Microsecond molecular dynamics studies of cholesterol-mediated myelin sheath degeneration in early Alzheimer's disease†
Mayuri
Gupta
 a and
Donald F.
Weaver
a and
Donald F.
Weaver
 *ab
*ab
aKrembil Research Institute, University Health Network, 60 Leonard Avenue, Toronto, M5T 0S8, Canada. E-mail: donald.weaver@uhnresearch.ca
bDepartments of Chemistry, Medicine and Pharmaceutical Sciences, University of Toronto, Toronto, ON M55 3H6, Canada
First published on 1st December 2021
Abstract
Cholesterol-mediated perturbations of membrane structural integrity are key early events in the molecular pathogenesis of Alzheimer's disease (AD). In AD, protein misfolding (proteopathy) and pro-inflammatory conditions (immunopathy) culminate in neuronal death, a process enabled by altered membrane biophysical properties which render neurons more susceptible to proteopathic and immunopathic cytotoxicities. Since cholesterol is a principal neuronal membrane lipid, normal cholesterol homeostasis is central to membrane health; also, since increased cholesterol composition is especially present in neuronal myelin sheath (i.e. brain “white matter”), recent studies have not surprisingly revealed that white matter atrophy precedes the conventional biomarkers of AD (amyloid plaques, tau tangles). Employing extensive microsecond all-atom molecular dynamics simulations, we investigated biophysical and mechanical properties of myelin sheath membrane as a function of cholesterol mole fraction (χCHL). Impaired χCHL modulates multiple bilayer properties, including surface area per lipid (APL), chain order, number and mass density profiles, area compressibility and bending moduli, bilayer thickness, lipid tilt angles, H-bonding interactions and tail interdigitation. The increased orientational ordering of both palmitoyl and oleoyl chains in model healthy myelin sheath (HMS) membranes illustrates the condensing effect of cholesterol. With an increase in χCHL, number density profiles of water tend to attain bulk water number density more quickly, indicating shrinkage in the interfacial region with increasing χCHL. The average tilt value is 11.5° for the C10–C13 angle in cholesterol and 64.2° for the P–N angle in POPC lipids in HMS. These calculations provide a molecular-level understanding of myelin sheath susceptibility to pathology as an early event in the pathogenesis of AD.
1. Introduction
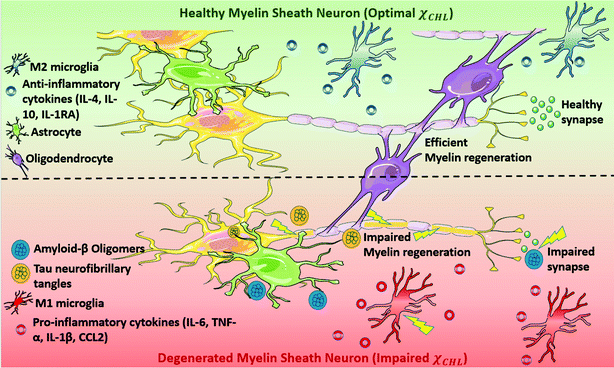
Alzheimer's disease (AD) is a devastating neurodegenerative disorder traditionally characterized by the accumulation of extracellular β-amyloid (Aβ) plaques and intracellular hyperphosphorylated tau protein neurofibrillary tangles.1 The underlying mechanisms of AD-related cellular damage and cognitive decline remain unelucidated despite decades of research.2,3 In recent years, increasing evidence has suggested that demyelination of neurons occurs very early in the pathogenesis of AD, even before plaque and tangle deposition.4–8 Understanding the biomolecular mechanism of this demyelination may provide insights regarding new druggable targets for AD.Myelin is a multilamellar membrane structure that functions as an electrical insulator surrounding the neuronal axon in the white matter of the central nervous system (CNS); it is critical for efficient and fast conduction of nerve impulses.9,10 Myelin sheath homeostasis in brain is preserved by the coordinated working of oligodendrocyte progenitor cells (OPCs), microglia and astrocytes. Microglia phagocytose myelin debris and secrete signaling factors to promote the differentiation of OPCs to oligodendrocytes, whose main function is to generate myelin.11,12 Astrocytes regulate myelination by supporting proliferation and migration of oligodendrocytes. However, due to various pathological, genetic and environmental factors, myelin regeneration–degeneration homeostasis is disrupted early during the pathogenesis of AD; in turn, this activates astrocytes and transforms microglia into a senescence phenotype.13,14 Activation of M1-phenotype microglia triggers release of pro-inflammatory cytokines (e.g. IL6, IL10, IL1R1, TNF-α, IFNγ) and increases cellular stress thereby further activating the innate immune response. Oligodendrocytes, the myelin forming glial cells of the CNS, are damaged due to coupled proteopathic (plaques and tangles) and immunopathic distress.15,16 Impaired oligodendrocytes result in decrease in myelin density and ultimately neuronal death leading to cognitive impairment (see Fig. 1).4,5,17,18
Accordingly, myelin damage initiates a cascade of molecular mechanisms which contribute to the ongoing AD pathology. Understanding the biochemical elements responsible for myelin degeneration is necessary to mitigate such effects. The major molecular constituent of myelin is cholesterol; cholesterol constitutes ∼10% of the dry weight of the whole brain with nearly 70% being in myelin.19–21 Thus, brain, particularly the white matter, contains a disproportionate amount of cholesterol relative to other organs, and in a healthy brain, cholesterol homeostasis is maintained tightly.22
An impaired cholesterol mole fraction (χCHL) in the myelin sheath adversely affects its biological function. High χCHL obstructs permeability of essential nutrients/ions across the myelin sheath, whereas low χCHL allows facile passage of unwanted molecules/impurities into the neuron. High cholesterol content released after myelin disruption also stimulates inflammasome activation resulting in a maladaptive immune response which initiates phagolysosomal-induced membrane rupture.23 Tong et al. have shown that high χCHL is a trigger of white matter alterations and cognitive decline in a mouse model.24 Low cholesterol likewise adversely impacts myelination since a relatively high cholesterol content is essential for normal myelin membrane growth.25 Therefore, the cholesterol content of a myelin sheath membrane is rigorously regulated in a healthy brain; an altered χCHL in the myelin sheath membrane adversely affects membrane biophysical properties such as fluidity and permeability.22,26
Published information regarding the effect of cholesterol content on the biophysical and structural properties of heterogeneous biological membranes is limited.27 The impact of cholesterol content on lipid bilayers composed of POPC/PSM or POPE/PSM, typical constituents of eukaryotic cell membranes, has been methodically studied by Klauda and co-workers.28–33 However, the eukaryotic bilayer models were limited to ternary lipid systems.28,30,31 Villa and co-workers modelled neuronal axon, myelin sheath and the node of Ranvier employing all-atom, coarse-grained molecular dynamics simulations and finite element simulations to understand structural and mechanical properties of these bilayers.34–36 However, they have not reported the effect of variation in cholesterol content on these biological bilayers.
Herein, we have investigated a series of model myelin sheath membranes containing six representative lipids based on experimental lipidomic data (as explained in the methods section), using all-atom molecular dynamics simulations to gain insights relevant to AD pathology. The effect of impaired membrane integrity from an altered cholesterol mole fraction in the heterogeneous myelin sheath membrane has also been investigated.
2. Methods
2.1 Model membranes
A typical biological membrane contains ∼2000 diverse lipids resulting from variations in the head-groups and aliphatic chains of the different lipid species.37–40 The most abundant lipids in a biological membrane are phosphatidylcholines (PC), phosphatidylethanolamines (PE), cholesterol (CHL), sphingomyelins (SM) and glycolipids. The model myelin sheath (MS) membrane systems studied in this work contain: cholesterol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), galactosylceramide (GalCer) and N-stearoyl sphingomyelin (SSM) as specific constituent lipids. The myelin sheath model has been developed based on experimental lipid composition data for cellular membranes provided by Alberts et al.41 and as justified by other literature studies.34,42,43 Since the myelin sheath membrane is high in cholesterol, the effect of fluctuations in the mole fraction of cholesterol (χCHL) was studied by varying χCHL in the model MS systems in this work. The variation in the composition of other lipids (PC, SM and glycolipids) also affects myelin sheath degeneration. The effect of deficiency in phosphatidylcholine synthesis on myelination has not been reported in the literature. However, recent literature shows that choline homeostasis may regulate myelination during development and myelin repair.44,45 No myelin degeneration was detected in sphingomyelin-deficient Sphingomyelin synthase 1 or Sphingomyelin synthase 2 knockout mice.46,47 However, significant myelin recovery was seen in animals with inhibition or depletion of the enzymes responsible for sphingomyelin hydrolysis into phosphatidylcholine and ceramide; this suggests that optimal sphingomyelin content is a component of myelin regeneration.48 Myelin sheath thinning with myelin splitting is observed in animals unable to synthesize galactosylceramide.49,50 Also, mice with myelin deficient in galactosylceramide (2-hydroxlyated galactosylceramide and sulfatide) exhibited myelin degeneration by 18 months of age; mice having a low concentration of galactosylceramide sulfatide demonstrated disorganization of the uncompact myelin sheath domains.51–55 Significantly decreased sphingomyelin, and increased phospholipid content have been demonstrated in the degenerated myelin sheath pathology of in multiple sclerosis.56 Composition of healthy myelin sheath membrane and different MS bilayer systems studied in this work are provided in Table 1. Homogeneous bilayer systems with pure lipids and having equal mole fraction of lipids were also modelled to compare against experimental data and to dissect the effects of specific lipid species on various properties. The atomistic structures of POPC, POPE, POPS, SSM, GalCer and CHL are given in ESI.†| Set | Bilayer system | N PC | N PE | N CHL | N SSM | N GalCer | N PS | n H2O | m H2O | χ CHL | Ions | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Na+ | Cl− | |||||||||||
| Where lipid name abbreviations used are as follows: POPC = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPE = 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; CHL = cholesterol; SSM = sphingosine-phosphorylcholine; GalCer = beta-D-galactosylceramide; POPS = 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine. | ||||||||||||
| 1 | Healthy myelin sheath membrane and reference bilayer systems | |||||||||||
| PC | 200 | 0 | 0 | 0 | 0 | 0 | 10139 | 50.69 | 0 | 25 | 25 | |
| PC:PE | 100 | 100 | 0 | 0 | 0 | 0 | 9477 | 47.38 | 0 | 23 | 23 | |
| PC:PE:CHL | 66 | 66 | 68 | 0 | 0 | 0 | 8350 | 41.75 | 0.34 | 21 | 21 | |
| PC:PE:CHL:SSM | 50 | 50 | 50 | 50 | 0 | 0 | 8117 | 40.58 | 0.25 | 20 | 20 | |
| PC:PE:CHL:GalCer | 50 | 50 | 50 | 0 | 50 | 0 | 9405 | 47.02 | 0.25 | 25 | 25 | |
| Healthy Myelin Sheath (HMS) | 18 | 28 | 78 | 14 | 46 | 16 | 8883 | 44.41 | 0.39 | 40 | 24 | |
| 2 | Cholesterol variations in myelin sheath membrane composition | |||||||||||
| MS_1 | 34 | 44 | 0 | 30 | 60 | 32 | 9961 | 49.81 | 0 | 58 | 26 | |
| MS_2 | 32 | 42 | 8 | 28 | 60 | 30 | 9947 | 49.74 | 0.04 | 56 | 26 | |
| MS_3 | 30 | 40 | 18 | 26 | 58 | 28 | 8939 | 44.70 | 0.09 | 51 | 23 | |
| MS_4 | 28 | 38 | 28 | 24 | 56 | 26 | 9609 | 48.05 | 0.14 | 51 | 25 | |
| MS_5 | 26 | 36 | 38 | 22 | 54 | 24 | 9560 | 47.80 | 0.19 | 49 | 25 | |
| MS_6 | 24 | 34 | 48 | 20 | 52 | 22 | 9400 | 47.00 | 0.24 | 47 | 25 | |
| MS_7 | 22 | 32 | 58 | 18 | 50 | 20 | 8969 | 44.85 | 0.29 | 44 | 24 | |
| MS_8 | 20 | 30 | 68 | 16 | 48 | 18 | 9095 | 45.48 | 0.34 | 38 | 24 | |
| MS_9 | 16 | 26 | 88 | 12 | 44 | 14 | 8620 | 43.10 | 0.44 | 37 | 23 | |
| MS_10 | 14 | 24 | 98 | 10 | 42 | 12 | 8484 | 42.42 | 0.49 | 34 | 22 | |
| MS_11 | 12 | 22 | 108 | 8 | 40 | 10 | 8322 | 41.61 | 0.54 | 32 | 22 | |
| MS_12 | 10 | 20 | 118 | 6 | 38 | 8 | 8214 | 41.07 | 0.59 | 30 | 22 | |
| MS_13 | 8 | 18 | 128 | 4 | 36 | 6 | 8014 | 40.07 | 0.64 | 28 | 22 | |
| MS_14 | 6 | 16 | 138 | 2 | 34 | 4 | 7895 | 39.48 | 0.69 | 25 | 21 | |
| MS_15 | 4 | 14 | 148 | 0 | 32 | 2 | 7673 | 38.37 | 0.74 | 23 | 21 | |
| MS_16 | 2 | 12 | 158 | 0 | 28 | 0 | 7596 | 37.98 | 0.79 | 20 | 20 | |
| MS_17 | 0 | 8 | 168 | 0 | 24 | 0 | 7404 | 37.02 | 0.84 | 20 | 20 | |
| MS_18 | 0 | 4 | 178 | 0 | 18 | 0 | 7280 | 36.40 | 0.89 | 20 | 20 | |
| MS_19 | 0 | 2 | 188 | 0 | 10 | 0 | 7202 | 36.01 | 0.94 | 20 | 20 | |
| MS_20 | 0 | 0 | 198 | 0 | 2 | 0 | 7108 | 35.54 | 0.99 | 19 | 19 | |
The systems were modelled as a lipid bilayer with both leaflets having the same lipid composition with constant number of lipids (200) to avoid bias due to system size. The initial coordinates of the atomistic bilayers were constructed using the membrane-only generation option of Membrane Builder in CHARMM-GUI.57,58 All lipids are predefined in the CHARMM-GUI. Galcer is the predefined glycolipid in the CHARMM-GUI within the Galactocerebrosides subgroup,59 characterized by having β-D-galactose as a head-group (see Fig. S1, ESI†). An ion concentration of 0.15 M NaCl was added to the bilayer systems using distance as the ion placing method to mimic physiological conditions.
