Open Access Article
Open Access ArticleDevelopment of ultra-high affinity bivalent ligands targeting the polo-like kinase 1†‡
Kohei Tsuji ab,
David Hymel§
a,
Buyong Ma¶
ab,
David Hymel§
a,
Buyong Ma¶
 c,
Hirokazu Tamamura
c,
Hirokazu Tamamura b,
Ruth Nussinov
b,
Ruth Nussinov c and
Terrence R. Burke Jr.
c and
Terrence R. Burke Jr. *a
*a
aChemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, MD 21702, USA. E-mail: burkete@nih.gov
bDepartment of Medicinal Chemistry, Institute of Biomaterials and Bioengineering, Tokyo Medical and Dental University, Tokyo 101-0062, Japan
cComputational Structural Biology Section, Laboratory of Immunometabolism, Frederick National Laboratory for Cancer Research, National Cancer Institute at Frederick, Frederick, MD 21702, USA
First published on 15th July 2022
Abstract
The polo-like kinase 1 (Plk1) is an important mediator of cell cycle regulation and a recognized anti-cancer molecular target. In addition to its catalytic kinase domain (KD), Plk1 contains a polo-box domain (PBD), which engages in protein–protein interactions (PPIs) essential to proper Plk1 function. We have developed a number of extremely high-affinity PBD-binding peptide inhibitors. However, we have reached an apparent limit to increasing the affinities of these monovalent ligands. Accordingly, we undertook an extensive investigation of bivalent ligands, designed to engage both KD and PBD regions of Plk1. This has resulted in bivalent constructs exhibiting more than 100-fold Plk1 affinity enhancement relative to the best monovalent PBD-binding ligands. Startlingly, and in contradiction to widely accepted notions of KD–PBD interactions, we have found that full affinities can be retained even with minimal linkers between KD and PBD-binding components. In addition to significantly advancing the development of PBD-binding ligands, our findings may cause a rethinking of the structure – function of Plk1.
Introduction
Multivalency can be a powerful means of achieving highly potent and selective ligand – protein interactions.1 Intermolecular multivalency describes the simultaneous binding of several ligands contained within a single scaffold to multiple binding sites on separate molecules, while intramolecular multivalency can be defined as the binding of multiple ligand components of a single entity with multiple binding sites on a single protein. In both cases, a number of disparate and not clearly understood factors can contribute to affinity enhancements that can be several orders-of-magnitude greater affinity than monovalent ligands.1 Protein kinases (PKs) are a family of enzymes where the principles of multivalency can be applied to design inhibitory constructs with particularly exquisite efficacies.2 This is because PKs are often modular in structure, having in addition to a catalytic domain, other protein domains that either direct the kinase to specific locations or modulate its catalytic activity.3,4 PKs function by transferring the γ-phosphate of adenosine triphosphate (ATP) to serine, threonine or tyrosine residues in substrate proteins. Productive catalysis requires the simultaneous binding of ATP and association with appropriate acceptor proteins.5 The selectivity and affinity of PK inhibitors can be greatly increased by linking an element that binds within the ATP-binding cleft together with a component that binds exterior to the cleft. When the secondary component is a pseudosubstrate, the construct may be viewed as being “bisubstrate.”6 When the secondary component accesses ancillary regulatory domains, the resulting ligand may be described as being intramolecular bivalent.2,7 Bivalent kinase inhibitors, which are referred to as “type V” inhibitors, are the subject of a recent review.8The serine/threonine specific polo-like kinase 1 (Plk1) is an important cell cycle regulator,9 which has been defined as a molecular target for anti-cancer therapy development.10–12 This protein is overexpressed in many cancers and its inhibition can result in antiproliferative effects. Plk1 requires the coordinated actions of both an N-terminal kinase domain (KD), which executes its catalytic function and a C-terminal polo-box domain (PBD), which engages in protein–protein interactions (PPIs) with phosphoserine (pS) and phosphothreonine (pT)-containing sequences.13 Although Plk1 KD-directed agents are currently in clinical trials for the treatment of cancers, issues related to cytotoxicity have arisen that may result from off-target effects.10–12 This is thought to be due in part to the fact that ATP is bound in a deep cleft that shares a high degree of homology among the approximately 500 members of the human kinome.14 Because PBDs are limited to the five members of the Plk family of kinases, targeting PBDs may afford attractive alternatives to ATP-competitive inhibitors for down-regulating Plk1 activity.15,16
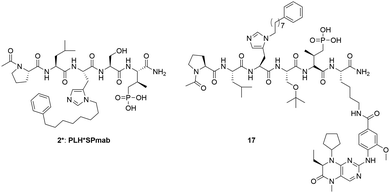
Over the past decade, we have exerted significant effort toward developing PBD-binding antagonists. Much of our work has employed peptides based on a region of the polo-box domain interacting protein 1 (PBIP1) proximal to the phosphorylated Thr78 (pT78) residue PLHSpT (1).17 We have found by tethering alkylphenyl groups from different positions on this sequence, that we can access a hydrophobic “cryptic binding pocket” formed by Y417, Y421, Y481, F482, Y485 and L478, which is revealed by a more than 100° rotation of the Y481 side chain.18–20 Occupying the cryptic binding pocket can provide up to three-orders-of-magnitude enhancement in PBD-binding affinity. We have been able to reach the pocket from the pT-2 His residue using peptides of the form PLH*SpT (2), where H* indicates the presence of a –(CH2)8Ph group on the His N3(π) nitrogen [i.e., H* = His-[N(π)–(CH2)8Ph]]. We have also found that tethering aryl and heteroaryl moieties from the His N3(π) nitrogen21,22 or introducing long-chain alkylphenyl groups into amino acids other than His at the pT-2 position, can also substantially improve binding affinities.23 Yet, despite intense effort, attempts to improve the binding affinities for peptides within this series has reached an apparent ceiling in the low nanomolar range, depending on the assay.
