. and A. J. W. conceived and designed the research program, S. S. designed studies and performed the research. The manuscript was written by S. S. and A. J. W. and edited into its final form by S. L. W. and A. J. W. with contributions from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by EPSRC (EP/N013573/1 and EP/K039202/1). This project has received funding from the EU Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie programme H2020-MSCA-ITN-2015 Grant number 675179 (TASPPI). We would like to thank Christian Ottmann, Madita Wolter (TU/e) and Gavin O’Mahony (AstraZeneca) for useful discussions and ongoing collaboration.Notes and references
- E. K. Grant, D. J. Fallon, M. M. Hann, K. G. M. Fantom, C. Quinn, F. Zappacosta, R. S. Annan, C.-W. Chung, P. Bamborough, D. P. Dixon, P. Stacey, D. House, V. K. Patel, N. C. O. Tomkinson and J. T. Bush, Angew. Chem., Int. Ed., 2020, 59, 21096–21105 CrossRef CAS PubMed.
- N. Favalli, G. Bassi, C. Pellegrino, J. Millul, R. De Luca, S. Cazzamalli, S. Yang, A. Trenner, N. L. Mozaffari, R. Myburgh, M. Moroglu, S. J. Conway, A. A. Sartori, M. G. Manz, R. A. Lerner, P. K. Vogt, J. Scheuermann and D. Neri, Nat. Chem., 2021, 13, 540–548 CrossRef CAS PubMed.
- M. Pelay-Gimeno, A. Glas, O. Koch and T. N. Grossmann, Angew. Chem., Int. Ed., 2015, 54, 8896–8927 CrossRef CAS PubMed.
- D. E. Scott, A. R. Bayly, C. Abell and J. Skidmore, Nat. Rev. Drug Discovery, 2016, 15, 533–550 CrossRef CAS PubMed.
- L. M. Stevers, E. Sijbesma, M. Botta, C. MacKintosh, T. Obsil, I. Landrieu, Y. Cau, A. J. Wilson, A. Karawajczyk, J. Eickhoff, J. Davis, M. Hann, G. O'Mahony, R. G. Doveston, L. Brunsveld and C. Ottmann, J. Med. Chem., 2018, 61, 3755–3778 CrossRef CAS PubMed.
- S. A. Andrei, E. Sijbesma, M. Hann, J. Davis, G. O’Mahony, M. W. D. Perry, A. Karawajczyk, J. Eickhoff, L. Brunsveld, R. G. Doveston, L.-G. Milroy and C. Ottmann, Expert Opin. Drug Discovery, 2017, 12, 925–940 CrossRef CAS PubMed.
- H. Jhoti, G. Williams, D. C. Rees and C. W. Murray, Nat. Rev. Drug Discovery, 2013, 12, 644 CrossRef CAS PubMed.
- D. A. Erlanson, S. W. Fesik, R. E. Hubbard, W. Jahnke and H. Jhoti, Nat. Rev. Drug Discovery, 2016, 15, 605–619 CrossRef CAS PubMed.
- A. W. Roberts, M. S. Davids, J. M. Pagel, B. S. Kahl, S. D. Puvvada, J. F. Gerecitano, T. J. Kipps, M. A. Anderson, J. R. Brown, L. Gressick, S. Wong, M. Dunbar, M. Zhu, M. B. Desai, E. Cerri, S. Heitner Enschede, R. A. Humerickhouse, W. G. Wierda and J. F. Seymour, N. Engl. J. Med., 2016, 374, 311–322 CrossRef CAS PubMed.
- P. Thiel, M. Kaiser and C. Ottmann, Angew. Chem., Int. Ed., 2012, 51, 2012–2018 CrossRef CAS PubMed.
- D. A. Erlanson, A. C. Braisted, D. R. Raphael, M. Randal, R. M. Stroud, E. M. Gordon and J. A. Wells, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 9367–9372 CrossRef CAS PubMed.
- D. A. Erlanson, J. W. Lam, C. Wiesmann, T. N. Luong, R. L. Simmons, W. L. DeLano, I. C. Choong, M. T. Burdett, W. M. Flanagan, D. Lee, E. M. Gordon and T. O'Brien, Nat. Biotech., 2003, 21, 308–314 CrossRef CAS PubMed.
- N. Wang, C. Y. Majmudar, W. C. Pomerantz, J. K. Gagnon, J. D. Sadowsky, J. L. Meagher, T. K. Johnson, J. A. Stuckey, C. L. Brooks, J. A. Wells and A. K. Mapp, J. Am. Chem. Soc., 2013, 135, 3363–3366 CrossRef CAS PubMed.