2.2 Molecular dynamics simulations
All MD simulations were performed using the NAMD package version 2.13 employing all-atom CHARMM36 lipid force field and the modified TIP3P model for explicitly representing solvent molecules.60–65 An aqueous environment was mimicked for these simulations by employing periodic boundary conditions. For constraining covalent bonds containing a hydrogen atom at their equilibrium bond length, the SETTLE algorithm66 for water and the SHAKE algorithm67 for phospholipids was used. Long-range electrostatic interactions were computed using the Particle Mesh Ewald (PME) algorithm.68 The van der Waals interactions were truncated at a cutoff distance of 12 Å and were modeled using a Lennard-Jones potential. The isobaric–isothermal (NPT) ensemble was generated with temperature and pressure maintained at 310 K (which is well above the melting points of phospholipids) and 1 atm, respectively. Constant temperature and pressure was maintained by employing Langevin dynamics with a damping coefficient of 1 ps-1 and the Langevin piston Nosé–Hoover method69,70 in which the pressure coupling is isotropic only in the x- and y directions, respectively. Different myelin sheath membrane and reference bilayer systems were first equilibrated using the CHARMM-GUI standard six-step equilibration process in Membrane Builder.57,58 Each MS and reference bilayer systems were further equilibrated for a minimum of 1 μs; the total simulation time was approximately 26 μs. The graphs of area per lipid of different bilayer systems as a function of simulation time are given in ESI,† confirming that each bilayer system reached equilibrium before running production simulation trajectories. All bilayer systems reached equilibrium after approximately 800 ns of simulation time. The velocity-Verlet algorithm71 with a time step of 2 fs was used to generate MD trajectories and VMD version 1.9.372 was used to visualize the systems. The last 200 ns part of equilibrated trajectory of each system was used for analysis.2.3 Simulation analysis
To understand structural and biophysical properties of different myelin sheath membrane systems, we have calculated ten properties for each model membrane lipid bilayer: [1] area per lipid (APL) in the membrane including APL of each specific lipid as a function of χCHL in different MS systems, [2] bilayer thickness (DHH), [3] number density profile of POPC, water (TIP3P) and cholesterol molecules, [4] mass density profiles of POPC, water (TIP3P) and cholesterol molecules, [5] Sn-1 and Sn-2 carbon order parameters, [6] area compressibility modulus (KA), [7] bending modulus (Kc), [8] POPC and cholesterol lipid tilt angles, [9] lipid interdigitation and [10] H-bonding. The results are reported as an average value for the last 0.2 μs of 1 μs atomistic simulation trajectory There was minimum deviation in the results for different properties for the last 0.2 μs of 1 μs atomistic simulation trajectory as bilayer systems had already reached equilibrium. Results of different structural and mechanical properties of reference bilayer systems (given in Table 1) were compared against available experimentally determined values to validate simulation results. The x and y dimensions of the simulation box are multiplied and divided by total number of lipids present in one leaflet of the bilayer to calculate area per lipid (APL). Component surface area of different lipids was calculated by determining the X and Y coordinates of representative atoms for each lipid (e.g. O3 of Cholesterol, C2, C21, and C31 for phospholipids; as shown in Fig. S5 of ESI†). The X and Y coordinates of these selected representative atoms are projected onto a plane delimited by the simulation box. This plane is subsequently divided into polygons through a Voronoi diagram using the qvoronoi program within the Quickhull73 package. The sum of all polygonal areas is the area of the simulation box, and summing areas of representative atoms gives the area per lipid for individual lipid components.The thickness of the bilayer is calculated as the distance between the phosphate atom peaks in the mass density profile. Traditionally, orientational mobility of C–H bonds in lipid bilayers is calculated by C–H bond order parameter available from 2H NMR experiments (as C–D bond in 2H NMR).74 Carbon order parameter (SCD) is calculated as follows:
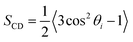 | (1) |
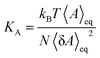 | (2) |
The bending modulus (Kc) was calculated using eqn (3).75
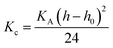 | (3) |
To identify lipid interdigitation and the mass overlap between the upper and lower leaflets of the bilayer as a function χCHL, the lipid interdigitation tool in Membplugin77 (an extension of VMD) was used. The degree of acyl chain interdigitation is calculated using a correlation-based fraction which gives the width of overlapping bilayer regions (Wρ) in nm, fraction of mass overlap (Iρ) and contacts between two leaflets (Ic). Iρ represents zero to complete mass overlap between lipid leaflets and varies within 0 ≤ Iρ ≤ 1. Ic gives number of heavy atoms in contact with opposite leaflet within a default cutoff range of 4 Å. To examine orientation of cholesterol molecules in the MS and reference membrane systems, we calculated the angle between the cholesterol vector (defined by the carbon atom bonded to the hydroxyl group (OH) and the carbon atom bonded to the acyl tail of the cholesterol) and the bilayer normal (z-axis). Further explanation of tilt angle definition employed for POPC and cholesterol is given in the results section. Analyses were performed using VMD and TCL scripts written in-house. VMD software was used for graphical representations.
3. Results and discussion
Myelin sheath composition is specialized and different from other types of biological membranes. One of the most distinct features of myelin sheath membrane is its enrichment with a variety of lipids such as cerebrosides, phospholipids and particularly cholesterol. The white matter concentration of cerebrosides is directly proportional to myelin content.41 In this work, a model myelin sheath membrane and different myelin sheath membrane bilayer systems with varying cholesterol mole fraction (χCHL) have been studied as discussed in methods section.A total of 26 model membrane systems with different lipid composition were studied in this work as given in Table 1. The mole fraction of cholesterol was varied in healthy myelin sheath membrane for studying the effect of altered cholesterol concentration. Reference bilayer systems composed of only phospholipids (PC, PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE) or phospholipids
PE) or phospholipids![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol (PC
cholesterol (PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE
PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHL, PC
CHL, PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE
PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHL
CHL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SSM, PC
SSM, PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PE
PE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHL
CHL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GalCer, where different lipids and cholesterol are in equal proportion in different bilayer systems), have been also studied in order to compare results against available experimental data. The lipid bilayer compositions for reference bilayer systems are also given in Table 1.
GalCer, where different lipids and cholesterol are in equal proportion in different bilayer systems), have been also studied in order to compare results against available experimental data. The lipid bilayer compositions for reference bilayer systems are also given in Table 1.
The final snapshots of 1 μs equilibrated reference bilayer systems, healthy myelin sheath (HMS), and myelin sheath bilayers with varying χCHL are shown in Fig. 2–4, respectively.