Plk1 presents an attractive target for the development of intramolecular bivalent ligands. Such agents could be constructed by combining elements that simultaneously bind to both the KD and PBD regions of the protein.10,12,16 Importantly, binding enthalpies could be as great as the sum of the energies associated with binding to each domain (i.e., potentially yielding binding constants that approach the product of individual affinities). In the case of Plk1, where ligand binding constants for both the KD and PBD are low nanomolar, the binding constants of bivalent ligands constructs could be well below nanomolar. Our current paper presents our extensive investigation of bivalent ligands, designed to simultaneously engage both KD and PBD regions of Plk1. As will be shown, this has resulted in bivalent constructs exhibiting more than 100-fold Plk1 affinity enhancement relative monovalent PBD-binding ligands, which had until this time, exhibited among the highest PBD-binding affinities yet reported. Startlingly, and in contradiction to widely accepted notions of KD–PBD interactions, we have found that extremely high affinities can be retained even with minimal linkers between KD and PBD-binding components. In addition to significantly advancing the development of PBD-binding ligands, our findings may cause a rethinking of the structure – function of Plk1 and potential implications for the physiological roles played by this kinase.
Results and discussion
Design of initial bivalent ligands
Bivalent ligands must be capable of simultaneously binding to both KD and PBD domains. While high affinity ligands for the individual domains are known, to date there have been no structures (either crystal, solution, or cryo-EM) of full-length Plk1, which would indicate the relative orientation of the two domains. However, the potential spatial organization of the domains has been informed by the recent crystal structure of a mixture of the isolated polo kinase KD and PBD from zebrafish (D. rerio), which have high homology to human Plk1 and the PBD-binding motif of Map205 (Map205PBM) from Drosophila (PDB accession code 4J7B).24 In this structure the KD appears to be held in a potentially relevant orientation relative to the PBD. By superimposing of the human Plk1 KD complexed to BI253625 (a potent Plk1 kinase inhibitor, exhibiting an in vitro IC50 value of 0.8 nM; PDB accession code 2RKU) onto the zebrafish KD and the human Plk1 PBD complexed to peptide 2 (PDB accession code 3RQ7) onto the zebrafish PBD, respectively, we were able to obtain a model from which to approximate distances between ligands binding to the two domains (Fig. 1). The 2RKU crystal structure shows that the 1-methylpiperidin-4-amide moiety of BI2536 extends out from the KD catalytic cleft into solvent, indicating that this portion of the molecule could potentially be removed without undue deleterious effect. In fact, we have already reported that deletion of this moiety does not affect Plk1 inhibitory activity of BI2536.26 It is less clear whether optimum geometry could be achieved by tethering the BI2536 portion to the PBD-binding peptide from its C- or N-terminus. From Fig. 1, it is apparent that C-terminal linkage would be the most direct. However, the protein inter-domain linker (IDL) joining the KD to the PBD is thought to be highly flexible and the relative orientations of the two domains could be quite variable (not restrained to what is shown in the 4J7B crystal structure). Therefore, examining N-terminal linkage in addition to C-terminal linkage is warranted. We have previously shown that PBD-binding peptides are highly tolerant of N-terminal PEG chains bearing bulky fluorophores, suggesting that an N-terminal linker should not dramatically impact PBD-binding affinities.18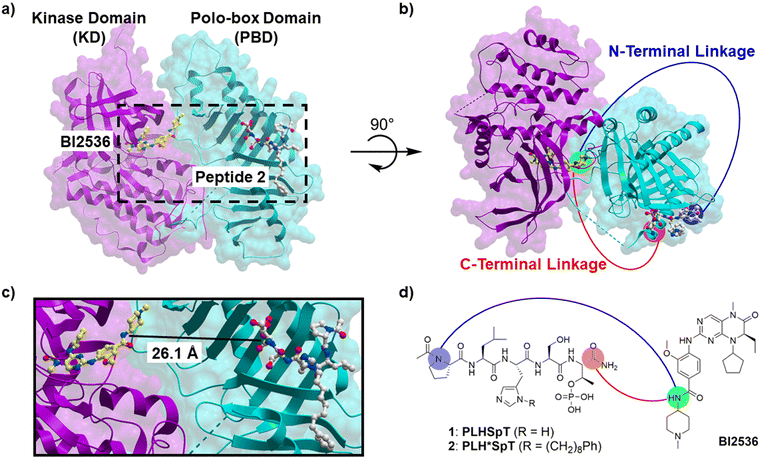 | ||
| Fig. 1 (a) Superimposed structure of zebrafish polo kinase isolated KD and PBD complex (4J7B) with BI2536-bound human Plk1 KD (2RKU, magenta) and peptide 2-bound human Plk1 PBD (3RQ7, cyan); (b) a different angle view of the structure in panel a. (c) An enlargement of the contents within the dashed line box in panel a. (d) Structures of BI2536 and peptides 1 and 2 showing proposed linking from the C-terminus (red) and the N-terminus (blue). | ||
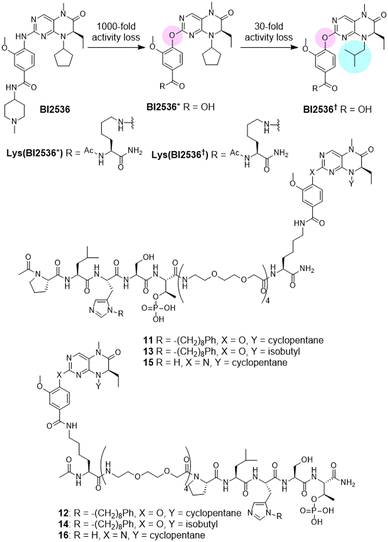
We designed a set of bivalent ligands, in which peptide 2 was linked to the core structure of BI2536 (Lys(BI2536), Fig. 2) by means of mini-PEG units, each of which spanned a maximum distance of approximately 9.4 Å (Fig. 2). These consisted of both C-terminal [3, 5, 7, and 9 (n = 4, 3, 1, and 0, respectively)] and N-terminal [4, 6, 8, and 10 (n = 4, 3, 1, and 0)] linkages (Fig. 2). The maximum spanning distance achievable was approximately 43 Å (n = 4). A C-terminal spanning distance of 26 Å was anticipated to be minimally acceptable based on the 4J7B crystal structure (Fig. 1). The syntheses of these peptides are shown in Scheme S1 (ESI†).
