Detection and structural analysis of pyrimidine-derived radicals generated on DNA using a profluorescent nitroxide probe†
Kosho
Yamauchi
a,
Yuta
Matsuoka
a,
Masatomo
Takahashi
b,
Yoshihiro
Izumi
 b,
Hideto
Naka
a,
Yosuke
Taniguchi
b,
Hideto
Naka
a,
Yosuke
Taniguchi
 c,
Kazuaki
Kawai
d,
Takeshi
Bamba
c,
Kazuaki
Kawai
d,
Takeshi
Bamba
 b and
Ken-ichi
Yamada
b and
Ken-ichi
Yamada
 *a
*a
aPhysical Chemistry for Life Science Laboratory, Faculty of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi Higashi-ku, Fukuoka 812-8582, Japan. E-mail: kenyamada@phar.kyushu-u.ac.jp; Fax: +81-92-642-6626; Tel: +81-92-642-6624
bMetabolomics Laboratory, Research Center for Transomics Medicine, Medical Institute of Bioregulation, Kyushu University, 3-1-1 Maidashi Higashi-ku, Fukuoka 812-8582, Japan
cFrontier in Biofunction of Nucleic Acid and Organic Chemistry, Faculty of Pharmaceutical Sciences, Kyushu University, 3-1-1 Maidashi Higashi-ku, Fukuoka 812-8582, Japan
dDepartment of Environmental Oncology, Institute of Industrial Ecological Sciences, University of Occupational and Environmental Health, 1-1 Iseigaoka, Yahatanishi-ku, Kitakyushu, 807-8555, Japan
First published on 2nd December 2021
Abstract
The oxidative damage of DNA is associated with aging and the development of various diseases. Although nucleoside-derived radicals play an important role in DNA oxidation, their analysis methods are limited. Herein, we propose a fluorometric detection and structural analysis of radicals on the surface of oxidatively damaged DNA using a profluorescent nitroxide probe combined with liquid chromatography–fluorometry and high-resolution tandem mass spectrometry.
The oxidative damage of DNA is associated with aging and the development of various diseases, such as cancer and neurodegenerative disorders.1,2 The basic moieties in nucleosides are highly susceptible to oxidation by reactive oxygen species or ionizing radiation, resulting in the formation of nucleoside-derived radicals, which are precursors of well-known oxidized nucleosides such as 8-oxoguanine (oxoG), thymine glycol (Tg), and 5-formyluracil (formylU).3,4 The nucleoside-derived radicals can initiate and propagate oxidative damage among nearby nucleosides in DNA and induce DNA-duplication mutations.5 Thus, the detection and identification of these radicals is essential to understand DNA oxidation mechanisms.
The spin-trapping method is the only approach to analyze nucleoside- and DNA-derived radicals.6,7 The reaction adducts between spin-trapping agents and radical species can be measured by electron spin resonance spectroscopy, mass spectrometry, or immunochemical detection. However, owing to the low reactivity of spin-trapping agents toward radical species (105–106 M−1 s−1),8,9 research on the structural analysis of nucleoside-derived radicals and the number of identified radicals is limited. Therefore, a more sensitive method to detect DNA radicals is required to elucidate the detailed mechanism of DNA oxidation.
In contrast, piperidine nitroxides are highly reactive toward carbon-centered radicals (108–109 M−1 s−1),10 which inspired us to develop a fluorescent detection probe for a lipid-derived radical, 2,2,6-trimethyl-6-pentyl-4-(4-nitrobenzo[1,2,5]oxadiazol-7-ylamino)piperidine-1-oxyl (NBD-Pen).11 Furthermore, the ability of NBD-Pen to form adducts with lipid-derived carbon-centered radicals enabled the structure of radical species to be analyzed by liquid chromatography–fluorometry and high-resolution tandem mass spectrometry (LC/FL/HRMS/MS). As a result, we identified 132 lipid-derived radicals, including 111 new species.12 Because the formation of carbon-centered radicals occurs in both lipids and DNA, we concluded that nucleoside-derived radicals generated in the DNA strand could also be analyzed using a profluorescent nitroxide probe combined with LC/FL/HRMS/MS.
In this study, we developed a method to detect and analyze nucleoside-derived radicals using a profluorescent nitroxide probe and an LC/FL/HRMS/MS system. First, we examined the structure of the radical species generated via oxidation of individual nucleosides. Subsequently, we analyzed the radicals generated from calf thymus DNA.