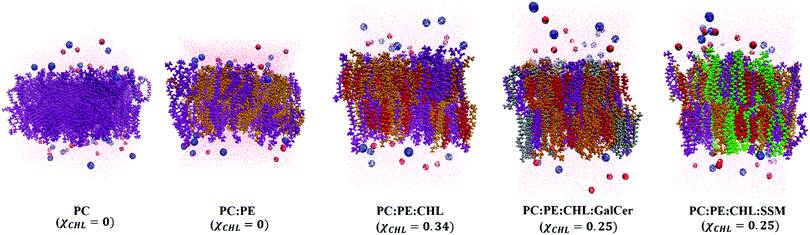 | ||
| Fig. 2 Final equilibration snapshots of reference lipid bilayer systems (as given in Table 1) having varying cholesterol mole fraction (χCHL), with POPC (purple), POPE (orange), POPS (cyan), cholesterol (red), SSM (green), GalCer (silver), sodium ions (blue spheres), chloride ions (red spheres) and water (pink dots). All lipid bilayers were equilibrated for a minimum of 1 μs. | ||
 | ||
| Fig. 3 Final equilibration snapshot of a healthy myelin sheath membrane (as given in Table 1) with POPC (purple), POPE (orange), POPS (cyan), cholesterol (red), SSM (green), GalCer (silver), sodium ions (blue spheres), chloride ions (red spheres) and water (pink dots). All lipid bilayers were equilibrated for a minimum of 1 μs. | ||
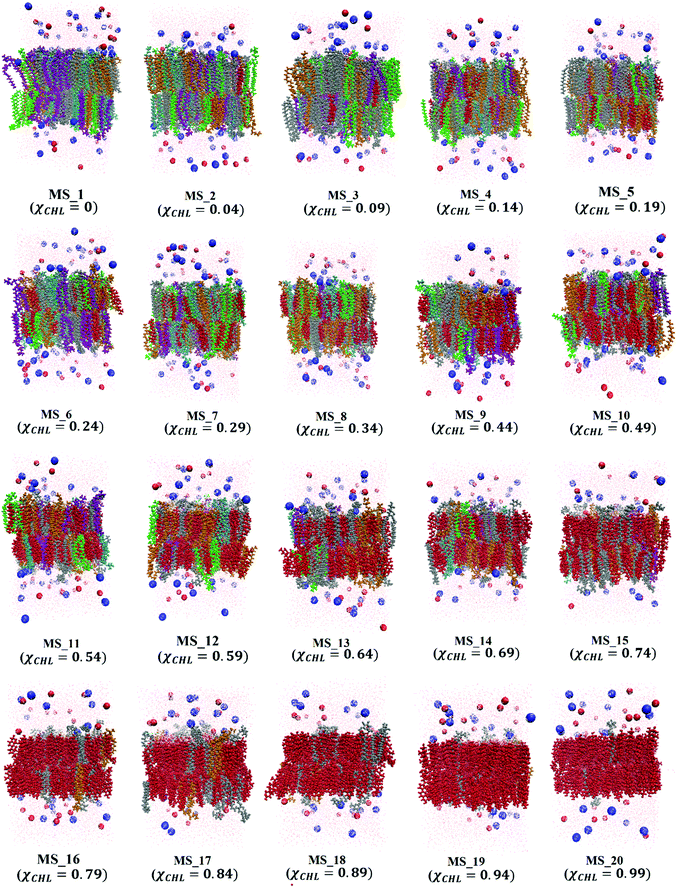 | ||
| Fig. 4 Final equilibration snapshots of different myelin sheath membrane systems (as given in Table 1) having varying cholesterol mole fraction (χCHL), with POPC (purple), POPE (orange), POPS (cyan), cholesterol (red), SSM (green), GalCer (silver), sodium ions (blue spheres), chloride ions (red spheres) and water (pink dots). All lipid bilayers were equilibrated for a minimum of 1 μs. | ||
3.1 Lateral bilayer area and bilayer thickness
The reference bilayer and myelin sheath bilayer systems converged at 800 ns equilibration time. Graphs depicting the convergence of area per lipid (APL) as a function of simulation time are provided in the ESI.† Average APL and thickness DHH for reference, HMS and different myelin sheath (MS) bilayer systems with varying χCHL are given in Table 2.| Set | Bilayer system | χ CHL | APL (Å2) | D HH (Å) |
|---|---|---|---|---|
| 1 | Reference bilayer systems | |||
| PC | 0 | 65.35 ± 1.10 | 38.10 ± 0.53 | |
| PC:PE | 0 | 60.49 ± 1.02 | 40.89 ± 0.57 | |
| PC:PE:CHL | 0.34 | 45.70 ± 0.54 | 45.75 ± 0.45 | |
| PC:PE:CHL:SSM | 0.25 | 44.96 ± 0.55 | 47.20 ± 0.40 | |
| PC:PE:CHL:GalCer | 0.25 | 43.74 ± 0.45 | 47.23 ± 0.45 | |
| 2 | Myelin sheath (MS) membrane systems | |||
| MS_1 | 0 | 46.17 ± 0.27 | 47.73 ± 0.24 | |
| MS_2 | 0.04 | 46.63 ± 0.29 | 47.67 ± 0.23 | |
| MS_3 | 0.09 | 46.70 ± 0.57 | 47.26 ± 0.38 | |
| MS_4 | 0.14 | 44.30 ± 0.26 | 48.04 ± 0.21 | |
| MS_5 | 0.19 | 44.42 ± 0.48 | 47.79 ± 0.33 | |
| MS_6 | 0.24 | 43.80 ± 0.33 | 47.69 ± 0.27 | |
| MS_7 | 0.29 | 42.26 ± 0.26 | 47.34 ± 0.23 | |
| MS_8 | 0.34 | 42.64 ± 0.29 | 46.76 ± 0.34 | |
| Healthy myelin sheath (HMS) | 0.39 | 42.33 ± 0.33 | 46.99 ± 0.30 | |
| MS_9 | 0.44 | 41.28 ± 0.22 | 47.03 ± 0.28 | |
| MS_10 | 0.49 | 41.04 ± 0.23 | 46.38 ± 0.28 | |
| MS_11 | 0.54 | 40.08 ± 0.19 | 45.62 ± 0.29 | |
| MS_12 | 0.59 | 40.18 ± 0.21 | 45.99 ± 0.40 | |
| MS_13 | 0.64 | 39.89 ± 0.22 | 45.04 ± 0.38 | |
| MS_14 | 0.69 | 39.59 ± 0.19 | 43.84 ± 0.54 | |
| MS_15 | 0.74 | 38.73 ± 0.18 | 43.58 ± 0.61 | |
| MS_16 | 0.79 | 38.78 ± 0.19 | 43.07 ± 0.79 | |
| MS_17 | 0.84 | 38.41 ± 0.15 | 43.01 ± 1.03 | |
| MS_18 | 0.89 | 38.19 ± 0.16 | 41.17 ± 1.39 | |
| MS_19 | 0.94 | 38.07 ± 0.16 | 39.47 ± 2.02 | |
| MS_20 | 0.99 | 37.80 ± 0.14 | 39.07 ± 0.89 | |
In general, APL decreases with an increase in χCHL in reference and myelin sheath membrane bilayer systems. For reference systems, APL follows the trend: PC:PE:CHL:GalCer ∼ PC:PE:CHL:SSM < PC:PE:CHL < PC:PE < PC; whereas bilayer thickness exhibits an opposite trend: PC:PE:CHL:GalCer ∼ PC:PE:CHL:SSM > PC:PE:CHL >PC:PE ∼ PC. In reference bilayers, increasing χCHL from 0 to 0.34, decreases APL by 33% and increases thickness by 23%, which is consistent with NMR data.78 The calculated PC bilayer APL is 65.35 ± 1.10 Å2 at 310 K which agrees with experimental APL values of 63, 62 and 66 Å2 at 297, 323 and 310 K, respectively.79–81 Also, the calculated pure PC bilayer thickness is 38.10 Å at 310 K as given in Table 2 which compares well with an experimental DHH of 37.0 Å at 303 K.82 PC:PE:CHL:SSM and PC:PE:CHL:GalCer systems have slightly lower APLs than PC:PE:CHL, which suggest that both SSM and GalCer lipids have similar condensing effects and this condensing effect is slightly higher than cholesterol molecules.
The effect of varying cholesterol content on APL and DHH in different myelin sheath membranes was also studied. Table 2 presents APL and DHH values for 21 different myelin sheath membrane systems containing a model healthy myelin sheath membrane system. In general, APL decreases with increasing χCHL. In particular, as we increase χCHL from 0 to 0.99 in different MS systems, APL decreases by approximately 18%. Previous experimental and theoretical investigations on the condensing effects of cholesterol also correlate with this finding.83–86DHH does not follow any discernible trend with increasing χCHL in the lower cholesterol range until χCHL becomes close to that of the HMS membrane. There is a slight decrease in DHH values on further increasing χCHL.
Fig. 5 visualizes the trends of APL and DHH with variation in χCHL in various bilayer systems. There is a sharp decrease in APL as we introduce cholesterol into the lipid bilayers as compared to PC and PC:PE reference bilayers, as shown in Fig. 5. APL decreases with an increase in χCHL, where HMS corresponds to the APL of a model healthy myelin sheath membrane. For the HMS bilayer, APL is 42.33 Å2. Similar to the sharp decrease in APL upon introduction of cholesterol into phospholipid bilayers, there is a sharp increase in thickness of bilayers as well with increasing χCHL. However, a clear monotonic trend is not observed in thickness of MS bilayers in lower ranges, but at higher χCHL, we observed a decrease in DHH values. The thickness of a healthy myelin sheath membrane is 46.99 Å, which is highlighted on the DHH curve presented in Fig. 5. The deviation of APL and DHH from HMS values corresponding to altered χCHL in MS bilayers impairs their biological functions (possibly leading to the implications observed in neurological disorders).
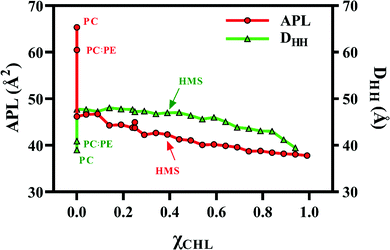 | ||
| Fig. 5 Area per lipid (APL) and bilayer thickness (DHH) for POPC bilayer (PC), healthy myelin sheath (HMS) and different model bilayer systems (given in Table 1) as a function of cholesterol composition at 310 K. | ||
3.2 Component surface areas
Table 3 presents APL values of individual lipids in modelling different reference, HMS and MS membrane systems. Phospholipids (i.e., POPC, POPE and POPS) have higher APLs compared to other lipids (i.e., sphingomyelin (SM) and ceramide (Cer)). In eukaryotic membranes, POPC lipids are the main phospholipid constituents (∼50%), whereas POPS lipids are the most abundant anionic phospholipid. A pure POPC bilayer has an APL of 65.17 Å2, which decreases to approximately 57–60 Å2 range in different reference and MS systems. Cholesterol has an usually low APL compared to phospholipids. For the MS_20 bilayer system, with an χCHL of 0.99, the APL value is 37.90 Å2, which agrees well with the experimental value of 40 Å2 for pure cholesterol. SSM and GalCer lipids also experience condensing effects at higher χCHL. Compared to other lipids in different bilayer systems, cholesterol molecules expand considerably with increasing χCHL fluctuating in a 30–38 Å2 range.| Set | Bilayer system | χ CHL | POPC (Å2) | POPE (Å2) | CHL (Å2) | SSM (Å2) | GalCer (Å2) | POPS (Å2) |
|---|---|---|---|---|---|---|---|---|
| 1 | Reference bilayer systems | |||||||
| PC | 0 | 65.168 | ||||||
| PC:PE | 0 | 61.159 | 59.879 | |||||
| PC:PE:CHL | 0.34 | 54.612 | 54.044 | 28.263 | ||||
| PC:PE:CHL:SSM | 0.25 | 59.668 | 57.557 | 30.518 | 31.043 | |||
| PC:PE:CHL:GalCer | 0.25 | 56.884 | 54.328 | 31.410 | 33.628 | |||
| 2 | Myelin sheath (MS) membrane systems | |||||||
| MS_1 | 0 | 56.404 | 57.993 | 29.716 | 33.750 | 57.872 | ||
| MS_2 | 0.04 | 59.160 | 57.310 | 29.033 | 34.186 | 35.341 | 57.374 | |
| MS_3 | 0.09 | 58.736 | 61.994 | 32.336 | 33.466 | 36.069 | 60.156 | |
| MS_4 | 0.14 | 57.690 | 57.473 | 30.325 | 31.109 | 32.596 | 60.826 | |
| MS_5 | 0.19 | 63.993 | 62.441 | 31.639 | 30.319 | 34.195 | 59.590 | |
| MS_6 | 0.24 | 58.136 | 59.697 | 31.931 | 31.608 | 37.333 | 59.756 | |
| MS_7 | 0.29 | 58.385 | 58.298 | 31.305 | 33.894 | 33.072 | 59.179 | |
| MS_8 | 0.34 | 57.980 | 55.610 | 33.886 | 34.875 | 34.743 | 59.030 | |
| Healthy Myelin Sheath (HMS) | 0.39 | 58.398 | 60.979 | 34.624 | 31.915 | 34.452 | 59.054 | |
| MS_9 | 0.44 | 63.100 | 64.239 | 32.778 | 32.845 | 32.385 | 59.936 | |
| MS_10 | 0.49 | 60.852 | 60.639 | 35.003 | 37.338 | 32.618 | 59.937 | |
| MS_11 | 0.54 | 59.526 | 60.803 | 34.798 | 36.648 | 33.425 | 63.229 | |
| MS_12 | 0.59 | 62.199 | 57.197 | 36.274 | 26.877 | 32.944 | 66.236 | |
| MS_13 | 0.64 | 61.064 | 60.929 | 36.430 | 32.954 | 34.491 | 60.235 | |
| MS_14 | 0.69 | 64.620 | 60.609 | 36.851 | 25.781 | 33.743 | 60.438 | |
| MS_15 | 0.74 | 64.883 | 62.268 | 36.775 | 32.926 | 60.149 | ||
| MS_16 | 0.79 | 64.887 | 64.028 | 37.382 | 33.882 | |||
| MS_17 | 0.84 | 63.793 | 37.893 | 33.144 | ||||
| MS_18 | 0.89 | 64.600 | 37.936 | 34.470 | ||||
| MS_19 | 0.94 | 64.782 | 37.894 | 36.095 | ||||
| MS_20 | 0.99 | 37.903 | 31.800 | |||||
Optimal cholesterol content in biological membranes facilitates both passive diffusion and active transmembrane transport of molecules.87–89 Alterations in the APL values of constituent membrane lipids alters their physical properties thereby altering optimal transmembrane molecular diffusion and related cellular functions.
