Novel peroxosolvates of tetraalkylammonium halides: the first case of layers containing hydrogen-bonded peroxide molecules†
Mger A.
Navasardyan
 ,
Stanislav I.
Bezzubov
,
Stanislav I.
Bezzubov
 ,
Alexander G.
Medvedev
,
Alexander G.
Medvedev
 ,
Petr V.
Prikhodchenko
,
Petr V.
Prikhodchenko
 and
Andrei V.
Churakov
and
Andrei V.
Churakov
 *
*
Kurnakov Institute of General and Inorganic Chemistry, Russian Academy of Sciences, Leninskii Prosp. 31, 119991 Moscow, Russia. E-mail: churakov@igic.ras.ru; Fax: +7 495 9541279
First published on 2nd December 2021
Abstract
Novel peroxosolvates of tetraalkylammonium halides Et4N+Cl−·2(H2O2) (1), Et4N+Br−·2(H2O2) (2), Me3(ClCH2CH2)N+Cl−·H2O2 (3) and Me3PhN+Cl−·H2O2 (4) were prepared from concentrated hydrogen peroxide and the corresponding structures were determined by X-ray crystallography. Structures 1 and 2 are formed by globose Et4N+ cations and are the first examples of layers containing H-bonded peroxide molecules. By contrast, 3 and 4 are formed by sufficiently aspherical Me3(ClCH2CH2)N+ and Me3PhN+ units and contain infinitely long hydrogen-bonded peroxide–halide chains.
Introduction
Peroxosolvates (crystalline adducts of hydrogen peroxide) were first discovered by Tanatar in the early 20th century.1 Two of these compounds, namely peroxosolvates of urea and sodium carbonate, have become widespread as bleaching and disinfection agents for industrial, housekeeping and pharmaceutical use and current production reaches up to several hundred tons a year.2 The main research direction for peroxosolvates during the twentieth century has been the preparation of novel stable hydrogen peroxide carriers.3–6 It has been proposed that highly stable peroxosolvates could be designed by maximizing the number of hydrogen bonds in structures.7 Several quite stable organic peroxosolvates were thus produced in which peroxide molecules form the largest possible number of hydrogen bonds – six (corresponding to 2 donor bonds and 4 acceptor bonds).8Over the last decade several entirely new perspective approaches in peroxosolvate chemistry have been developed. A) Hydrogen peroxide adducts of some organic compounds have been proposed as mild, selective and stoichiometric oxidizing agents for organic synthesis.9,10 B) Several peroxosolvates of highly energetic materials have been prepared in an attempt to enhance properties like oxygen balance, and detonation velocities and pressures.11–13 C) Drug–drug cocrystals containing hydrogen peroxide have been developed because H2O2 exhibits wide spectrum of antimicrobial activity.14 Recently, a promising peroxosolvate of antifungal drug miconazole was structurally characterized.15 D) Particular attention has been given to H2O2 cocrystals containing hydrogen-bonded peroxide and peroxide–water clusters8 due to the importance of hydrogen peroxide as a signalling biological molecule that undergoes cooperative transport across cellular membranes.16,17 Recently, large 0D-hydrogen peroxide insular dodecameric and pentameric clusters have been reported in the structures of 2-aminonicotinic acid and lidocaine N-oxide peroxosolvates, and it appears that no special conditions are required for hydrogen peroxide cluster formation.18 Structures containing 1D-chains of H-bonded peroxide19–22 and water–peroxide23,24 have been intensively studied. Peroxide–halide and peroxide–carbonate chains have also been investigated.25,26 To the best of our knowledge, peroxosolvates with hydrogen-bonded 2D and 3D peroxide motifs have not been reported to date.