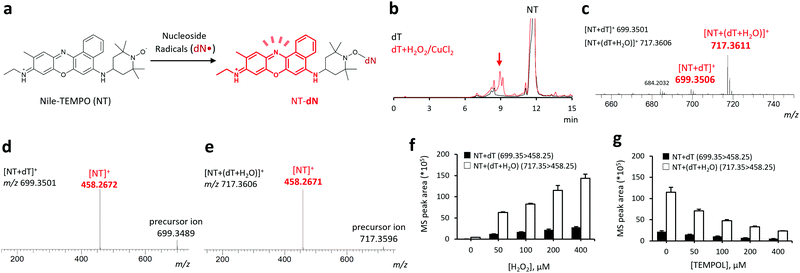
Both nucleosides and DNA are water-soluble molecules. Therefore, we were able to use our previously developed profluorescent nitroxide probe, 9-(ethylimino)-N-(2,2,6,6-tetramethylpiperidin-1-oxyl)-10-methyl-9H-benzo[a]phenoxazin-5-amine hydrochloride (Nile-TEMPO; NT),13 containing Nile blue as a hydrophilic fluorophore. Typically, NT reacts with free radicals to form fluorescent adducts (Fig. 1a).
First, we examined the ability of NT to capture nucleoside-derived radicals and form reaction adducts, using 2-deoxythymidine (dT). The dT radical is formed via either hydrogen atom abstraction or radical addition by the hydroxyl radical.3,14 Oxidation of dT using the H2O2 + CuCl2 mixture in the presence of NT led to the formation of multiple new peaks at tR = 7–10 min observed in the fluorescence chromatogram (Fig. 1b). Major signals at m/z = 699.3506 and 717.3611, corresponding to protonated molecular ions of NT + dT and NT + (dT + H2O) adducts (calculated m/z: 699.3501 for [NT + dT]+ and 717.3606 for [NT + (dT + H2O)]+), were observed in the HRMS analysis of the fraction eluted at tR = 7–10 min, as shown in Fig. 1c. These ions were further analyzed by LC/HRMS/MS that detected a common fragment (m/z = 458.267), consistent with the NT moiety (calculated m/z = 458.2671, Fig. 1d). Furthermore, MS/MS analysis confirmed that the addition of H2O2 increased the peak area of each adduct in a concentration-dependent manner (Fig. 1f) and that this increase can be inhibited by the co-addition of 2,2,6,6-tetramethyl-4-hydroxypiperidine-1-oxyl (TEMPOL, Fig. 1g). Additionally, LC/FL/HRMS/MS analysis allowed us to detect and structurally analyze two dT-derived radicals, ˙dT + O and ˙dT + H2O2 (ESI,† Fig. S1), the presence of which suggests that NT can efficiently react with dT-derived radicals. Furthermore, when we used a conventional radical trapping probe, 3,4-dihydro-2,2-dimethyl-2H-pyrrole 1-oxide (DMPO),6,7 instead of NT and conducted experiments under the same conditions employed in this study, the amounts of several probe-radical adducts were smaller than those obtained using NT as a radical trapping probe (ESI,† Fig. S2).
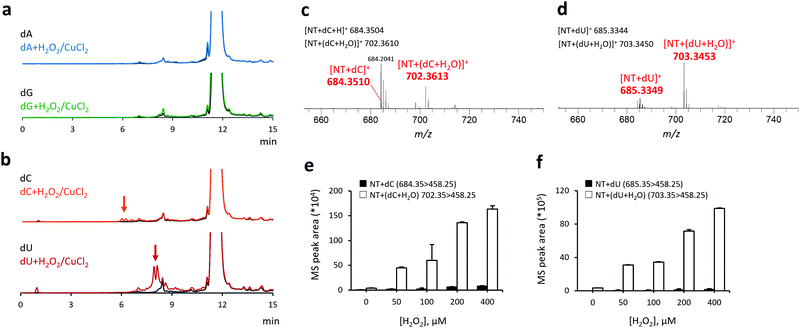
Next, we investigated the reactivity of NT toward other nucleoside-derived radical species (Fig. 2), including 2-deoxyadenosine (dA), 2-deoxyguanosine (dG), 2-deoxycytidine (dC), and 2-deoxyuridine (dU). No fluorescence enhancement was observed in the reaction of purine nucleosides (dA and dG) with H2O2 + CuCl2 + NT (Fig. 2a). Conversely, the reaction of pyrimidine nucleosides (dC and dU) with H2O2 + CuCl2 and NT generated multiple fluorescence peaks at tR: 6–8 and 7–10 min, respectively (Fig. 2b). The average HRMS spectral peak areas for each fraction (eluted at the aforementioned retention times) are shown in Fig. 2c and d. In both cases, we observed the HRMS peaks derived from the reaction adducts with non-oxygenated (calculated m/z: 684.3504 for [NT + dC]+ and 685.3344 for [NT + dU]+) and oxygenated radicals (calculated m/z: 702.3610 for [NT + (dC + H2O)]+ and 703.3450 for [NT + (dU + H2O)]+). Analogous to the results obtained with dT, the MS/MS peak areas of dC and dU adducts increased with the addition of H2O2 in a concentration-dependent manner (Fig. 2e and f), indicating that NT can selectively react with pyrimidine-, but not purine-derived radicals. It should be noted that the radicals of purine nucleosides have a lone electron at the nitrogen atom,5,6 whereas the pyrimidine nucleosides mainly form carbon-centered radicals (ESI,† Scheme S1). The lack of reactivity between NT and purine-derived radicals can be attributed to the low reactivity of these aminyl radicals toward nitroxyl radicals, rendering them more difficult to capture by NT. Ultimately, we identified nine pyrimidine-derived radicals, seven of which (highlighted in ESI,† Table S1) have not been reported previously.15