- K. K. Hallenbeck, J. L. Davies, C. Merron, P. Ogden, E. Sijbesma, C. Ottmann, A. R. Renslo, C. Wilson and M. R. Arkin, SLAS Discovery, 2018, 23, 183–192 CrossRef CAS PubMed.
- J. M. Ostrem, U. Peters, M. L. Sos, J. A. Wells and K. M. Shokat, Nature, 2013, 503, 548–551 CrossRef CAS PubMed.
- J. D. Sadowsky, M. A. Burlingame, D. W. Wolan, C. L. McClendon, M. P. Jacobson and J. A. Wells, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6056–6061 CrossRef CAS PubMed.
- E. E. Cawood, N. Guthertz, J. S. Ebo, T. K. Karamanos, S. E. Radford and A. J. Wilson, J. Am. Chem. Soc., 2020, 142, 20845–20854 CrossRef CAS PubMed.
- E. Sijbesma, K. K. Hallenbeck, S. Leysen, P. J. de Vink, L. Skóra, W. Jahnke, L. Brunsveld, M. R. Arkin and C. Ottmann, J. Am. Chem. Soc., 2019, 141, 3524–3531 CrossRef CAS PubMed.
- E. Sijbesma, B. A. Somsen, G. P. Miley, I. A. Leijten-van de Gevel, L. Brunsveld, M. R. Arkin and C. Ottmann, ACS Chem. Biol., 2020, 15, 3143–3148 CrossRef CAS PubMed.
- M. Wolter, D. Valenti, P. J. Cossar, L. M. Levy, S. Hristeva, T. Genski, T. Hoffmann, L. Brunsveld, D. Tzalis and C. Ottmann, Angew. Chem., Int. Ed., 2020, 59, 21520–21524 CrossRef CAS PubMed.
- M. Wolter, D. Valenti, P. J. Cossar, S. Hristeva, L. M. Levy, T. Genski, T. Hoffmann, L. Brunsveld, D. Tzalis and C. Ottmann, J. Med. Chem., 2021, 64, 8423–8436 CrossRef CAS PubMed.
- A. M. Hartman, W. A. M. Elgaher, N. Hertrich, S. A. Andrei, C. Ottmann and A. K. H. Hirsch, ACS Med. Chem. Lett., 2020, 11, 1041–1046 CrossRef CAS PubMed.
- Z. Hegedüs, F. Hóbor, D. K. Shoemark, S. Celis, L.-Y. Lian, C. H. Trinh, R. B. Sessions, T. A. Edwards and A. J. Wilson, Chem. Sci., 2021, 12, 2286–2293 RSC.
- V. T. Bhat, A. M. Caniard, T. Luksch, R. Brenk, D. J. Campopiano and M. F. Greaney, Nat. Chem., 2010, 2, 490–497 CrossRef CAS PubMed.
- D. Larsen, A. M. Kietrys, S. A. Clark, H. S. Park, A. Ekebergh and E. T. Kool, Chem. Sci., 2018, 9, 5252–5259 RSC.
- P. Parvatkar, N. Kato, M. Uesugi, S.-I. Sato and J. Ohkanda, J. Am. Chem. Soc., 2015, 137, 15624–15627 CrossRef CAS PubMed.
- S. Srdanovic, M. Wolter, C. H. Trinh, C. Ottmann, S. L. Warriner and A. J. Wilson, FEBS J., 2022 DOI:10.1111/febs.16433.
- S. J. Rowan, S. J. Cantrill, G. R. L. Cousins, J. K. M. Sanders and J. F. Stoddart, Angew. Chem., Int. Ed., 2002, 41, 898–952 CrossRef PubMed.
- L. M. Stevers, E. Sijbesma, M. Botta, C. MacKintosh, T. Obsil, I. Landrieu, Y. Cau, A. J. Wilson, A. Karawajczyk, J. Eickhoff, J. Davis, M. Hann, G. O’Mahony, R. G. Doveston, L. Brunsveld and C. Ottmann, J. Med. Chem., 2018, 61, 3755–3778 CrossRef CAS PubMed.
- B. S. Zerbe, D. R. Hall, S. Vajda, A. Whitty and D. Kozakov, J. Chem. Inf. Model., 2012, 52, 2236–2244 CrossRef CAS PubMed.
- D. Kozakov, L. E. Grove, D. R. Hall, T. Bohnuud, S. E. Mottarella, L. Luo, B. Xia, D. Beglov and S. Vajda, Nat. Protoc., 2015, 10, 733–755 CrossRef CAS PubMed.
- P. S. Chelushkin, K. V. Polyanichko, M. V. Leko, M. Y. Dorosh, T. Bruckdorfer and S. V. Burov, Tetrahedron Lett., 2015, 56, 619–622 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2cb00025c |
| This journal is © The Royal Society of Chemistry 2022 |