Results and discussion
Recently, the usage of organic coformers without donor H-atoms has been suggested for the targeted formation of peroxosolvates with an infinite 1D-hydrogen peroxide clusters.27 It has been shown that hydrogen bonds in Hal−⋯HO2H units can act as templates for the supramolecular organization of tetraphenylphosphonium halide peroxosolvates.25 Thus tetraalkylammonium salts appear to have good prospects due to their availability and multiplicity. Herein we report four novel structures of peroxosolvates of tetraalkylammonium salts.Structures 1–4 comprise Et4N+ or Me3RN+ cations (R = ClCH2CH2–, and Ph), halide counterions and one or two crystallographically independent hydrogen peroxide molecules (Fig. 1–3). The R4N+ cations adopt an ordinary tetrahedral geometry.28 The halide anions and all the peroxide molecules occupy general positions. The O–O distances lie within normal range for solvent H2O2 molecules (1.431(2)–1.460(1) Å).22 The torsion H–O–O–H angles (94(2)–159(5)°) are also typical for hydrogen peroxide molecules without imposed crystallographic symmetry.8
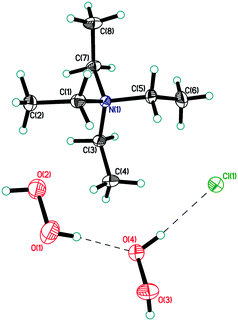 | ||
| Fig. 1 Asymmetric unit in structure 1. Displacement ellipsoids are shown at the 50% probability level. Hydrogen bonds are shown by dashed lines. | ||
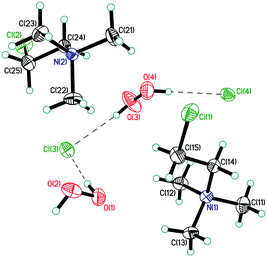 | ||
| Fig. 2 Asymmetric unit in structure 3. Displacement ellipsoids are shown at the 50% probability level. Hydrogen bonds are shown by dashed lines. | ||
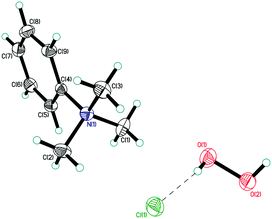 | ||
| Fig. 3 Asymmetric unit in structure 4. Displacement ellipsoids are shown at the 50% probability level. Hydrogen bonds are shown by dashed lines. | ||
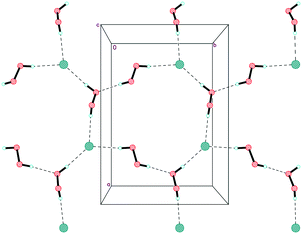
In structure 1, the peroxide molecule H1–O1–O2–H2 forms two moderate donor hydrogen bonds with chlorine anions and a neighbouring peroxide molecule H3–O3–O4–H4 (Fig. S1 in ESI,†Table 1). H3–O3–O4–H4 forms three almost linear hydrogen bonds (2 donor bonds and 1 acceptor bond) (Fig. S2 in ESI†). As a result, unprecedented 2D-layers containing hydrogen-bonded peroxides are formed (Fig. 4 and 5). Within these layers, chloride anions participate in three hydrogen bonds for a total Cl−/H2O2 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Note that only 1D peroxide–halide clusters of ⋯Cl−⋯(H2O2)n⋯Cl−⋯ (n = 1, 2) are reported to date.25 Of interest, exactly the same 2D clusters of water⋯Cl− are well-known.29 According to Infantes–Motherwell notation, they belong to L8(6) extended motif.30 By contrast to structure 1, these 2D water⋯halogen clusters are usually involved in additional hydrogen bonding with the cation layers.31 Moreover, the similar N–H⋯Hal− bonded layers were reported for some simple amine hydrohalides, namely anilinium chloride,32 cyclohexylammonium chloride,33 and 1-decylammonium bromide.34
2. Note that only 1D peroxide–halide clusters of ⋯Cl−⋯(H2O2)n⋯Cl−⋯ (n = 1, 2) are reported to date.25 Of interest, exactly the same 2D clusters of water⋯Cl− are well-known.29 According to Infantes–Motherwell notation, they belong to L8(6) extended motif.30 By contrast to structure 1, these 2D water⋯halogen clusters are usually involved in additional hydrogen bonding with the cation layers.31 Moreover, the similar N–H⋯Hal− bonded layers were reported for some simple amine hydrohalides, namely anilinium chloride,32 cyclohexylammonium chloride,33 and 1-decylammonium bromide.34
| d(O–H), Å | d(H⋯A), Å | d(O⋯A), Å | ∠(O–H⋯A), ° | ||
|---|---|---|---|---|---|
| O1–H1⋯O4 | 1 | 0.86(3) | 1.97(3) | 2.833(2) | 173(3) |
| 2 | 0.76(4) | 2.07(4) | 2.818(3) | 168(5) | |
| O2–H2⋯Cl1 | 1 | 0.89(3) | 2.26(3) | 3.1517(15) | 172(3) |
| Br1 | 2 | 0.84(4) | 2.48(4) | 3.2881(19) | 161(6) |
| O3–H3⋯Cl1 | 1 | 0.82(3) | 2.30(3) | 3.1179(13) | 170(3) |
| Br1 | 2 | 0.85(4) | 2.40(4) | 3.238(2) | 166(3) |
| O4–H4⋯Cl1 | 1 | 0.89(2) | 2.21(2) | 3.0951(12) | 173(3) |
| Br1 | 2 | 0.81(3) | 2.43(3) | 3.2353(18) | 175(3) |
| O1–H1⋯Cl3 | 3 | 0.826(18) | 2.223(19) | 3.0438(9) | 172.4(17) |
| O2–H2⋯Cl4 | 0.868(18) | 2.207(18) | 3.0659(9) | 170.1(14) | |
| O3–H3⋯Cl3 | 0.810(19) | 2.27(2) | 3.0710(10) | 169.2(17) | |
| O4–H4⋯Cl4 | 0.803(17) | 2.273(18) | 3.0768(9) | 180(2) | |