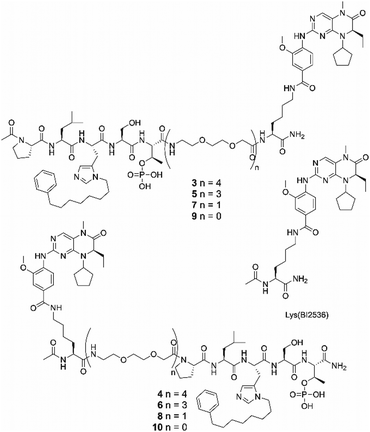 | ||
| Fig. 2 Structure of Lys(BI2536) and bivalent ligands with different linker lengths conjugated the PBD-binding peptide from C-terminus (3, 5, 7, and 9) and N-terminus (4, 6, 8, and 10). | ||
Biological evaluation of initial bivalent ligands having four mini-PEG units as linker
We evaluated the binding affinities of the synthetic ligands using fluorescence polarization (FP) assays, which measured the ability of ligands to inhibit the binding of FITC-labelled 2 (FITC-2, Scheme S2, ESI†) to either full-length Plk1 (having both KD and PBD components) or to isolated PBD (lacking a KD component). It is worth noting that IC50 values measured against the isolated PBD are lower than those measured against full-length Plk1. This is consistent with a literature report that PBD-dependent binding of full-length Plk1 to pS/pT epitopes is significantly reduced in comparison to the binding of isolated PBD.27 This difference is attributed to inhibitory interactions between the PBD and KD that affect equilibrium binding to phosphopeptides. The KD-binding “BI2536 motif” [Lys(BI2536)] was tethered to peptide 2 by four mini-PEG units from either the C-terminus or the N-terminus, designated as peptides 3 and 4, respectively. It is important to note that our previously reported IC50 value for 2 was extremely low (IC50 = 17 nM). This ELISA data was generated using a plate-bound biotinylated pT78 peptide derived from the PBIP1 sequence and cell lysate containing GFP-labelled full-length Plk1. In contrast, the FP assays used in our current work employed the higher affinity FITC-2 as a probe and purified myc-tag fused full-length Plk1 without the GFP fusion. Under these conditions the parent monovalent peptide 2 gave an IC50 value of 340 ± 15 nM and the bivalent peptides 3 and 4 gave IC50 values that were approximately two orders-of-magnitude lower than for 2 (IC50 = 2.3 ± 0.22 nM and 2.2 ± 0.24 nM, respectively, Table 1 and Fig. S2a, ESI†). This data indicates that tethering Lys(BI2536) by four mini-PEG units to either the C-terminal or N-terminal positions of 2 results in significant enhancement of binding affinities. Moreover, in the presence of 100 μM BI2536, the bivalent constructs 3 and 4 decreased their affinities by more than one order-of-magnitude (IC50 = 37 ± 1.8 nM and IC50 = 79 ± 4.9 nM, respectively). These affinities are similar to the affinity of monovalent 2 in the presence of 1 μM BI2536. Repeating the above assays using the isolated PBD showed that BI2536 had no effect on the binding affinities of the bivalent constructs. Since the isolated PBD lacks a KD, this supports a key role for the KD in the enhanced affinity of bivalent constructs against full-length Plk1 (Fig. S2b, ESI†).| Ligand | PBD-binding affinitya (nM) (full-length Plk1) | PBD-binding affinitya (nM) (isolated PBD) |
|---|---|---|
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Not determined. | ||
| BI2536 | No inhibition | N.d.b |
| 1 | No inhibition | N.d. |
| 2 | 340 ± 15 | 34 ± 1.2 |
| 3 | 2.3 ± 0.22 | 12 ± 0.76 |
| 4 | 2.2 ± 0.24 | 18 ± 7.9 |
| 2 + 1 μM of BI2536 | 130 ± 4.9 | 41 ± 2.9 |
| 3 + 100 μM of BI2536 | 37 ± 1.8 | 16 ± 1.7 |
| 4 + 100 μM of BI2536 | 79 ± 4.9 | 22 ± 0.36 |
Potencies of the compounds were also evaluated for inhibition of catalytic phosphorylation by the KD using a FRET-based kinase assay (Z′-LYTE kinase assay kit Ser/Thr 16 peptide, Invitrogen).28 In addition, we evaluated the constructs using our recently reported fluorescence recovery-based binding assay, which measures KD-affinity using full-length Plk1 (Table 2 and Fig. S3, S4, ESI†).26 The results of the kinase assay showed that the bivalent constructs stoichiometrically inhibit Plk1 kinase activity, as evidence by the steep Hill slopes associated with the dose-response curves. Addition of excess monovalent 2 dramatically decreased bivalent binding so that results observed were nearly the same as co-incubation of Lys(BI2536) with 1 μM of 2 (Fig. S3, ESI†). We interpret this data as supporting binding of the bivalent construct to both the Plk1 KD and PBD components. The results of the fluorescence recovery assays also show dose-response curves with steep Hill slopes, consistent with the binding affinity of the bivalent ligands being higher than that of parent BI2536 (Fig. S4, ESI†). These data also support binding of the bivalent construct to Plk1 KD and PBD.