3.3 Orientation ordering of lipid tails
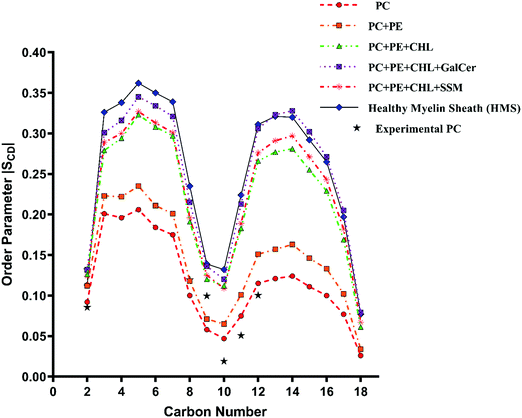
The variation of deuterium order parameter values for the carbon atoms of the palmitoyl (sn-1) and unsaturated oleoyl (sn-2) tails of POPC lipids for reference and myelin sheath systems at different χCHL is shown in Fig. 6–9.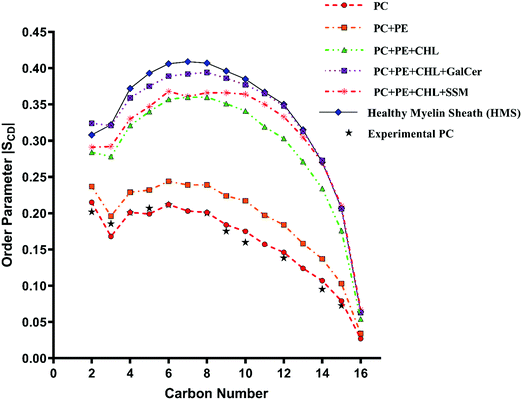 | ||
| Fig. 6 Change in orientation ordering of the carbon atoms of the palmitoyl tail of POPC lipid molecules in pure POPC, healthy myelin sheath (HMS) membrane and reference bilayer systems as given in Table 1. The experimental order parameters for a pure POPC bilayer at 310 K from Seelig et al.74 are plotted for comparison against calculated results. | ||
 | ||
| Fig. 7 Change in orientation ordering of the carbon atoms of the oleoyl tail of POPC lipid molecules in pure POPC, healthy myelin sheath (HMS) membrane and reference bilayer systems as given in Table 1. The experimental order parameters for pure POPC bilayer at 310 K from Seelig et al.74 are also plotted for comparison against calculated results. | ||
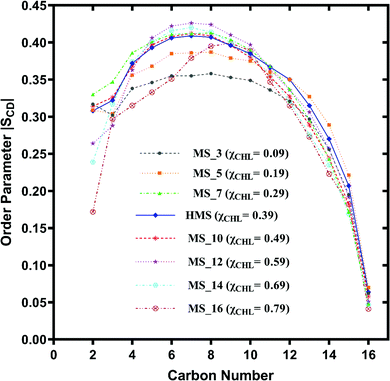 | ||
| Fig. 8 Change in orientation ordering of the carbon atoms of the palmitoyl tail of POPC lipid molecules in healthy myelin sheath (HMS) membrane and different myelin sheath membrane systems with varying composition of cholesterol as given in Table 1. | ||
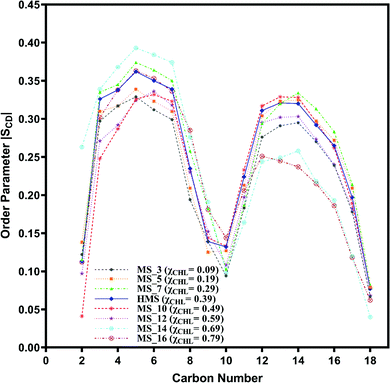 | ||
| Fig. 9 Change in orientation ordering of the carbon atoms of the oleoyl tail of POPC lipid molecules in axonal plasma (AP) membrane and different axonal membrane systems with varying composition of cholesterol as given in Table 1. | ||
The experimental sn-1 and sn-2 SCD results from Seelig et al.74 for POPC have been reproduced with the CHARMM36m force field computed POPC bilayer SCD, as shown in Fig. 6 and 7, respectively. The order parameter profiles become higher for both palmitoyl (sn-1) and unsaturated oleoyl (sn-2) tails of POPC lipids with the addition of cholesterol in reference and healthy myelin sheath (HMS) membrane systems, as shown in Fig. 6 and 7 respectively. The ordering effect of GalCer lipids is higher than SSM lipids. Noticeably, myelin sheath membranes have a higher content of GalCer lipids as compared to SSM lipids; therefore, they are inherently biased towards attaining higher order and structure compared to other biological lipid membranes. High content of GalCer lipids in myelin sheath membrane increases its structural order, thereby reducing its permeability which helps in maintaining its crucial function of providing insulation for electrical signal propagation along neuronal axons. The order parameter profile for HMS (with χCHL = 0.39) is highest for C4–C10 carbon atoms for palmitoyl tails and C2–C12 for oleoyl tails as compared to other reference bilayers systems, as shown in Fig. 6 and 7. In other words, the increase in the orientational ordering of both palmitoyl and oleoyl chains is higher in HMS membranes compared to other reference bilayers, which illustrates the condensing effect of cholesterol. Lipid bilayer systems with higher χCHL tend to pack more tightly resulting in elevated thickness. These finding are also supported with the decrease in the lateral APL and increase in thickness of the reference and HMS with addition of cholesterol, as discussed in Table 2.
The palmitoyl (sn-1) and oleoyl (sn-2) POPC SCD parameters for HMS and different myelin sheath membranes, as a function of χCHL′ are presented in Fig. 8 and 9, respectively, which show that altering χCHL of MS membranes enables palmitoyl and oleoyl chains of POPC to assume either higher or lower order than HMS. In general, both palmitoyl and oleoyl POPC SCD parameters increase with an increase of χCHL in different MS membrane systems. However, substantial fluctuations in order parameters are observed for C2–C12 carbon atoms in palmitoyl chain and C3–C8 and C12–C16 of oleoyl chains. Modified order of lipid chains as a function of χCHL in different myelin sheath membranes compared to normal myelin sheath (HMS) adversely affects both its structural and functional properties. Lower order in membrane lipids increases fluidity and permeability, thereby enabling transmembrane permeation of toxic or unwanted molecules. Conversely, higher than normal order deleteriously hinders diffusion of essential ions/nutrients through the lipid bilayer.
3.4 Distribution of water, POPC and CHL along bilayer normal axis
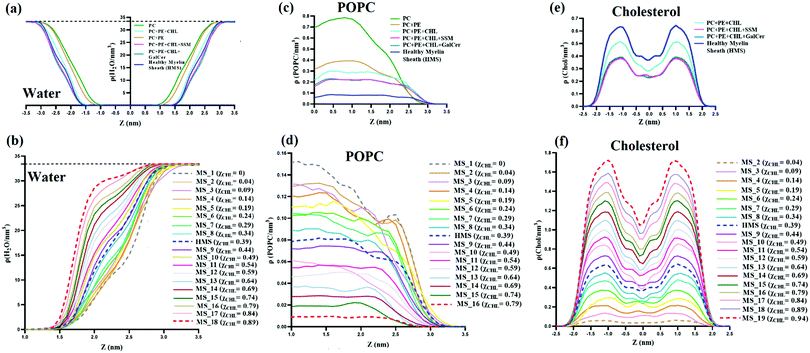
Distribution of water, POPC and cholesterol along the bilayer normal (z-axis) for reference systems, model healthy myelin sheath (HMS) membrane, and different myelin sheath (MS) systems was studied by computing atomic number density profiles, as shown in Fig. 10. Fig. 10(a and b), present number density profiles of water molecules for reference and MS systems, respectively. The bilayers can be divided into three regions. First, the hydrophobic region rich in lipid tails in which no water molecule is present. Secondly, the bulk region at which the density of water meets the water bulk density (i.e., ∼0.9979 g cm−3 = 33.4 H2O nm−3); the bulk water density is represented by a dotted black line in Fig. 10(a and b), for both myelin sheath and reference systems. Thirdly, the interfacial region, sandwiched between the hydrophobic and bulk regions; in the interfacial region, the number density of water is characterized by 0 < ρ < 33.4. In Fig. 10(a), both pure PC and PC:PE bilayers have sigmoidal number density curves. However, with the introduction of cholesterol in pure PC and PC:PE bilayers, the number density curves start to deviate from their sigmoidal shape and to show a slight curvature. This slight curvature in the middle of the water number density curves is also observed in health myelin sheath and other MS bilayer systems. The number density profiles of water tend to attain bulk water number density much more quickly with an increase in mole fraction of cholesterol in lipid bilayers; for example, Δρ (within 1.0 < z < 2.0) for MS_1 (χCHL = 0), healthy myelin sheath membrane (χCHL = 0.39) and MS_18 (χCHL = 0.89) are 6.27, 11.86, 28.63 H2O nm−3 respectively. This suggests that there is a shrinkage in the interfacial region with increasing χCHL, thereby potentially impacting various biophysical processes occurring at the lipid–water interface of bilayers. Therefore, the increase of cholesterol results in higher water density and the interfacial region endeavours to attain a bulk-like environment. In other words, high cholesterol content decreases APL of lipids and increases water number density in the interfacial region. Magnetic resonance imaging (MRI) provides clinical images of cerebral atrophy for assessing neurodegeneration in AD as a function of brain volume/cerebrospinal fluid [CSF] index.90 The atomistic details of the increased number density of water in the cholesterol rich membrane presented in this work is compatible with this MRI-based cerebral atrophy characterization.Fig. 10(c and d) represent POPC number density profiles of reference, model healthy myelin sheath and different myelin sheath membranes. In both reference and MS systems bilayers, the number density profiles shift towards the interfacial region having higher ‘z’ values on the x-axis. This indicates that lipids are more exposed to the interfacial water region as we decrease the mole fraction of cholesterol in the bilayer, possibly interrupting interfacial biochemical processes occurring at the membrane–water interface. Fig. 10(e and f) show that with an increase in the cholesterol mole fraction in both MS and reference systems, the number density profile peaks become sharper and higher. The number density profiles for different MS systems with mole fractions of cholesterol below 0.34 exhibit a comparatively smoother profile. This indicates that with increasing χCHL, the lipid membrane is compressed in the lateral direction (i.e. APL decreases) and elongated in the transverse direction (i.e. DHH increases).