| O1–H1⋯Cl1 | 4 | 0.873(19) | 2.197(19) | 3.0548(11) | 167.3(16) |
| O2–H2⋯Cl1 | 0.87(2) | 2.22(2) | 3.0733(10) | 169.3(18) |
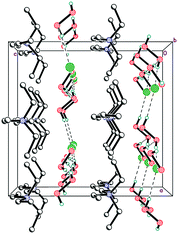
As expected, Et4N+ cations also form layers perpendicular to the c-axis. The shortest interaction separations (7.24 and 7.31 Å) for these layers are observed along both ab-diagonals (Fig. 6). The globose shape of the Et4N+ cations causes the aforementioned separations to be very close to each other. However, these separations are also very close to the halogen⋯halogen distances along the ab-diagonal within the Cl−⋯(H2O2)n⋯Cl− units (both of which are 7.28 Å). The adequate aforementioned bilateral separations appear to be the main cause of peroxide layered crystal packing. Bromide 2 is isomorphous to 1, with very similar H-bond parameters. However, Hal−⋯(H2O2)n⋯Hal− units are easily tunable25 due to the low barriers to rotation for peroxide molecules.35
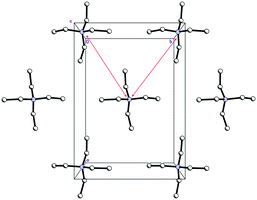 | ||
| Fig. 6 Cationic layers in the structure 1 (view along c-axis). The shortest interaction separations are shown by red narrows. | ||
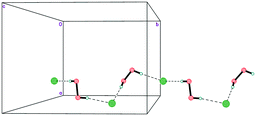
The mixed-substituted cation Me3(ClCH2CH2)N+ is essentially nonspherical (Fig. 2). Thus, it is highly unlikely that hydrogen-bonded peroxide layers form in the corresponding peroxosolvate structures. In fact, only 1D chains of ⋯Cl−⋯H2O2⋯Cl−⋯ were observed in the crystals of 3 (Fig. 7). Both peroxide molecules and both chlorine anions only form two donor hydrogen bonds. These bonds are somewhat shorter than those in 1, with O⋯Cl− distances in 3.0438(9)–3.0768(9) Å range.
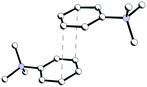
In structure 4, two Me3PhN+ cations combined into elongated centrosymmetric dimers via π⋯π interactions with an interplanar separation of 3.430 Å (Fig. 8). A CSD search showed that the same dimers were recently observed in the structure of 2(Me3PhN+)·(Cl122−).36 H-Bonded 1D chains of ⋯Cl−⋯H2O2⋯Cl−⋯ similar to those in 3 were found for peroxosolvate 4.
Conclusions
The structures of Et4N+Cl−·2(H2O2) and Et4N+Br−·2(H2O2) formed by globose Et4N+ cations are the first examples of peroxosolvates with 2D layers containing H-bonded peroxides. By contrast, the structures Me3(ClCH2CH2)N+Cl−·H2O2 and Me3PhN+Cl−·H2O2 formed by sufficiently aspherical Me3(ClCH2CH2)N+ and Me3PhN+ units contain infinitely long 1D hydrogen-bonded peroxide–halide chains. These compounds appear to be promising for applications because both components, hydrogen peroxide14 and Et4N+ salts,37 demonstrate high antimicrobial activity.Experimental
Tetraalkylammonium salts and 50% hydrogen peroxide were purchased from Aldrich. 94% hydrogen peroxide was prepared by an extraction method.38 Handling procedures for concentrated hydrogen peroxide are described in detail (danger of explosion!).39,40 Colourless crystals of 1–4 were obtained by cooling to −21 °C saturated solutions (rt) of respective anhydrous salts in 94% hydrogen peroxide. Unfortunately, all crystalline samples 1–4 were unstable without mother liquors and completely degrade in a few minutes when exposed to open air. The latter prevented carrying-out powder diffraction and DSC experiments.Experimental data sets were collected on a Bruker SMART APEX II diffractometer (for 1, 3 and 4) and Bruker D8 Venture machine for 2 using graphite monochromatized Mo-Kα radiation (λ = 0.71073 Å). Absorption corrections based on measurements of equivalent reflections were applied.41 The structures were solved by direct methods and refined by full matrix least-squares on F2 with anisotropic thermal parameters for all non-hydrogen atoms.42 All hydrogen atoms were found from difference Fourier synthesis and refined with isotropic thermal parameters. In all structures, partial substitutional disorder of hydrogen peroxide by water molecules43–46 was not observed since no residual peaks with an intensity more than 0.17 e A−3 were seen in the hydrogen peroxide molecule regions. Crystal data and details of X-ray analysis are given in Table S1.† The single crystal X-ray diffraction analysis were performed at the Centre of Shared Equipment of IGIC RAS within the State Assignment on Fundamental Research to the Kurnakov Institute of General and Inorganic Chemistry.