| Ligand | Kinase inhibitory potencya (nM) | KD affinitya (nM) |
|---|---|---|
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Reported IC50 value.26 c Not determined. | ||
| BI2536 | 19 ± 1.0b | 47 ± 0.79 |
| Lys(BI2536) | 17 ± 2.2b | 19 ± 0.53 |
| 3 | 55 ± 5.4 | 16 ± 0.58 |
| 4 | 83 ± 4.7 | 17 ± 0.20 |
| Lys(BI2536) + 1 μM of 2 | 5.5 ± 0.27 | N.d.c |
| 3 + 100 μM of 2 | 7.3 ± 0.48 | N.d. |
| 4 + 100 μM of 2 | 5.0 ± 0.27 | N.d. |
Structure-activity relationship (SAR) studies examining bivalent ligands with different linker lengths
The results described above show that bivalent ligands having tethers composed of four mini-PEG units exhibit significantly enhanced Plk1 binding affinities in KD-dependent fashion. However, the distances separating the PBD and KD ligand binding sites in Plk1 were not known. The fact that these two domains are joined by a highly flexible IDL, (residues 326–368) further emphasizes uncertainties regarding optimum bivalent linker composition and length. A C-terminal spanning distance of 26 Å was anticipated to be minimally acceptable based on the 4J7B crystal structure (Fig. 1). Each mini-PEG unit can span a maximum distance of approximately 9.4 Å (Fig. 2). The bivalent constructs reported above having four mini-PEG units in their linkers should be able to span approximately 37 Å. Therefore, we explored the effects of shortening the linkers first to three mini-PEG units, then to one and finally to zero mini-PEG units (Table 3 and Fig. S5, ESI†). Somewhat to our surprise, we found that irrespective of linker length, all bivalent constructs examined (3–10 having linkers with 4, 3, 1 and 0 mini-PEG units, respectively) showed similar single-digit nanomolar affinities. These affinities were more than two orders-of-magnitude greater than the parent monovalent penta-peptide 2 (Table 3). It is important to note that due to a bottoming out of the assay sensitivity, these IC50 values probably represent an upper limit. Actual affinities among ligands having different linker lengths could potentially show significant variation at sub-nanomolar levels. We also performed FP and fluorescence recovery-based binding assays with excess non-labelled probes (Fig. S6, ESI†). However, in both assays, there is no significant attenuation of the affinities of the bivalent compounds 3 (IC50 = 1.4 ± 0.084, 1.4 ± 0.21, 1.5 ± 0.23, and 1.6 ± 0.057 nM in the presence of 0, 10, 100, and 1000 nM of 2, respectively for FP assays, and IC50 = 5.7 ± 0.16, 5.1 ± 0.10, 3.1 ± 1.5 nM in the presence of 0, 10, and 100 nM of BI2536, respectively for fluorescence recovery-based binding assays) and 9 (IC50 = 2.6 ± 0.082, 3.0 ± 0.27, 2.8 ± 0.14, and 3.1 ± 0.20 nM in the presence of 0, 10, 100, and 1000 nM of 2, respectively for FP assay, and IC50 = 14 ± 0.94, 8.5 ± 0.33, 7.2 ± 3.3 nM in the presence of 0, 10, and 100 nM of BI2536, respectively for fluorescence recovery-based binding assays). These results also support the extremely high binding potencies of the bivalent compounds.| Ligand | PBD affinitya (nM) (full-length Plk1) |
|---|---|
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Number of mini-PEG units in linker. | |
| 2 | 550 ± 23 |
| 3 (n = 4)b | 1.8 ± 0.20 |
| 4 (n = 4) | 1.9 ± 0.095 |
| 5 (n = 3) | 1.7 ± 0.097 |
| 6 (n = 3) | 1.3 ± 0.13 |
| 7 (n = 1) | 1.2 ± 0.40 |
| 8 (n = 1) | 1.9 ± 0.089 |
| 9 (n = 0) | 3.1 ± 0.59 |
| 10 (n = 0) | 3.6 ± 0.35 |
Examination of bivalent ligands having BI2536 motifs with attenuated KD affinities
We wanted to confirm that the KD and PBD-binding components of the bivalent constructs were binding as intended in their respective target domains. This was important, since a conceptually similar study by Berg et al. using a BI2536 analog tethered to the N-terminus of the PBD-binding peptide GPLHSpTA, concluded that affinity enhancement of the bifunctional construct was not due to the BI2536 moiety binding in the KD active site.29 Accordingly, in order to provide evidence that the BI2536 motif in our bivalent ligands is binding in the KD active site, we attenuated the KD-binding affinity of the BI2536 component by making relatively minor structural changes known to have dramatic effects on KD affinities but which should not greatly affect binding at sites other than the KD. First, we replaced a key nitrogen atom with oxygen (BI2536*, Fig. 3). For the parent BI2536, this simple change results in an approximate 1000-fold loss of potency (Plk1 Ki = 220 nM for BI2536* as compared to Ki = 0.2 nM for BI2536).30 We attenuated the affinity of BI2536* even further by replacing its N-cyclopentyl group with an i-butyl group to yield BI2536† based on the fact that this change in BI2536 results in a 30-fold loss of potency (Fig. 3).30 The syntheses of BI2536 derivatives and their bivalent ligands are shown in Schemes S3–S5 (ESI†).As anticipated, the bivalent constructs incorporating either BI2536* (11 and 12) or BI2536† (13 and 14) showed significantly less affinity for the PBD in full-length Plk1 than the parent BI2536-containing compounds (Table 4 and Fig. S6, ESI†). Again, it is important to note that the actual IC50 values of parent 3 and 4 could potentially be significantly lower than measured. This would mean that the apparent binding attenuation is probably much greater than indicated by these numbers. This tendency was also observed in the kinase assays and fluorescence recovery assays that evaluated inhibitory potency and affinity against the KD, respectively (Table 4 and Fig. S4, S7, ESI†). These data showing that modified bivalent constructs 11 and 12 having reduced KD-affinities also exhibit concomitant reduced PBD-binding affinities are consistent with binding in the KD active site playing an important role in overall enhanced PBD-binding affinity.
| Ligand | PDB affinitya (nM) | Kinase inhibitory potencya (nM) | KD affinitya (nM) |
|---|---|---|---|
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Not determined. c IC50 value shown in Table 2. | |||
| BI2536 | N.d.b | 0.93 ± 0.15 | 47 ± 0.79c |
| Lys(BI2536) | N.d. | 0.065 ± 0.0017 | 19 ± 0.53c |
| 2 | 310 ± 82 | N.d. | N.d. |
| 3 | 2.0 ± 0.045 | 9.0 ± 0.99 | 16 ± 0.58c |
| 4 | 3.4 ± 0.27 | 8.7 ± 0.49 | 17 ± 0.20c |
| Lys(BI2536*) | N.d. | 960 ± 360 | No inhibition |
| 11 | 33 ± 3.5 | 710 ± 230 | No inhibition |
| 12 | 60 ± 1.2 | 1000 ± 600 | No inhibition |
| Lys(BI2536†) | N.d. | No inhibition | No inhibition |
| 13 | 60 ± 2.9 | ≫10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
No inhibition |
| 14 | 100 ± 4.3 | ≫10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
No inhibition |
In order to compare our results with recently reported bifunctional inhibitors targeting the Plk1 KD and PBD,29 we modified our bivalent construct design by using PLHSpT (1), which we first reported as a preferred minimal PBD-binding peptide and which is routinely used as a reference PBD-binding peptide (Table 5 and Fig. S8, ESI†).17 Peptide 1 shows approximately 1000-fold less PBD-binding affinity than peptide 2.18 We found that bivalent constructs utilizing PLHSpT (15 and 16) failed to show inhibitory potencies in our assay (Table 5). These results are consistent with previously reported bifunctional ligands formed by conjugating the lower affinity PBD-binding sequence, GPLHSpTA with BI2536.29 These latter constructs employed long alkyl chain linkers arising solely from the N-terminus of the peptide. No advantage was achieved by bivalent conjugation, although the affinity of the FP probe used to measure binding (5-CF-GPMQSpTPLNG-OH) was significantly lower than the affinity of the probe (FITC-2) used in our assays. We attribute the results obtained with 15 and 16 as being due to their inability to compete for binding to the PBD with the much higher affinity fluorescent probe (FITC-2, Scheme S2, ESI†). This was regardless of the fact that they retained as their KD-binding component, a nanomolar-affinity BI2536 analog.