3.5 Mass density profile
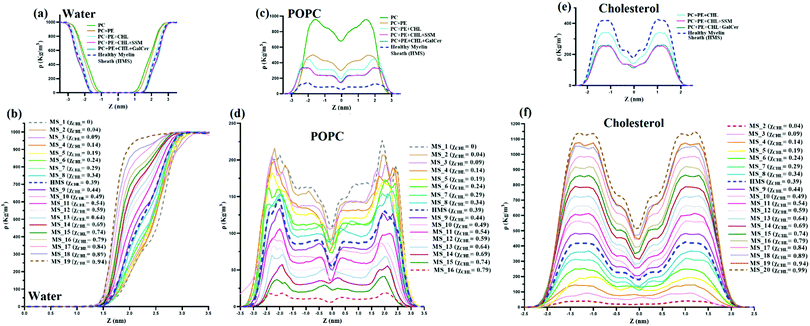
Mass density profiles of water, POPC and cholesterol are plotted in Fig. 11. The distribution of mass of various molecules along the membrane z axis is evaluated through mass density profiles for various species. Fig. 11(a) and (b) show that with an increase in χCHL the mass density profile of water attains bulk water density much sooner as compared to HMS or bilayers with higher χCHL. The presence of a high volume of water molecules at bulk water density (∼1000 kg m−3) in cholesterol rich bilayer systems indicates that the mass distribution of water molecules is substantially perturbed as the cholesterol content is altered within the myelin sheath membrane.Fig. 11(c and d) show mass density profiles of POPC along the bilayer normal for reference and different myelin sheath systems. As χCHL is increased, the peaks in the POPC mass density profiles are diminished and become smoother. This shows that at lower χCHL, POPC head groups become more exposed at the lipid-water interface. Also, the width and height of the minima at the center of the bilayer corresponding to the low density region is dependent upon the χCHL. The mass density profiles of cholesterol, for both reference and MS systems, are given in Fig. 11(e and f) respectively. The density profiles are smoother for the MS bilayer with lower χCHL and have two distinct peaks near interfacial regions as χCHL is increased. Mass density profile peaks also shift slightly towards the interfacial region at higher. This clearly shows that alterations in the cholesterol content in the myelin sheath have significant impact on the intensities of the peaks. For the MS bilayers with χCHL higher than 0.34, these peaks become more prominent, indicating that the introduction of cholesterol yields enhanced structuring and bilayer stiffness.
3.6 Area compressibility modulus (KA) and bending rigidity modulus (Kc)
The results for the area compressibility modulus (KA) and for bending rigidity modulus (Kc), as a function of χCHL′ for reference and healthy myelin sheath membrane bilayer systems are presented in Table 4. The experimental data for KA and Kc for pure POPC and POPC:CHL employing different measuring techniques are also summarized in Table 4 for comparison. The calculated area compressibility modulus of pure POPC is 251 mN m−1; experimentally, KA is reported to be 180 – 330 mN m−1 at 25 °C using infrared measurements and 208–237 mN m−1 at 21 °C using micropipette aspiration methods.75,91 The area compressibility modulus increases with an increase in χCHL in reference bilayer systems. Different experimental studies also report an increase in KA values with an increase of cholesterol content in lipid bilayers.92,93 POPC bilayer experimental bending rigidity data (shown in Table 4) demonstrate large variations in experimental Kc values depending upon measurement technique and experimental conditions. The calculated Kc value for a pure POPC bilayer employing the polymer-brush model proposed by Evans et al.76 is 15.16 × 10−20 J (as given in Table 4), which is compatible with an experimental Kc value at 310 K.42,75 With increasing χCHL in reference bilayer systems, bending rigidity increases and follows the trend: PC:PE:CHL:GalCer > PC:PE:CHL:SSM ∼ PC:PE:CHL > PC:PE ∼ PC. Galcer lipids induce more bending rigidity as compared to SSM lipids. Experimentally, POPC/cholesterol bilayers exhibit strong stiffening with increasing χCHL as determined via micropipette aspiration94 and neutron spin echo and dynamic light scattering95 techniques. Fig. 12 shows that both the area compressibility modulus (KA) and bending rigidity modulus (Kc) increase with an increase in cholesterol mole fraction for different myelin sheath membrane systems. Chakraborty et al.96 have noted similar observations as an effect of increasing cholesterol mole fraction. Cholesterol increases the compressibility and bending modulus by reducing average area per lipid. The points corresponding to area compressibility modulus and bending rigidity modulus for health myelin sheath bilayer system are labelled as HMS in Fig. 12. In summary, higher χCHL increases the compressibility modulus and bending rigidity of myelin sheath membranes. The tilt and interdigitation deformations of lipids along with fluctuations in membrane thickness modulates the overall elasticity of cholesterol-doped membranes.| System | χ CHL | K A (mN m−1) | K c × 10−20 (J) | ||||
|---|---|---|---|---|---|---|---|
| This work | Exp. literature data | This work | Exp. literature data | ||||
| PC | 0 | 251.19 | 180–33091 | 15.160 | Temp. | Experimental technique | K c × 10−20 (J) |
| 24 | Fluctuation analysis | 14.697 | |||||
| 20 | Fluctuation analysis | 15.99 ± 0.3198 | |||||
| 25 | Fluctuation analysis | 21.1 ± 0.499 | |||||
| 30 | X-ray scattering | 8.5100 | |||||
| 25 | Micropipette aspiration | 21.1 ± 0.494 | |||||
| 30 | Fluctuation analysis | 10101 | |||||
| 22 | Fluctuation analysis | 12.9 ± 0.4102 | |||||
| 24 | Fluctuation analysis | 3.9 ± 0.9103 | |||||
| 22 | Neutron spin echo and dynamic light scattering | 7.7 ± 0.895 | |||||
| 20 | Fluctuation analysis | 14.48 ± 0.1998 | |||||
| 20 | Fluctuation analysis | 12.55 ± 0.3398 | |||||
| 24 | Electro-deformation | 5.8 ± 1.2103 | |||||
| PC:PE | 0 | 253.81 | — | 16.825 | |||
| PC:PE:CHL:GalCer | 0.25 | 418.84 | — | 37.291 | |||
| PC:PE:CHL:SSM | 0.25 | 351.77 | — | 31.280 | |||
| PC:PE:CHL | 0.34 | 364.26 | — | 30.392 | |||
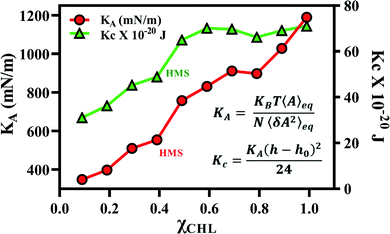 | ||
| Fig. 12 Area compressibility modulus (KA) and bending modulus (Kc) for different myelin sheath membrane bilayer systems as a function of χCHL at 310 K. The results are calculated using average area at equilibrium and its variance following eqn (2) and (3) given in methods section. | ||
3.7 P–N vector angle distribution for POPC, tilt angle of cholesterol and lipid interdigitation in model myelin sheath membranes
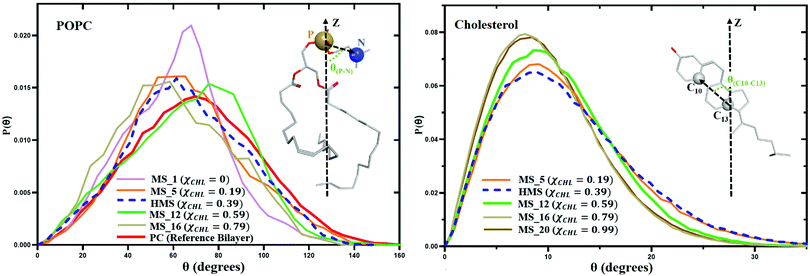
Changing cholesterol concentration in lipid bilayer membranes modulates the angular orientation of the various lipids and cholesterol. Fig. 13 represents variation in the P–N angle distribution function P(θ) for POPC lipid molecules and variation of the C10–C13 angle distribution function P(θ) for cholesterol molecules for different myelin sheath membrane and reference bilayer systems as a function of χCHL at 310 K. With the increase of mole fraction of cholesterol in different myelin sheath membrane model systems, the tilt angles of POPC and cholesterol are shifted towards lower values. The angle distribution for the phosphorous–nitrogen (P–N) vector of the phosphocholine head group and the C10–C13 vector for cholesterol become narrower as χCHL increases, indicating that cholesterol molecules become more ordered and align more towards the bilayer normal.The average P–N angle for POPC lipids and average C10–C13 cholesterol tilt angle as a function of cholesterol mole function for different model myelin sheath membrane systems are given in Table 5. Table 5 data show that increasing χCHL is inversely correlated with POPC and cholesterol tilt angles. Decrease in POPC and cholesterol tilt angles with higher χCHL is primarily a result of the decrease in APL and corresponding increase in lipid order and thickness. Increased APL and reduced chain order at lower χCHL allow more space for angular tilting. The average tilt value of the C10–C13 angle in cholesterol is 11.5° and 64.2° for the P–N angle in POPC lipids in a healthy myelin sheath membrane.
| S. no. | Bilayer | χ CHL | P–N angle (deg) | C13-C10 angle (deg) | Interdigitation of bilayer leaflets i.e. width of overlapping regions Wρ (nm) | Fraction of mass overlap (Iρ) | Fraction of contacts (Ic) |
|---|---|---|---|---|---|---|---|
| PC | Cholesterol | ||||||
| 1 | MS_1 | 0 | 59.775 ± 3.845 | — | 0.311 ± 0.037 | 0.174 ± 0.012 | 0.026 ± 0.002 |
| 2 | MS_3 | 0.11 | 66.327 ± 4.628 | 11.786 ± 0.714 | 0.337 ± 0.037 | 0.169 ± 0.012 | 0.025 ± 0.002 |
| 3 | MS_5 | 0.19 | 63.925 ± 4.884 | 11.291 ± 1.014 | 0.335 ± 0.046 | 0.168 ± 0.017 | 0.024 ± 0.003 |
| 4 | HMS | 0.39 | 64.225 ± 5.923 | 11.516 ± 0.741 | 0.262 ± 0.036 | 0.150 ± 0.013 | 0.026 ± 0.003 |
| 5 | MS_12 | 0.59 | 64.579 ± 7.656 | 10.510 ± 0.502 | 0.268 ± 0.033 | 0.158 ± 0.014 | 0.028 ± 0.003 |
| 6 | MS_16 | 0.79 | 57.417 ± 12.075 | 9.603 ± 0.413 | 0.324 ± 0.042 | 0.177 ± 0.017 | 0.033 ± 0.004 |
| 7 | MS_20 | 0.99 | — | 9.782 ± 0.374 | 0.326 ± 0.026 | 0.207 ± 0.011 | 0.046 ± 0.004 |
At lower χCHL, interdigitation of lipid bilayers decreases with an increase in cholesterol content. The interdigitation of bilayer leaflets increases as we further increase χCHL. The increase in thickness at lower concentrations is driven by acyl-chain ordering, whereas the decrease at higher concentrations is caused by an increase in interdigitation between the ordered lipid tails.