Author contributions
Mger A. Navasardyan – crystallization, data collection; Stanislav I. Bezzubov – data collection, visualization; Alexander G. Medvedev – crystallization, data collection; Petr V. Prikhodchenko – conceptualization, formal analysis; Andrei V. Churakov – funding acquisition; project administration, writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank RFBR for financial support (grants 18-29-19119 and 20-03-00449).Notes and references
- S. Tanatar, Zh. Russ. Fiz.-Khim. O-va., 1908, 40, 376 Search PubMed.
- H. Jakob, S. Leininger, T. Lehmann, S. Jacobi and S. Gutewort, Peroxo Compounds, Inorganic, Ullmann's encyclopedia of industrial chemistry, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2012, vol. 26 Search PubMed.
- T. Wagner-Jauregg, J. Am. Chem. Soc., 1952, 74, 1358 CrossRef CAS.
- J. M. Adams, R. G. Pritchard and J. M. Thomas, J. Chem. Soc., Chem. Commun., 1976, 358 RSC.
- D. Thierbach, F. Huber and H. Preut, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1980, 36, 974 CrossRef.
- A. Adam and M. Mehta, Angew. Chem., Int. Ed., 1998, 37, 1387 CrossRef CAS.
- J. M. Adams and V. Ramdas, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1978, 34, 2150 CrossRef.
- A. G. Medvedev, A. V. Churakov, P. V. Prikhodchenko, O. Lev and M. V. Vener, Molecules, 2021, 26, 26 CrossRef CAS PubMed.
- S. H. Ahn, K. J. Cluff, N. Bhuvanesh and J. Blümel, Angew. Chem., Int. Ed., 2015, 54, 13341 CrossRef CAS PubMed.
- G. Chehardoli and M. A. Zolfigol, Phosphorus, Sulfur Silicon Relat. Elem., 2010, 185, 193 CrossRef CAS.
- J. C. Bennion, N. Chowdhury, J. W. Kampf and A. J. Matzger, Angew. Chem., Int. Ed., 2016, 55, 1 CrossRef PubMed.
- R. A. Wiscons, M. Bellas, J. C. Bennion and A. J. Matzger, Cryst. Growth Des., 2018, 18, 7701 CrossRef CAS.
- J. Luo, H. Xia, W. Zhang, S. Song and Q. Zhang, J. Mater. Chem. A, 2020, 8, 12334 RSC.
- E. Linley, S. P. Denyer, G. McDonnell, C. Simons and J. Y. Maillard, J. Antimicrob. Chemother., 2012, 67, 1589 CrossRef CAS PubMed.
- K. M. Kersten, M. E. Breen, A. K. Mapp and A. J. Matzger, Chem. Commun., 2018, 54, 9286 RSC.
- O. Rodrigues, G. Reshetnyak, A. Grondin, Y. Saijo, N. Leonhardt, C. Maurel and L. Verdoucq, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9200 CrossRef CAS PubMed.
- G. P. Bienert and F. Chaumont, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 1596 CrossRef CAS PubMed.
- D. A. Grishanov, M. A. Navasardyan, A. G. Medvedev, O. Lev, P. V. Prikhodchenko and A. V. Churakov, Angew. Chem., Int. Ed., 2017, 56, 15241 CrossRef CAS PubMed.
- M. A. Navasardyan, D. A. Grishanov, P. V. Prikhodchenko and A. V. Churakov, Acta Crystallogr., Sect. E: Crystallogr. Commun., 2020, 76, 1331 CrossRef CAS PubMed.