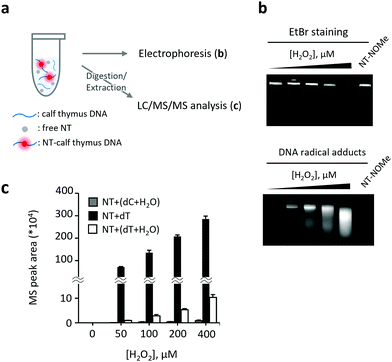
After confirming the applicability of the analytical method to pure nucleosides, we proceeded to the detection and structural analysis of DNA-derived radicals (Fig. 3). In this experiment, calf thymus DNA was oxidized by H2O2 + CuCl2 in the presence of NT. The DNA was first separated by agarose gel electrophoresis and then the intensity of fluorescence was measured in the separated fractions. Additionally, the NT-radical adducts were analyzed by LC/MS/MS following DNA digestion (Fig. 3a). Interestingly, the DNA bands stained with ethidium bromide (EtBr) moved faster on the agarose gel after the addition of H2O2 + CuCl2 to the sample, possibly due to the oxidative fragmentation of DNA strands. The fluorescent bands derived from NT-radical adducts had a comparable mobility on the agarose gel as DNA bands, and their intensities increased with H2O2 in a concentration-dependent manner (Fig. 3b). In contrast, when we replaced NT with (9-(ethylimino)-N-(1-methoxy-2,2,6,6-tetramethylpiperidin-1-oxyl)-10-methyl-9H-benzo[a]phenoxazin-5-amine hydrochloride (NT-NOMe), which does not have the capacity to capture free radicals, as a fluorescent probe, the fluorescent bands derived from radical adducts were not observed, suggesting that NT is capable of capturing DNA-derived radicals generated at oxidatively damaged sites. Subsequently, we analyzed the structures of DNA-generated nucleoside-derived radicals and observed an increase in the level of three pyrimidine-derived reaction adducts, namely, ˙dC + H2O, ˙dT, and ˙dT + H2O (Fig. 3c). Among them, the MS/MS peak area of NT–dT showed the most significant increase.
The C5-methyl group of dT, protruding into the major groove of the DNA strand, is highly susceptible to hydrogen atom abstraction by hydroxyl radicals, resulting in the selective formation of ˙dT. Therefore, we concluded that NT selectively captures ˙dT in the sample of oxidized DNA. Notably, oxidation of monomeric dT using H2O2 + CuCl2 generates a large amount of ˙dT + H2O, suggesting that the reaction conditions strongly affect the ability of hydroxyl radicals to access various reaction sites (Fig. 1 and ESI,† Scheme S1a) and profiles of nucleoside radicals in general. Based on these results, we concluded that our method is suitable for fluorometric detection and structural analysis of pyrimidine-derived radicals, especially ˙dT, generated on the surface of oxidatively damaged DNA. This method represents a valuable tool to elucidate the oxidation mechanisms in DNA.
The oxidation of dT plays a major role in the oxidative damage of DNA, accounting for approximately 20% of the total oxidative damage caused by ionizing radiation.16 Robert et al. reported that ˙dT can initiate and propagate a radical chain reaction on DNA strands, resulting in the formation of oxoG.17 While methods to generate ˙dT in the DNA strands using photolabile derivatives are known,17,18 only a few methods are available for the analysis of nucleoside-derived radicals. Therefore, our newly developed method, which allowed the identification of seven novel pyrimidine-derived radicals and is applicable to the oxidatively damaged DNA, will help the role of dT-derived radicals to be clarified in future studies. To further verify the validity of our method, additional studies, using a probe with improved sensitivity, should be carried out in cultured cells and animal models.