3.8 Hydrogen bonds
To understand the local interactions between different lipid molecules, we have studied hydrogen bonding for different lipids in MS and reference atomistic bilayers. The average number of H-bonds between water and a corresponding lipid or cholesterol molecule is given in Table 6. A POPC bilayer forms an average of 7.35 H-bonds with water molecules; little variation is observed in POPC–water H-bonding in the PC:PE:CHL bilayer (∼7 H-bonds) despite the addition of 35% cholesterol mole fraction. This indicates that the incorporation of hydrophobic cholesterol into the zwitterionic POPC bilayer brings only slight change to the surface chemistry of the bilayer.| Bilayer system | χ CHL | POPC–water | POPE–water | CHOL–water | GalCer–water | SSM–water | POPS–water |
|---|---|---|---|---|---|---|---|
| Reference bilayer systems | |||||||
| PC | 0 | 7.35 | |||||
| PC:PE | 0 | 6.91 | 5.65 | ||||
| PC:PE:CHL | 0.34 | 7.02 | 5.93 | 1.30 | |||
| PC:PE:CHL:GalCer | 0.25 | 6.42 | 5.46 | 1.19 | 5.26 | ||
| PC:PE:CHL:SSM | 0.25 | 6.78 | 5.45 | 1.05 | 7.38 | ||
| Myelin sheath (MS) membrane systems | |||||||
| MS_1 | 0 | 10.35 | 9.10 | 9.68 | 12.78 | 19.51 | |
| MS_5 | 0.19 | 13.79 | 11.33 | 1.91 | 10.33 | 14.02 | 19.83 |
| HMS | 0.39 | 14.35 | 12.06 | 2.41 | 11.88 | 12.28 | 19.06 |
| MS_12 | 0.59 | 16.14 | 12.53 | 2.90 | 13.32 | 13.55 | 20.58 |
| MS_16 | 0.79 | 17.52 | 12.71 | 3.29 | 14.60 | ||
| MS_20 | 0.99 | 3.72 | 16.49 | ||||
POPC contains only H-bond accepting groups (i.e. phosphoryl oxygens and ester group oxygens), whereas POPE, SSM and GalCer lipids have both H-bond acceptor and donor groups. SSM and PC lipids have a similar phosphocholine head group; however, due to the presence of a sphingo chain, SSM lipids contain an amide group, a free hydroxyl group at the C3 carbon, and a trans double bond between C4 and C5. The OH and NH groups of SSM lipids can act both as a H-bond donor and acceptor, whereas the carbonyl oxygen in the acyl chain acts as a H-bond acceptor. The sugar hydroxyl groups of GalCer can also be involved in hydrogen-bond formation with water.
Cholesterol participates in H-bonding with water at the lipid–water interface via its hydroxyl group. In different myelin sheath membrane systems, the number of H-bonds per lipid or cholesterol molecule increases with an increase in the cholesterol mole fraction. Cholesterol acts effectively as a spacer between lipid molecules, enabling more hydrated lipid headgroups.104,105 This increase in H-bonding contributes to the rise in order parameters and compressibility moduli in different MS systems. Water–cholesterol and water–lipid interactions therefore become important in the dynamic balance of forces within the membrane. Finally, it plays a role in determining one of the key structural features of a membrane – namely elasto-mechanical properties. This alteration in H-bonding interaction of lipids and cholesterol with water, as χCHL increases in MS bilayer systems, leads to the formation of local defects in the lipid−water interfacial region which allows restriction of permeation of water/ions into the interior of the bilayer – ultimately disturbing molecular integration within the myelin sheath membrane.
4. Conclusions
Cholesterol-mediated perturbations in neuronal membrane homeostasis and structural integrity are key early events in the molecular pathogenesis of AD. Protein misfolding (proteopathy) and pro-inflammatory conditions (immunopathy) culminate in neuronal death by necrotic and apoptotic mechanisms, both of which are facilitated by altered membrane properties which render neurons more susceptible to cytotoxic insult. Since cholesterol is one of the principal neuronal membrane lipids, normal cholesterol biochemistry is central to membrane health; also, since increased cholesterol composition is present in the myelin sheath (i.e. brain “white matter”), it is not surprising that recent studies have revealed subtle white matter atrophy preceding the conventional pathological biomarkers of AD (amyloid plaques and tau tangles) – contradicting the historical misconception that AD is exclusively a grey matter disease.To permit an improved understanding of the early molecular events of AD and to enable a biophysical understanding of the role of altered cholesterol concentration upon neuronal membrane homeostasis, we performed an extensive series of molecular modelling experiments of varying cholesterol concentrations in model myelin sheath membranes. Specifically, molecular dynamics simulations of model myelin sheath membranes with varying concentrations of cholesterol were performed to understand the effects of impaired χCHL on myelin sheath biophysical and mechanical properties to ascertain whether these altered properties increase vulnerability of myelin membranes to proteopathic and immunopathic attack. High χCHL introduces substantial myelin sheath thickening concurrent with a decrease in area per lipid molecule. This effect is a result of a cholesterol induced straightening of lipid hydrocarbon tails in the direction normal to the membrane plane. Altered cholesterol content not only modulates the tilting angles of different membrane lipids (e.g. POPC) but also changes their bilayer orientation, thereby altering multiple biophysical processes at the membrane interface including increased permeability to oligopeptide insertion. This molecular level understanding of myelin sheath membrane properties as a function of cholesterol content provides useful insights into the preliminary stages of neuronal susceptibility during the early pre-clinical phases of AD.
Author contributions
The manuscript was written through contributions of both authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors thank the Krembil Foundation for financial support. D. F. W. thanks the Canada Research Chair program for support as a Tier I CRC in Protein Misfolding Diseases. The authors acknowledge SciNet and Compute Canada for providing additional computing facilities.References
- W. Jagust, Imaging the evolution and pathophysiology of Alzheimer disease, Nat. Rev. Neurosci., 2018, 19(11), 687–700 CrossRef CAS PubMed.
- P. T. Nelson, I. Alafuzoff, E. H. Bigio, C. Bouras, H. Braak, N. J. Cairns, R. J. Castellani, B. J. Crain, P. Davies, K. Del Tredici, C. Duyckaerts, M. P. Frosch, V. Haroutunian, P. R. Hof, C. M. Hulette, B. T. Hyman, T. Iwatsubo, K. A. Jellinger, G. A. Jicha, E. Kövari, W. A. Kukull, J. B. Leverenz, S. Love, I. R. Mackenzie, D. M. Mann, E. Masliah, A. C. McKee, T. J. Montine, J. C. Morris, J. A. Schneider, J. A. Sonnen, D. R. Thal, J. Q. Trojanowski, J. C. Troncoso, T. Wisniewski, R. L. Woltjer and T. G. Beach, Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature, J. Neuropathol. Exp. Neurol., 2012, 71(5), 362–381 CrossRef PubMed.
- L. M. Ittner and J. Götz, Amyloid-β and tau – A toxic pas de deux in Alzheimer's disease, Nat. Rev. Neurosci., 2011, 12(2), 65–72 CrossRef CAS PubMed.
- M. K. Desai, K. L. Sudol, M. C. Janelsins, M. A. Mastrangelo, M. E. Frazer and W. J. Bowers, Triple-transgenic Alzheimer's disease mice exhibit region-specific abnormalities in brain myelination patterns prior to appearance of amyloid and tau pathology, Glia, 2009, 57(1), 54–65 CrossRef.
- X. Zhan, G. C. Jickling, B. P. Ander, D. Liu, B. Stamova, C. Cox, L. W. Jin, C. DeCarli and F. R. Sharp, Myelin injury and degraded myelin vesicles in Alzheimer's disease, Curr. Alzheimer Res., 2014, 11(3), 232–238 CrossRef CAS PubMed.
- G. Behrendt, K. Baer, A. Buffo, M. A. Curtis, R. L. Faull, M. I. Rees, M. Götz and L. Dimou, Dynamic changes in myelin aberrations and oligodendrocyte generation in chronic amyloidosis in mice and men, Glia, 2013, 61(2), 273–286 CrossRef PubMed.
- Y.-X. Dong, H.-Y. Zhang, H.-Y. Li, P.-H. Liu, Y. Sui and X.-H. Sun, Association between Alzheimer's disease pathogenesis and early demyelination and oligodendrocyte dysfunction, Neural Regen. Res., 2018, 13(5), 908–914 CrossRef PubMed.
- F. Caso, F. Agosta and M. Filippi, Insights into white matter damage in Alzheimer's disease: From postmortem to in vivo diffusion tensor MRI studies, Neuro-degener. Dis., 2016, 16(1–2), 26–33 CrossRef.
- K. J. Chang, S. A. Redmond and J. R. Chan, Remodeling myelination: implications for mechanisms of neural plasticity, Nat. Neurosci., 2016, 19(2), 190–197 CrossRef CAS PubMed.
- Y. Tsukada and T. Kurihara, Myelin: Biology and chemistry, CRC Press, Inc., Boca Raton, FL, 1992 Search PubMed.
- R. J. M. Franklin and C. Ffrench-Constant, Regenerating CNS myelin – From mechanisms to experimental medicines, Nat. Rev., 2017, 18(12), 753–769 CrossRef CAS PubMed.
- A. F. Lloyd and V. E. Miron, The pro-remyelination properties of microglia in the central nervous system, Nat. Rev. Neurol., 2019, 15(8), 447–458 CrossRef PubMed.
- B. E. Flanary, N. W. Sammons, C. Nguyen, D. Walker and W. J. Streit, Evidence that aging and amyloid promote microglial cell senescence, Rejuvenation Res., 2007, 10(1), 61–74 CrossRef CAS PubMed.
- S. Saez-Atienzar and E. Masliah, Cellular senescence and Alzheimer disease: The egg and the chicken scenario, Nat. Rev. Neurosci., 2020, 21(8), 433–444 CrossRef CAS PubMed.
- P. Zhang, Y. Kishimoto, I. Grammatikakis, K. Gottimukkala, R. G. Cutler, S. Zhang, K. Abdelmohsen, V. A. Bohr, J. Misra Sen, M. Gorospe and M. P. Mattson, Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer's disease model, Nat. Neurosci., 2019, 22(5), 719–728 CrossRef CAS.
- J. Xu, S. Chen, S. H. Ahmed, H. Chen, G. Ku, M. P. Goldberg and C. Y. Hsu, Amyloid-β peptides are cytotoxic to oligodendrocytes, J. Neurosci., 2001, 21(1), RC118–RC118 CrossRef CAS PubMed.