Molecular dynamics simulations
In our SAR study examining the effects of varying linker length (Table 3 and Fig. S5, ESI†), we observed that all bivalent compounds (3–10) conjugated to the BI2536 motif whether from the C- or N-terminus of the peptide 2, exhibited high Plk1-binding affinities that were at least two orders-of-magnitude greater than the monovalent PBD-binding parent penta-peptide 2. This was in spite of the fact that the mini-PEG linkers (n = 4, 3, 1, or 0) showed extreme variation in lengths (Fig. 2). These results appear to be inconsistent with the initial model structure, which suggested that at least 26 Å would be required for simultaneous binding to the KD and PBD (Fig. 1). This suggested that the initial model based on the PBD 4J7B-derived structure, may not accurately describe the orientations of the KD and PBD in full-length Plk1. In order to explore possible alternate conformations of full-length Plk1 that would be more consistent with the SAR results, we performed molecular dynamics (MD) simulations on fully hydrated protein using the NIH HPC cluster. The protein model was constructed using the structure of isolated zebrafish Polo-kinase 1 KD and PBD with drosophila MAP205 peptide (4J7B) with addition of a homology-modelled 35-residue IDL (331–366). The ligands employed were monovalent BI2536 and PLHSpT (1) as the KD and PBD-binding components, respectively. The structure of the Plk1 construct with BI2536 and PLHSpT bound before and after MD simulations showed the relative motions of the KD (Fig. 4(a)–(c)). The simulations suggest that the structure of 4J7B was reasonable as one possible stable conformation for the apo form. However, the binding of ligands in the KD and PBD domains allowed a conformation with larger separation between the two respective binding pockets. Binding of the peptide alone stabilized both the structure of the PBD as well as the entire complex. However, simultaneous binding of BI2536 and the peptide destabilized the complex (Fig. 4(d)).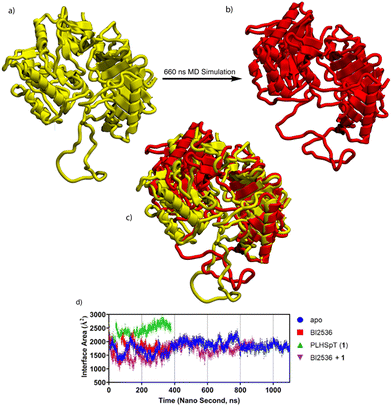 | ||
| Fig. 4 MD simulations on fully hydrated protein. The structure was modelled based on the structure of isolated zebrafish Polo kinase 1 KD and PBD with drosophila MAP 205 peptide (4J7B) with addition of a homology modelled 35 residue (331–366) inter domain linker (IDL). Ligands in this modelling are monovalent BI2536 and PLHSpT (1) for KD and PBD, respectively. Full length Plk1 with BI2536 bound (before (a) and after (b)) MD simulations showing relative motion of the KD. The superimposed structure of (a) and (b) is shown in (c). (d) Calculated contact surface area between KD and PBD during MD simulations. | ||
These results indicate that large variation of distances between two inhibitor-binding pockets is possible. Based on these MD simulation results, we hypothesized that additional KD–PBD binding interfaces could exist in an ensemble of conformations that would be consistent with the bivalent binding of constructs having various linker lengths. In order to find new reasonable conformations of full-length Plk1 allowing close contact between KD and PBD ligand-binding pockets, we docked the KD and PBD to accommodate the binding of bivalent peptides 9 and 10, which have the shortest linker (n = 0) conjugated from C- or N-terminus, respectively, and performed MD simulations (Fig. 5). The newly simulated conformations have smaller surface contacts than that of the structure of 4J7B. However, short pocket distances were stably maintained for long simulation times (>1000 ns).
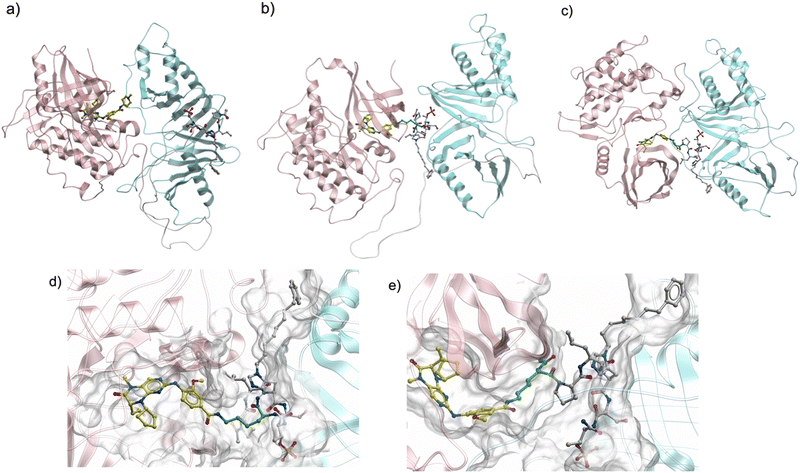 | ||
| Fig. 5 MD simulation of Plk1 with the KD and PBD arranged to allow binding of the bivalent ligands with the shortest linker (9 and 10). (a) Initial model structure based on 4J7B from Fig. 1(a). Simulated structures of full-length Plk1 with close contact of two ligand binding pockets with 9 (b) and 10 (c). Enlarged structures of (b) and (c) are shown in (d) and (e), respectively. Pink: KD, light blue: PBD, yellow: BI2536, white: peptide 2, green: linker (Lys), grey: binding surface of KD and PBD with 9 or 10. | ||
The MD simulations indicate that stable orientations of the KD with the PBD are possible, such that the respective ligand-binding pockets can be in close proximity. This permits binding of bivalent ligands with even the shortest linker (n = 0) from either the C- or N-terminus of 2 (peptides 9 and 10, respectively, Fig. 2). This is due in part to the high conformational flexibility of the 35 amino acid IDL joining the two domains. Although counter-intuitive based on the 4J7B crystal structure, these results are consistent with recently reported work that combined structural data from several different sources to define the overall relationship of the KD with the PBD in a multi-domain Plk1 model.31 These authors found that the KD uses its C-lobe to interact with the PBD near its ligand binding site and that full-length Plk1 is highly dynamic, making a variety of domain–domain interfaces in solution. This recent report provides strong validation of our own conclusions regarding the permissibility of close proximity of the KD and PBD ligand binding sites. It should be noted that these MD simulations examined bivalent ligands with very short linker segments. We did not perform additional simulations to determine whether ligands with longer linkers could potentially bind in intermolecular fashions. Although such intermolecular binding may be a distinct possibility, our primary interests focused on developing intramolecular bivalent ligands.