In the present study, we developed a method to analyze pyrimidine nucleoside-derived radicals and identified nine radical species, including seven new radical species. In addition, we succeeded in the fluorometric detection and structural analysis of radicals generated on the oxidatively damaged DNA. DNA oxidation is known to be associated with the development of various diseases, but its oxidation mechanism is still unclear. One of the major limitations in elucidating its mechanism is the lack of analytical methods for DNA-derived radicals and critical molecules to initiate and propagate DNA oxidation. Therefore, in future studies, the developed analytical system will be a valuable tool to reveal the detailed mechanism of DNA oxidation and its significance in pathological events.
This work was partially supported by the AMED-CREST grant (JP21gm0910013 to K. Y.), and the JSPS KAKENHI grants (18K14884 to Y. M.; 17H03977, 18K19405, and 20H00493 to K. Y.; 17H06304 to Y. I. and T. B.; and 17H06299 to T. B.), Japan. This work was also supported by the Platform Project for Supporting Drug Discovery and Life Science Research from AMED. We appreciate technical support provided by the Research Support Center of the Graduate School of Medical Sciences, Kyushu University.
Conflicts of interest
There are no conflicts to declare.References
- Y. Xie, H. Yang, C. Cunanan, K. Okamoto, D. Shibata, J. Pan, D. E. Barnes, T. Lindahl, M. McIlhatton, R. Fishel and J. H. Miller, Cancer Res., 2004, 64, 3096–3102 CrossRef CAS PubMed.
- S. P. Gabbita, M. A. Lovell and W. R. Markesbery, J. Neurochem., 1998, 71, 2034–2040 CrossRef CAS PubMed.
- J. Cadet, T. Douki and J. L. Ravanat, Free Radical Biol. Med., 2010, 49, 9–21 CrossRef CAS PubMed.
- M. Dizdaroglu, P. Jaruga, M. Birincioglu and H. Rodriguez, Free Radical Biol. Med., 2002, 32, 1102–1115 CrossRef CAS PubMed.
- L. Zheng and M. M. Greenberg, J. Am. Chem. Soc., 2018, 140, 6400–6407 CrossRef CAS PubMed.
- S. Bhattacharjee, L. J. Deterding, S. Chatterjee, J. Jiang, M. Ehrenshaft, O. Lardinois, D. C. Ramirez, K. B. Tomer and R. P. Mason, Free Radical Biol. Med., 2011, 50, 1536–1545 CrossRef CAS PubMed.
- D. C. Ramirez, S. E. Mejiba and R. P. Mason, Nat. Methods, 2006, 3, 123–127 CrossRef CAS PubMed.
- P. Schmid and K. U. Ingold, J. Am. Chem. Soc., 1978, 100, 2493–2500 CrossRef CAS.
- Y. Maeda and K. U. Ingold, J. Am. Chem. Soc., 1979, 101, 4975–4981 CrossRef CAS.
- E. G. Bagryanskaya and S. R. Marque, Chem. Rev., 2014, 114, 5011–5056 CrossRef CAS PubMed.
- K. Yamada, F. Mito, Y. Matsuoka, S. Ide, K. Shikimachi, A. Fujiki, D. Kusakabe, Y. Ishida, M. Enoki, A. Tada, M. Ariyoshi, T. Yamasaki and M. Yamato, Nat. Chem. Biol., 2016, 12, 608–613 CrossRef CAS PubMed.
- Y. Matsuoka, Y. Izumi, M. Takahashi, T. Bamba and K. I. Yamada, Anal. Chem., 2020, 92, 6993–7002 CrossRef CAS PubMed.
- Y. Matsuoka, K. Ohkubo, T. Yamasaki, M. Yamato, H. Ohtabu, T. Shirouzu, S. Fukuzumi and K. I. Yamada, RSC Adv., 2016, 6, 60907–60915 RSC.
- M. Dizdaroglu and P. Jaruga, Free Radical Res., 2012, 46, 382–419 CrossRef CAS PubMed.
- V. Maurel, J. L. Ravanat and S. Gambarelli, Rapid Commun. Mass Spectrom., 2006, 20, 2235–2242 CrossRef CAS PubMed.
- S. Frelon, T. Douki, J. L. Ravanat, J. P. Pouget, C. Tornabene and J. Cadet, Chem. Res. Toxicol., 2000, 13, 1002–1010 Search PubMed.
- G. Robert and J. R. Wagner, Chem. Res. Toxicol., 2020, 33, 565–575 Search PubMed.
- G. Lin and L. Li, Angew. Chem., Int. Ed., 2013, 52, 5594–5598 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc04998d |
| This journal is © The Royal Society of Chemistry 2022 |