- M. A. Navasardyan, D. A. Grishanov, T. A. Tripol'skaya, L. G. Kuzmina, P. V. Prikhodchenko and A. V. Churakov, CrystEngComm, 2018, 20, 7413 RSC.
- P. V. Prikhodchenko, A. G. Medvedev, T. A. Tripol'skaya, A. V. Churakov, Y. Wolanov, J. A. K. Howard and O. Lev, CrystEngComm, 2011, 13, 2399 RSC.
- I. Yu. Chernyshov, M. V. Vener, P. V. Prikhodchenko, A. G. Medvedev, O. Lev and A. V. Churakov, Cryst. Growth Des., 2017, 17, 214 CrossRef CAS.
- A. V. Churakov and J. A. K. Howard, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2007, 63, o4483 CrossRef CAS.
- G. Laus, V. Kahlenberg, K. Wurst, T. Lörting and H. Schottenberger, CrystEngComm, 2008, 10, 1638 RSC.
- A. V. Churakov, P. V. Prikhodchenko and J. A. K. Howard, CrystEngComm, 2005, 7, 664 RSC.
- A. G. Medvedev, A. A. Mikhaylov, A. V. Churakov, P. V. Prikhodchenko and O. Lev, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2012, 68, i20 CrossRef CAS PubMed.
- M. A. Navasardyan, S. I. Bezzubov, L. G. Kuz'mina, P. V. Prikhodchenko and A. V. Churakov, Acta Crystallogr., Sect. E: Crystallogr. Commun., 2017, 73, 1793 CrossRef CAS PubMed.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171 CrossRef CAS PubMed.
- L. Infantes, J. Chisholm and S. Motherwell, CrystEngComm, 2003, 5, 480 RSC.
- L. Infantes and S. Motherwell, CrystEngComm, 2002, 4, 454 RSC.
- H. Bock, H. Schödel, T. T. H. Van, R. Dienelt and M. Gluth, J. Prakt. Chem./Chem.-Ztg., 1998, 340, 722 CrossRef CAS.
- E. López-Duplá, P. G. Jones and F. Vancea, Z. Naturforsch., B: J. Chem. Sci., 2003, 58, 191 CrossRef.
- S. T. Rao and M. Sundaralingam, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1969, 25, 2509 CrossRef CAS.
- L.-J. Zhang, Y.-Y. Di and D.-F. Lu, J. Chem. Thermodyn., 2011, 43, 1591 CrossRef CAS.
- R. H. Hunt, R. A. Leacock, W. Peters and K. T. Hecht, J. Chem. Phys., 1965, 42, 1931 CrossRef CAS.
- K. Sonnenberg, P. Pröhm, N. Schwarze, C. Müller, H. Beckers and S. Riedel, Angew. Chem., Int. Ed., 2018, 57, 9136 CrossRef CAS PubMed.
- F. M. Dolgushin, A. S. Goloveshkin, I. V. Ananyev, S. V. Osintseva, Yu. V. Torubaev, S. S. Krylov and A. S. Golub, Acta Crystallogr., Sect. C: Struct. Chem., 2019, 75, 402 CrossRef CAS PubMed.
- Y. Wolanov, O. Lev, A. V. Churakov, A. G. Medvedev, V. M. Novotortsev and P. V. Prikhodchenko, Tetrahedron, 2010, 66, 5130 CrossRef CAS.
- W. C. Schumb, C. N. Satterfield and R. P. Wentworth, Hydrogen peroxide, Reinhold publishing corp., New York, 1955 Search PubMed.
- O. Maass and W. H. Hatcher, J. Am. Chem. Soc., 1920, 42, 2569 CrossRef CAS.
- G. M. Sheldrick, SADABS. Program for scaling and correction of area detector data, University of Göttingen, Germany, 1997.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
- B. F. Pedersen, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1972, 28, 746 CrossRef CAS.
- B. F. Pedersen, Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem., 1972, 28, 1014 CrossRef CAS.
- G. Laus, V. Kahlenberg, K. Wurst, T. Lörting and H. Schottenberger, CrystEngComm, 2008, 10, 1638 RSC.
- A. V. Churakov, P. V. Prikhodchenko and J. A. K. Howard, CrystEngComm, 2005, 7, 664 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1. Crystal data, data collection and refinement parameters for 1–4. Fig. S1 and S2. CCDC 2116891 (1), 2116893 (2), 2116892 (3) and 2116894 (4). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ce01476e |
| This journal is © The Royal Society of Chemistry 2022 |