Evaluation of inhibitory potencies against a panel of kinases
We also evaluated kinase selectivity of the bivalent ligands 3 and 9. The kinase assays were performed by Reaction Biology Corporation using a panel of 8 kinases (Table 6). The kinases used were selected from the Reaction Biology Corporation inventory based on those showing the highest inhibitory potencies by BI2536 as reported by Boehringer Ingelheim.32 The compounds were tested in a 10-dose IC50 mode with a 3-fold serial dilution starting at 10 μM. The bivalent compound 9, which had the shortest C-terminal linker (Fig. 2, n = 0), showed from moderate to good selectivity among the kinases compared to BI2536. The bivalent compound 3, having a long C-terminal linker (Fig. 2, n = 4) showed slightly less selectivity than 9 against the panel of Plk kinases (Plk1, Plk2 and Plk3) (Table 6).Synthesis and evaluation of a bivalent compound possessing a hydrolytically stable pT mimetic
In order to overcome instability of phosphate ester moiety in the bivalent compounds against phosphatases, (2S,3R)-2-amino-3-methyl-4-phosphono-butanoic acid (Pmab) was used instead of pT to synthesized compound 17 (Fig. 6). Pmab has been utilized as a non-hydrolyzable pT mimetic in a variety of biologically active peptides, including peptidomimetic ligands that bind to the Plk1 PBD. These constructs typically retain binding affinities comparable to the corresponding pT-containing peptides. As the results of the assays (Table 7, Fig. S10 and S11, ESI†), compound 17 retained high Plk1 PBD and KD affinity (IC50 = 3.2 ± 0.039 nM for PBD and 17 ± 1.3 nM for KD), comparable to that of 9 (IC50 = 3.0 ± 0.20 nM for PBD and 19 ± 0.94 nM for KD). Next, we performed MTT assays to evaluate cellular potency of the bivalent compounds 9 and 17 (Table 7, Fig. S12, ESI†). These assays showed that BI2536 exhibits high potency (IC50 = 32 ± 1.6 nM) against HeLa cells. However, 9 (IC50 = 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 1700 nM), 17 (12
000 ± 1700 nM), 17 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 630 nM), 2 (IC50 ≫ 200
000 ± 630 nM), 2 (IC50 ≫ 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM), 2* (PLH*SPmab, IC50 ≫ 200
000 nM), 2* (PLH*SPmab, IC50 ≫ 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nM), Lys(BI2536) (IC50 = 600 ± 29 nM) showed much lower cellular potencies against HeLa cells. These results can potentially reflect low cell membrane permeability, and they are consistent with the results of cell membrane permeability assays (Caco-2 assays) performed by Charles River Laboratories (Table S3, ESI†).
000 nM), Lys(BI2536) (IC50 = 600 ± 29 nM) showed much lower cellular potencies against HeLa cells. These results can potentially reflect low cell membrane permeability, and they are consistent with the results of cell membrane permeability assays (Caco-2 assays) performed by Charles River Laboratories (Table S3, ESI†).
Immunostaining experiments of HeLa cells treated with Plk1 inhibitors
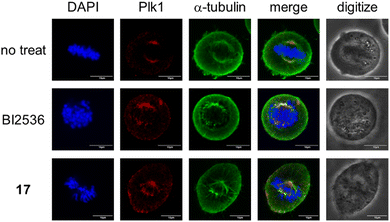
Finally, we conducted immunostaining experiments to visualize the disruption of spindle assembly and chromosome alignment as well as abnormal localization of Plk1 incurred by the treatment with the bivalent compound 17 (Fig. 7). As compared with the bivalent compounds 3 and 9, the control compound BI2536 has less Plk1 selectivity. Therefore, treatment with BI2536 affects a variety of cell signaling pathways leading to cytotoxicity due to multi-kinase inhibition including Plk1 (Table 6).| Ligand | PDB affinitya (nM) (full-length Plk1) | KD affinitya (nM) (full-length Plk1) | Cytotoxicitya (nM) (HeLa cells) |
|---|---|---|---|
| a Numbers represent the average IC50 value ± SEM (nM) from three independent experiments. b Not determined. | |||
| BI2536 | N.d.b | 53 ± 6.0 | 32 ± 1.6 |
| Lys(BI2536) | N.d. | N.d. | 600 ± 29 |
| 2 | 450 ± 37 | N.d. | ≫200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 2* | 1900 ± 160 | N.d. | ≫200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 9 | 3.0 ± 0.20 | 19 ± 0.94 | 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 1700 000 ± 1700 |
| 17 | 3.2 ± 0.039 | 17 ± 1.3 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 630 000 ± 630 |
Conclusions
In our development of PBD-binding peptides, we had previously achieved some of the highest affinity ligands yet reported. This is exemplified by the pentapeptide 2. Yet, in spite of continued efforts, we have not been able to significantly increase affinities further. In our current paper, we present an extensive investigation of ligands designed to engage both the KD and PBD regions of Plk1 in an intramolecular bivalent fashion. For this purpose, we used peptide 2 as the PBD-binding component and a truncated BI2536 motif, designated as Lys(BI2536) (Fig. 2), as the KD-binding construct. Since there have been no structures of full-length Plk1 yet reported, we took 26 Å as a reasonable minimum length for the bivalent linker based on the 4J7B crystal structure, which shows a potentially relevant orientation of the KD relative to the PBD. We constructed our bivalent ligands with linkers composed of multiples of “mini-PEG” units, each spanning approximately 9 Å and tethering the BI2536 motif from both the N- and C-terminus of peptide 2. We began with bivalent constructs having a maximum linker length of approximately 37 Å (n = 4). We found that regardless of tethering from the N- or C-terminus, these bivalent constructs showed enhancements in PBD-binding affinity that were more than two orders-of-magnitude higher than for the parent monovalent 2. Shortening the linker length first to approximately 28 Å (n = 3), then to 9 Å (n = 1) and finally deleting the linker entirely, showed that in all cases very high affinities were maintained. From FP assays measuring PBD-binding inhibition, kinase assays measuring inhibition of catalytic phosphorylation, and fluorescence