- T.-H. Chu, K. Cummins, J. S. Sparling, S. Tsutsui, C. Brideau, K. P. R. Nilsson, J. T. Joseph and P. K. Stys, Axonal and myelinic pathology in 5xFAD Alzheimer's mouse spinal cord, PLoS One, 2017, 12(11), e0188218 CrossRef PubMed.
- L. Gu, D. Wu, X. Tang, X. Qi, X. Li, F. Bai, X. Chen, Q. Ren and Z. Zhang, Myelin changes at the early stage of 5XFAD mice, Brain Res. Bull., 2018, 137, 285–293 CrossRef CAS PubMed.
- A. N. Davison and M. Wajda, Cerebral lipids in multiple sclerosis, J. Neurochem., 1962, 9, 427–432 CrossRef CAS PubMed.
- A. N. Davison, Brain sterol metabolism, Adv. Lipid Res., 1965, 3, 171–196 CrossRef CAS PubMed.
- R. H. Laatsch, M. W. Kies, S. Gordon and E. C. Alvord, Jr., The encephalomyelitic activity of myelin isolated by ultracentrifugation, J. Exp. Med., 1962, 115(4), 777–788 CrossRef CAS PubMed.
- L. Anchisi, S. Dessi, A. Pani and A. Mandas, Cholesterol homeostasis: A key to prevent or slow down neurodegeneration, Front. Physiol., 2013, 3, 486 Search PubMed.
- L. Cantuti-Castelvetri, D. Fitzner, M. Bosch-Queralt, M.-T. Weil, M. Su, P. Sen, T. Ruhwedel, M. Mitkovski, G. Trendelenburg, D. Lütjohann, W. Möbius and M. Simons, Defective cholesterol clearance limits remyelination in the aged central nervous system, Science, 2018, 359(6376), 684–688 CrossRef CAS PubMed.
- X.-K. Tong, L. J. Trigiani and E. Hamel, High cholesterol triggers white matter alterations and cognitive deficits in a mouse model of cerebrovascular disease: Benefits of simvastatin, Cell Death Dis., 2019, 10(2), 89 CrossRef PubMed.
- G. Saher, B. Brügger, C. Lappe-Siefke, W. Möbius, R.-i. Tozawa, M. C. Wehr, F. Wieland, S. Ishibashi and K.-A. Nave, High cholesterol level is essential for myelin membrane growth, Nat. Neurosci., 2005, 8(4), 468–475 CrossRef CAS PubMed.
- L. Dai, L. Zou, L. Meng, G. Qiang, M. Yan and Z. Zhang, Cholesterol metabolism in neurodegenerative diseases: Molecular mechanisms and therapeutic targets, Mol. Neurobiol., 2021, 58(5), 2183–2201 CrossRef CAS PubMed.
- T. L. Steck and Y. Lange, Transverse distribution of plasma membrane bilayer cholesterol: Picking sides, Traffic, 2018, 19(10), 750–760 CrossRef CAS PubMed.
- I. Bera and J. B. Klauda, Molecular simulations of mixed lipid bilayers with sphingomyelin, glycerophospholipids, and cholesterol, J. Phys. Chem. B, 2017, 121(20), 5197–5208 CrossRef CAS PubMed.
- M. Adams, E. Wang, X. Zhuang and J. B. Klauda, Simulations of simple Bovine and Homo sapiens outer cortex ocular lens membrane models with a majority concentration of cholesterol, Biochim. Biophys. Acta, Biomembr., 2018, 1860(10), 2134–2144 CrossRef CAS.
- C. T. Boughter, V. Monje-Galvan, W. Im and J. B. Klauda, Influence of cholesterol on phospholipid bilayer structure and dynamics, J. Phys. Chem. B, 2016, 120(45), 11761–11772 CrossRef CAS.
- E. Wang and J. B. Klauda, Examination of mixtures containing sphingomyelin and cholesterol by molecular dynamics simulations, J. Phys. Chem. B, 2017, 121(18), 4833–4844 CrossRef CAS PubMed.
- P. Khakbaz and J. B. Klauda, Probing the importance of lipid diversity in cell membranes via molecular simulation, Chem. Phys. Lipids, 2015, 192, 12–22 CrossRef CAS PubMed.
- E. Wang and J. B. Klauda, Structure and permeability of ceramide bilayers and multilayers, J. Phys. Chem. B, 2019, 123(11), 2525–2535 CrossRef PubMed.
- M. Saeedimasine, A. Montanino, S. Kleiven and A. Villa, Elucidating axonal injuries through molecular modelling of myelin sheaths and nodes of Ranvier, Frontiers in Molecular Biosciences, 2021, 8, 669897, DOI:10.3389/fmolb.2021.669897.
- A. Montanino, M. Saeedimasine, A. Villa and S. Kleiven, Localized axolemma deformations suggest mechanoporation as axonal injury trigger, Front Neurol, 2020, 11, 25, DOI:10.3389/fneur.2020.00025.
- M. Saeedimasine, A. Montanino, S. Kleiven and A. Villa, Role of lipid composition on the structural and mechanical features of axonal membranes: A molecular simulation study, Sci. Rep., 2019, 9, 8000, DOI:10.1038/s41598-019-44318-9.
- G. van Meer, D. R. Voelker and G. W. Feigenson, Membrane lipids: Where they are and how they behave, Nat. Rev. Mol. Cell Biol., 2008, 9(2), 112–124 CrossRef CAS PubMed.
- J. L. Sampaio, M. J. Gerl, C. Klose, C. S. Ejsing, H. Beug, K. Simons and A. Shevchenko, Membrane lipidome of an epithelial cell line, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(5), 1903–1907 CrossRef CAS PubMed.
- E. Fahy, S. Subramaniam, R. C. Murphy, M. Nishijima, C. R. H. Raetz, T. Shimizu, F. Spener, G. van Meer, M. J. O. Wakelam and E. A. Dennis, Update of the LIPID MAPS comprehensive classification system for lipids, J. Lipid Res., 2009, 50(Suppl), S9–S14 CrossRef PubMed.
- M. Gupta and D. F. Weaver, Axonal plasma membrane-mediated toxicity of cholesterol in Alzheimer’s disease: A microsecond molecular dynamics study, Biophys. Chem., 2022, 281, 106718, DOI:10.1016/j.bpc.2021.106718.
- B. Alberts, Molecular Biology of the Cell, 6th edn, 2015 Search PubMed.
- M. Saeedimasine, A. Montanino, S. Kleiven and A. Villa, Role of lipid composition on the structural and mechanical features of axonal membranes: A molecular simulation study, Sci. Rep., 2019, 9(1), 8000 CrossRef PubMed.
- M. N. Rasband and W. B. Macklin, Chapter 10 – Myelin structure and biochemistry, in Basic Neurochemistry (Eighth Edition), ed. S. T. Brady, G. J. Siegel, R. W. Albers and D. L. Price, Academic Press, New York, 2012, pp. 180–199 Search PubMed.
- T. Skripuletz, R. A. Linker and M. Stangel, The choline pathway as a strategy to promote central nervous system (CNS) remyelination, Neural Regen. Res., 2015, 10(9), 1369–1370 CrossRef CAS PubMed.
- C. Heffernan, M. R. Jain, T. Liu, H. Kim, K. Barretto, H. Li and P. Maurel, Nectin-like 4 complexes with choline transporter-like protein-1 and regulates schwann cell choline homeostasis and lipid biogenesis in vitro, J. Biol. Chem., 2017, 292(11), 4484–4498 CrossRef CAS PubMed.
- Z. Li, Y. Fan, J. Liu, Y. Li, C. Huan, H. H. Bui, M. S. Kuo, T. S. Park, G. Cao and X. C. Jiang, Impact of sphingomyelin synthase 1 deficiency on sphingolipid metabolism and atherosclerosis in mice, Arterioscler., Thromb., Vasc. Biol., 2012, 32(7), 1577–1584 CrossRef CAS PubMed.
- J. Xue, Y. Yu, X. Zhang, C. Zhang, Y. Zhao, B. Liu, L. Zhang, L. Wang, R. Chen, X. Gao, P. Jiao, G. Song, X. C. Jiang and S. Qin, Sphingomyelin synthase 2 inhibition ameliorates cerebral ischemic reperfusion injury through reducing the recruitment of toll-like receptor 4 to lipid rafts, J. Am. Heart Assoc., 2019, 8(22), e012885 Search PubMed.
- M. Chami, R. Halmer, L. Schnoeder, K. Anne Becker, C. Meier, K. Fassbender, E. Gulbins and S. Walter, Acid sphingomyelinase deficiency enhances myelin repair after acute and chronic demyelination, PLoS One, 2017, 12(6), e0178622 CrossRef PubMed.
- T. Coetzee, N. Fujita, J. Dupree, R. Shi, A. Blight, K. Suzuki, K. Suzuki and B. Popko, Myelination in the absence of galactocerebroside and sulfatide: Normal structure with abnormal function and regional instability, Cell, 1996, 86(2), 209–219 CrossRef CAS PubMed.
- A. Bosio, E. Binczek and W. Stoffel, Functional breakdown of the lipid bilayer of the myelin membrane in central and peripheral nervous system by disrupted galactocerebroside synthesis, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(23), 13280–13285 CrossRef CAS.
- I. Zöller, M. Meixner, D. Hartmann, H. Büssow, R. Meyer, V. Gieselmann and M. Eckhardt, Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration, J. Neurosci., 2008, 28(39), 9741–9754 CrossRef PubMed.
- K. A. Potter, M. J. Kern, G. Fullbright, J. Bielawski, S. S. Scherer, S. W. Yum, J. J. Li, H. Cheng, X. Han, J. K. Venkata, P. A. A. Khan, B. Rohrer and H. Hama, Central nervous system dysfunction in a mouse model of FA2H deficiency, Glia, 2011, 59(7), 1009–1021 CrossRef PubMed.
- J. Marcus, S. Honigbaum, S. Shroff, K. Honke, J. Rosenbluth and J. L. Dupree, Sulfatide is essential for the maintenance of CNS myelin and axon structure, Glia, 2006, 53(4), 372–381 CrossRef CAS PubMed.
- T. Hoshi, A. Suzuki, S. Hayashi, K. Tohyama, A. Hayashi, Y. Yamaguchi, K. Takeuchi and H. Baba, Nodal protrusions, increased Schmidt–Lanterman incisures, and paranodal disorganization are characteristic features of sulfatide-deficient peripheral nerves, Glia, 2007, 55(6), 584–594 CrossRef PubMed.
- A. Hayashi, N. Kaneko, C. Tomihira and H. Baba, Sulfatide decrease in myelin influences formation of the paranodal axo-glial junction and conduction velocity in the sciatic nerve, Glia, 2013, 61(4), 466–474 CrossRef PubMed.
- D. Wheeler, V. V. R. Bandaru, P. A. Calabresi, A. Nath and N. J. Haughey, A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis, Brain, 2008, 131(Pt 11), 3092–3102 CrossRef PubMed.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes, Biophys. J., 2009, 97(1), 50–58 CrossRef CAS PubMed.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, CHARMM-GUI membrane builder toward realistic biological membrane simulations, J. Comput. Chem., 2014, 35(27), 1997–2004 CrossRef CAS PubMed.