recovery assays measuring KD-binding inhibition, our data suggest that our constructs (3–10) exhibit bivalent binding that engages both the KD and PBD components.The apparent insensitivity in the PBD-binding affinities of bivalent ligands to internal linker lengths was unexpected. This may be partially explained by the fact that binding affinities of the bivalent ligands exceed the detection limits of the assays. Consequently, the observed IC50 values may be viewed as upper limits. In this light, the measured more than 100-fold improvement in bivalent PBD-binding affinities relative to the previous most potent monovalent PBD-binding affinities, could represent an under-estimate of the actual degree of enhancement. The actual IC50 values of the bivalent ligands potentially could be significantly below the single digit nanomolar figures generated by the assays. By exceeding the lower detection limits of the assays, the observed insensitivity of affinities based on either linker length or origination from the N- or C-terminus of the PBD-binding peptide, may be misleading. It is possible that these structural parameters may have pronounced effects on actual bivalent ligand affinities. Nonetheless, the central findings of our study concerning the dramatic effects of bivalency on PBD-binding affinity remain unaltered.
Independent of our inability to accurately ascertain the relative effects of linker length on bivalent affinities, it remains particularly surprising that extremely high affinities of bivalent ligands can be maintained even after removal of the linker entirely. This has a number of consequences. First, it suggests that the KD and PBD ligand binding sites can stably exist in extremely close proximity. This is inconsistent with our original assumptions based on the 4J7B crystal structure. However, our MD simulations show that such orientations can stably exist. This is further supported by a recent report finding that full-length Plk1 is highly dynamic and capable of making a variety of domain–domain interfaces in solution, including ones that place the KD and PBD binding sites in close proximity.31 The biological processes and physiological significance of Plk1 and the roles played by the KD and PBD are only incompletely understood.15,33 Our findings are supported by others, that the ligand-binding pockets for these two domains can stably exist in extremely close proximity. This provides important insights and extends the possible boundaries of how the KD and PBD may interact in promoting overall Plk1 function.
A second important finding of our current study is related to the development of PBD-binding inhibitors, which had reached an apparent stasis point. We now show that simply adding a KD-binding moiety directly onto a PBD-binding ligand can dramatically increase PBD-binding affinities. For peptides, this may be viewed as merely introducing an elaborate C-terminal amide group. These findings dramatically alter and expand the field of PBD-binding inhibitor development.
Our current results from cellular assays reveal that regardless of Plk1-binding affinities, the cellular efficacies of peptide-based Plk1 PBD inhibitors can be significantly attenuated by low cell membrane permeability. Separately, we have shown that issues related to cell membrane penetration of PBD-binding peptides can be at least partially addressed by attaching cell penetrating peptides such as NMΔ1.34
In summary our current work discloses bivalent constructs that exhibit PBD-binding affinities, which at a minimum, are more than two orders-of-magnitude greater than the best ligands previously reported. Our results support the findings of others that the KD and PBD of Plk1 can stably exist in orientations, which place the respective ligand binding sites in extremely close proximity. This may have important implications on the physiological functions of Plk1 and perhaps, other members of the Plk family. Finally, our bivalent constructs 9 and 10 may be viewed as variants of parent peptide 2 functionalized at either the C- or N-terminus, respectively. Analog 17, which is a non-hydrolysable variant of 9, may represent a promising lead for further development of Plk1 inhibitors. Our work provides new and valuable insights into the design of minimally sized bivalent ligands with exceptional PBD-binding affinities.
Conflicts of interest
Aspects of the work presented are contained within one or more patent applications.Acknowledgements
This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (Z01-BC 006198), by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at the NIH (K. T.), and by JSPS KAKENHI Grant Numbers 22K15243 (K. T.), and by federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261201500003I. We also thank to Dr. Pankaj Mahajan (CBL, NCI, NIH) for his support on NMR measurements, Dr. Ramesh M. Chingle (CBL, NCI, NIH) for providing Fmoc-His*-OH, and Prof. Nobuhiko Yui and Assoc. Prof. Atsushi Tamura for their support on immunostaining experiments. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov/). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.Notes and references
- C. Chittasupho, Ther. Delivery, 2012, 3, 1171–1187 CrossRef CAS PubMed.
- C. M. Gower, M. E. K. Chang and D. J. Maly, Crit. Rev. Biochem. Mol. Biol., 2014, 49, 102–115 CrossRef CAS PubMed.
- B. J. Mayer, Methods Mol. Biol., 2006, 332, 79–99 CAS.
- T. Pawson and P. Nash, in Handbook of Cell Signaling, ed. R. A. Bradshaw and E. A. Dennis, Academic Press, San Diego, 2nd edn, 2010, pp. 399–411 DOI:10.1016/B978-0-12-374145-5.00057-7.
- T. Hunter, Cell, 2000, 100, 113–127 CrossRef CAS.
- D. Lavogina, E. Enkvist and A. Uri, ChemMedChem, 2010, 5, 23–34 CrossRef CAS PubMed.
- K. J. Cox, C. D. Shomin and I. Ghosh, Future Med. Chem., 2011, 3, 29–43 CrossRef CAS PubMed.
- S. Lee, J. Kim, J. Jo, J. W. Chang, J. Sim and H. Yun, Eur. J. Med. Chem., 2021, 216, 113318 CrossRef CAS.
- R. F. Lera, G. K. Potts, A. Suzuki, J. M. Johnson, E. D. Salmon, J. J. Coon and M. E. Burkard, Nat. Chem. Biol., 2016, 12, 411–418 CrossRef CAS PubMed.
- K. S. Lee, T. R. Burke, Jr., J.-E. Park, J. K. Bang and E. Lee, Trends Pharmacol. Sci., 2015, 36, 858–877 CrossRef CAS PubMed.
- R. E. A. Gutteridge, M. A. Ndiaye, X. Liu and N. Ahmad, Mol. Cancer Ther., 2016, 15, 1427–1435 CrossRef CAS PubMed.
- J. E. Park, D. Hymel, T. R. Burke, Jr. and K. S. Lee, F1000 Res., 2017, 6, 1024, DOI:10.12688/f1000research.11398.1.
- V. Archambault, G. Lépine and D. Kachaner, Oncogene, 2015, 34, 4799–4807 CrossRef CAS PubMed.
- G. Manning, D. B. Whyte, R. Martinez, T. Hunter and S. Sudarsanam, Science, 2002, 298, 1912–1934 CrossRef CAS PubMed.
- S. Zitouni, C. Nabais, S. C. Jana, A. Guerrero and M. Bettencourt-Dias, Nat. Rev. Mol. Cell Biol., 2014, 15, 433–452 CrossRef CAS PubMed.
- A. Berg and T. Berg, ChemBioChem, 2016, 17, 650–656 CrossRef CAS PubMed.
- S.-M. Yun, T. Moulaei, D. Lim, J. K. Bang, J.-E. Park, S. R. Shenoy, F. Liu, Y. H. Kang, C. Liao, N.-K. Soung, S. Lee, D.-Y. Yoon, Y. Lim, D.-H. Lee, A. Otaka, E. Appella, J. B. McMahon, M. C. Nicklaus, T. R. Burke, Jr., M. B. Yaffe, A. Wlodawer and K. S. Lee, Nat. Struct. Mol. Biol., 2009, 16, 876–882 CrossRef CAS PubMed.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, M. Graber, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., Nat. Chem. Biol., 2011, 7, 595–601 CrossRef CAS PubMed.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, A. Scharow, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., ACS Chem. Biol., 2012, 7, 805–810 CrossRef CAS PubMed.
- F. Liu, J.-E. Park, W.-J. Qian, D. Lim, A. Scharow, T. Berg, M. B. Yaffe, K. S. Lee and T. R. Burke, Jr., ChemBioChem, 2012, 13, 1291–1296 CrossRef CAS PubMed.
- X. Z. Zhao, D. Hymel and T. R. Burke, Jr., Bioorg. Med. Chem. Lett., 2016, 26, 5009–5012 CrossRef CAS PubMed.
- X. Z. Zhao, D. Hymel and T. R. Burke, Jr., Bioorg. Med. Chem., 2017, 25, 5041–5049 CrossRef CAS PubMed.
- W.-J. Qian, J.-E. Park, K. S. Lee and T. R. Burke, Jr., Bioorg. Med. Chem. Lett., 2012, 22, 7306–7308 CrossRef CAS PubMed.
- J. Xu, C. Shen, T. Wang and J. Quan, Nat. Struct. Mol. Biol., 2013, 20, 1047–1053 CrossRef CAS PubMed.
- M. Steegmaier, M. Hoffmann, A. Baum, P. Lenart, M. Petronczki, M. Krssak, U. Guertler, P. Garin-Chesa, S. Lieb, J. Quant, M. Grauert, G. R. Adolf, N. Kraut, J.-M. Peters and W. J. Rettig, Curr. Biol., 2007, 17, 316–322 CrossRef CAS PubMed.
- K. Tsuji, D. Hymel and T. R. Burke, Jr., Anal. Methods, 2020, 12, 4418–4421 RSC.
- A. E. Elia, P. Rellos, L. F. Haire, J. W. Chao, F. J. Ivins, K. Hoepker, D. Mohammad, L. C. Cantley, S. J. Smerdon and M. B. Yaffe, Cell, 2003, 115, 83–95 CrossRef CAS PubMed.
- S.-B. Shin, S.-U. Woo, Y.-J. Lee and H. Yim, Anticancer Res., 2017, 37, 1177–1183 CrossRef CAS PubMed.
- A. Scharow, D. Knappe, W. Reindl, R. Hoffmann and T. Berg, ChemBioChem, 2016, 17, 759–767 CrossRef CAS PubMed.
- L. Chen, J. L. Yap, M. Yoshioka, M. E. Lanning, R. N. Fountain, M. Raje, J. A. Scheenstra, J. W. Strovel and S. Fletcher, ACS Med. Chem. Lett., 2015, 6, 764–769 CrossRef CAS PubMed.
- H. Ruan, J. Kiselar, W. Zhang, S. Li, R. Xiong, Y. Liu, S. Yang and L. Lai, Phys. Chem. Chem. Phys., 2020, 22, 27581–27589 RSC.
- https://opnme.com/molecules/plk1-bi-2536 .
- K. S. Lee, J.-E. Park, Y. H. Kang, T.-S. Kim and J. K. Bang, Mol. Cells, 2014, 37, 286–294 CrossRef PubMed.
- S. E. Miller, K. Tsuji, R. P. M. Abrams, T. R. Burke and J. P. Schneider, J. Am. Chem. Soc., 2020, 142, 19950–19955 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cb00153e |
| ‡ Preliminary accounts of the work has been reported: Tsuji, K.; Hymel, D.; Burke, T. R., Jr. Exploration of inhibitors targeting intramolecular protein–protein interaction.” 25th American Peptide Symposium, Whistler, Canada (June 17-22, 2017) poster P229 and Tsuji, K.; Hymel, D.; Ma, B.; Nussinov, R.; T. R. Burke, J. Unexpected findings in the development of bivalent polo-like kinase 1-binding ligands with potentially important implications. 26th American Peptide Symposium, Monterey, CA (June 22–27, 2019) poster YI-P261. |
| § Current address: Discovery Chemistry, Novo Nordisk Research Center Seattle, Seattle, WA 98109, USA. |
| ¶ Current address: Engineering Research Center of Cell & Therapeutic Antibody (MOE), School of Pharmacy, Shanghai Jiaotong University, Shanghai, 200240, China. |
| This journal is © The Royal Society of Chemistry 2022 |