- J. Lee, D. S. Patel, J. Ståhle, S.-J. Park, N. R. Kern, S. Kim, J. Lee, X. Cheng, M. A. Valvano, O. Holst, Y. A. Knirel, Y. Qi, S. Jo, J. B. Klauda, G. Widmalm and W. Im, CHARMM-GUI membrane builder for complex biological membrane simulations with glycolipids and lipoglycans, J. Chem. Theory Comput., 2019, 15(1), 775–786 CrossRef CAS PubMed.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O’Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell and R. W. Pastor, Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types, J. Phys. Chem. B, 2010, 114(23), 7830–7843 CrossRef CAS PubMed.
- R. M. Venable, A. J. Sodt, B. Rogaski, H. Rui, E. Hatcher, A. D. MacKerell Jr, R. W. Pastor and J. B. Klauda, CHARMM all-atom additive force field for sphingomyelin: Elucidation of hydrogen bonding and of positive curvature, Biophys. J., 2014, 107(1), 134–145 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, Scalable molecular dynamics with NAMD, J. Comput. Chem., 2005, 26(16), 1781–1802 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79(2), 926–935 CrossRef CAS.
- M. Gupta, E. F. Da Silva and H. F. Svendsen, Explicit solvation shell model and continuum solvation models for solvation energy and pKa determination of amino acids, J. Chem. Theory Comput., 2013, 9(11), 5021–5037 CrossRef CAS PubMed.
- M. Gupta, E. F. Da Silva and H. F. Svendsen, Postcombustion CO2 capture solvent characterization employing the explicit solvation shell model and continuum solvation models, J. Phys. Chem. B, 2016, 120(34), 9034–9050 CrossRef CAS PubMed.
- S. Miyamoto and P. A. Kollman, Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models, J. Comput. Chem., 1992, 13(8), 952–962 CrossRef CAS.
- J. P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, J. Comput. Phys., 1977, 23(3), 327–341 CrossRef CAS.
- P. P. Ewald, Die Berechnung optischer und elektrostatischer Gitterpotentiale, Ann. Phys., 1921, 369(3), 253–287 CrossRef.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, Constant pressure molecular dynamics simulation: The Langevin piston method, J. Chem. Phys., 1995, 103(11), 4613–4621 CrossRef CAS.
- W. G. Hoover, Canonical dynamics: Equilibrium phase-space distributions, Phys. Rev. A: At., Mol., Opt. Phys., 1985, 31(3), 1695–1697 CrossRef PubMed.
- M. P. Allen and D. J. Tildesley, Computer Simulation of Liquids, 2nd edn, Oxford University Press, Oxford, 2017, p. 640 Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14(1), 33–38 CrossRef CAS PubMed.
- C. B. Barber, D. P. Dobkin and H. Huhdanpaa, The quickhull algorithm for convex hulls, ACM Trans. Math. Softw, 1996, 22(4), 469–483 CrossRef.
- J. Seelig and N. Waespe-Sarcevic, Molecular order in cis and trans unsaturated phospholipid bilayers, Biochemistry, 1978, 17(16), 3310–3315 CrossRef CAS PubMed.
- W. Rawicz, K. C. Olbrich, T. McIntosh, D. Needham and E. Evans, Effect of chain length and unsaturation on elasticity of lipid bilayers, Biophys. J., 2000, 79(1), 328–339 CrossRef CAS PubMed.
- E. A. Evans, R. Waugh and L. Melnik, Elastic area compressibility modulus of red cell membrane, Biophys. J., 1976, 16(6), 585–595 CrossRef CAS PubMed.
- R. Guixà-González, I. Rodriguez-Espigares, J. M. Ramírez-Anguita, P. Carrió-Gaspar, H. Martinez-Seara, T. Giorgino and J. Selent, MEMBPLUGIN: Studying membrane complexity in VMD, Bioinformatics, 2014, 30(10), 1478–1480 CrossRef PubMed.
- G. W. Stockton and C. P. Smith, A deuterium nuclear magnetic resonance study of the condensing effect of cholesterol on egg phosphatidylcholine bilayer membranes. I. Perdeuterated fatty acid probes, Chem. Phys. Lipids, 1976, 17(2), 251–263 CrossRef CAS.
- P. A. Hyslop, B. Morel and R. D. Sauerheber, Organization and interaction of cholesterol and phosphatidylcholine in model bilayer membranes, Biochemistry, 1990, 29(4), 1025–1038 CrossRef CAS PubMed.
- G. Pabst, M. Rappolt, H. Amenitsch and P. Laggner, Structural information from multilamellar liposomes at full hydration: Full q-range fitting with high quality X-ray data, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62(3B), 4000–4009 CrossRef CAS PubMed.
- J. M. Smaby, M. M. Momsen, H. L. Brockman and R. E. Brown, Phosphatidylcholine acyl unsaturation modulates the decrease in interfacial elasticity induced by cholesterol, Biophys. J., 1997, 73(3), 1492–1505 CrossRef CAS PubMed.
- N. Kučerka, S. Tristram-Nagle and J. F. Nagle, Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains, J. Membr. Biol., 2006, 208(3), 193–202 CrossRef PubMed.
- T. J. McIntosh, The effect of cholesterol on the structure of phosphatidylcholine bilayers, Biochem. Biophys. Acta, 1978, 513(1), 43–58 CAS.
- F. de Meyer and B. Smit, Effect of cholesterol on the structure of a phospholipid bilayer, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(10), 3654–3658 CrossRef CAS PubMed.
- W. C. Hung, M. T. Lee, F. Y. Chen and H. W. Huang, The condensing effect of cholesterol in lipid bilayers, Biophys. J., 2007, 92(11), 3960–3967 CrossRef CAS PubMed.
- M. I. Oh, M. Gupta, C. I. Oh and D. F. Weaver, Understanding the effect of nanoconfinement on the structure of water hydrogen bond networks, Phys. Chem. Chem. Phys., 2019, 21(47), 26237–26250 RSC.
- M. Gupta, H. J. Lee, C. J. Barden and D. F. Weaver, The blood–brain barrier (BBB) score, J. Med. Chem., 2019, 62(21), 9824–9836 CrossRef CAS PubMed.
- M. Zhernenkov, D. Bolmatov, D. Soloviov, K. Zhernenkov, B. P. Toperverg, A. Cunsolo, A. Bosak and Y. Q. Cai, Revealing the mechanism of passive transport in lipid bilayers via phonon-mediated nanometre-scale density fluctuations, Nat. Commun., 2016, 7(1), 11575 CrossRef CAS PubMed.
- M. Gupta, T. Bogdanowicz, M. A. Reed, C. J. Barden and D. F. Weaver, The brain exposure efficiency (BEE) score, ACS Chem. Neurosci., 2020, 11(2), 205–224 CrossRef CAS PubMed.
- L. Pini, M. Pievani, M. Bocchetta, D. Altomare, P. Bosco, E. Cavedo, S. Galluzzi, M. Marizzoni and G. B. Frisoni, Brain atrophy in Alzheimer's disease and aging, Ageing Res. Rev., 2016, 30, 25–48 CrossRef PubMed.
- H. Binder and K. Gawrisch, Effect of unsaturated lipid chains on dimensions, molecular order and hydration of membranes, J. Phys. Chem. B, 2001, 105(49), 12378–12390 CrossRef CAS.
- D. Needham and R. S. Nunn, Elastic deformation and failure of lipid bilayer membranes containing cholesterol, Biophys. J., 1990, 58(4), 997–1009 CrossRef CAS PubMed.
- E. Evans and D. Needham, Physical properties of surfactant bilayer membranes: thermal transitions, elasticity, rigidity, cohesion and colloidal interactions, J. Phys. Chem., 1987, 91(16), 4219–4228 CrossRef CAS.
- J. Henriksen, A. C. Rowat, E. Brief, Y. W. Hsueh, J. L. Thewalt, M. J. Zuckermann and J. H. Ipsen, Universal behavior of membranes with sterols, Biophys. J., 2006, 90(5), 1639–1649 CrossRef CAS PubMed.
- L. R. Arriaga, I. López-Montero, F. Monroy, G. Orts-Gil, B. Farago and T. Hellweg, Stiffening effect of cholesterol on disordered lipid phases: A combined neutron spin echo + dynamic light scattering analysis of the bending elasticity of large unilamellar vesicles, Biophys. J., 2009, 96(9), 3629–3637 CrossRef CAS PubMed.
- S. Chakraborty, M. Doktorova, T. R. Molugu, F. A. Heberle, H. L. Scott, B. Dzikovski, M. Nagao, L.-R. Stingaciu, R. F. Standaert, F. N. Barrera, J. Katsaras, G. Khelashvili, M. F. Brown and R. Ashkar, How cholesterol stiffens unsaturated lipid membranes, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(36), 21896–21905 CrossRef CAS PubMed.
- K. I. Pakkanen, L. Duelund, K. Qvortrup, J. S. Pedersen and J. H. Ipsen, Mechanics and dynamics of triglyceride-phospholipid model membranes: Implications for cellular properties and function, Biochim. Biophys. Acta, Biomembr., 2011, 1808(8), 1947–1956 CrossRef CAS PubMed.
- H. Bouvrais, L. Duelund and J. H. Ipsen, Buffers affect the bending rigidity of model lipid membranes, Langmuir, 2014, 30(1), 13–16 CrossRef CAS PubMed.
- J. Henriksen, A. C. Rowat and J. H. Ipsen, Vesicle fluctuation analysis of the effects of sterols on membrane bending rigidity, Eur. Biophys. J., 2004, 33(8), 732–741 CrossRef CAS PubMed.
- N. Kucerka, S. Tristram-Nagle and J. F. Nagle, J. Membr. Biol., 2005, 205, 193 Search PubMed.
- H. Bouvrais, P. Méléard, T. Pott, K. J. Jensen, J. Brask and J. H. Ipsen, Softening of POPC membranes by magainin, Biophys. Chem., 2008, 137(1), 7–12 CrossRef CAS PubMed.
- H. Bouvrais, T. Pott, L. A. Bagatolli, J. H. Ipsen and P. Méléard, Impact of membrane-anchored fluorescent probes on the mechanical properties of lipid bilayers, Biochim. Biophys. Acta, Biomembr., 2010, 1798(7), 1333–1337 CrossRef CAS PubMed.
- G. Niggemann, M. Kummrow and W. Helfrich, The bending rigidity of phosphatidylcholine bilayers: Dependences on experimental method, sample cell sealing and temperature, J. Phys. II, 1995, 5(3), 413–425 CrossRef CAS.
- J. Gallová, D. Uhríková, N. Kučerka, S. Doktorovová, S. S. Funari, J. Teixeira and P. Balgavý, The effects of cholesterol and β-sitosterol on the structure of saturated diacylphosphatidylcholine bilayers, Eur. Biophys. J., 2011, 40(2), 153–163 CrossRef PubMed.
- N. Kučerka, J. Pencer, M. P. Nieh and J. Katsaras, Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles, Eur. Phys. J. E: Soft Matter Biol. Phys., 2007, 23(3), 247–254 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S4 of APL, component APL and bilayer thickness for healthy myelin sheath membrane (HMS) as a function of equilibration and production time; the average P–N angle for POPC, POPE, POPS, SSM lipids and average cholesterol tilt angle (C13–C10) of reference membrane systems at 310 K; the average fraction of contacts and mass overlap, interdigitation within lipid bilayers systems of reference membrane systems; Structure of various lipids and cholesterol molecules used in bilayer systems. See DOI: 10.1039/d1cp03844c |
| This journal is © the Owner Societies 2022 |