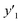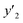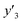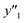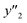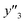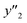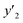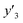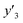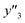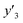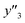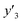Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sparse modeling for small data: case studies in controlled synthesis of 2D materials†
Yuri
Haraguchi
a,
Yasuhiko
Igarashi
bc,
Hiroaki
Imai
 a and
Yuya
Oaki
a and
Yuya
Oaki
 *ac
*ac
aDepartment of Applied Chemistry, Faculty of Science and Technology, Keio University, 3-14-1 Hiyoshi, Kohoku-ku, Yokohama 223-8522, Japan. E-mail: oakiyuya@applc.keio.ac.jp
bFaculty of Engineering, Information and Systems, University of Tsukuba, 1-1-1 Tennodai, Tsukuba 305-8573, Japan
cJST, PRESTO, 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
First published on 18th December 2021
Abstract
Data-scientific approaches have permeated into chemistry and materials science. In general, these approaches are not easily applied to small data, such as experimental data in laboratories. Our group has focused on sparse modeling (SpM) for small data in materials science and chemistry. The controlled synthesis of 2D materials, involving improvement of the yield and control of the size, was achieved by SpM coupled with our chemical perspectives for small data (SpM-S). In the present work, the conceptual and methodological advantages of SpM-S were studied using real experimental datasets to enable comparison with other machine learning (ML) methods, such as neural networks. The training datasets consisted of ca. 40 explanatory variables (xn) and 50 objective variables (y) regarding the yield, size, and size-distribution of exfoliated nanosheets. SpM-S provided more straightforward, generalizable, and interpretable prediction models and better prediction accuracy for new experiments as an unknown test dataset. The results indicate that machine learning coupled with our experience, intuition, and perspective can be applied to small data in a variety of fields.
Introduction
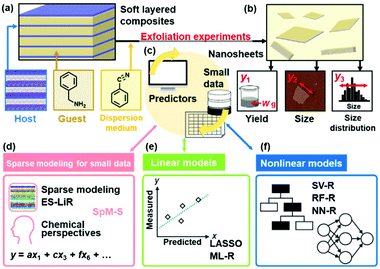
Machine learning (ML) on big data, such as deep learning, has been a powerful tool in our daily life.1,2 In materials science, discovery of new materials, optimization of processes, and enhancement of performances have been achieved by data-driven methods.3–15 In chemistry, new functional molecules and catalysts have been found using ML. Combination of ML and robotic equipment further accelerates discovery.16–19 However, the quantity of data is not always sufficient for effective use of ML. Small data generally causes problems,20 such as overtraining. While a well-trained predictor with high correlation between the estimated and actual values is prepared on small training data, the prediction accuracy lowers on unknown test data. Therefore, a variety of small data have been left with development of artificial intelligence. For example, experimental scientists have their own small data including successes and failures. If such small data is utilized by ML, research projects can be accelerated without wasting time, money, and effort. Specific methods, such as transfer learning, are developed to address the lack of data.21 However, additional data is eventually required to improve the prediction accuracy. Although data augmentation is used for image data to construct robust predictors,22 the method is not easily applied to small data in chemistry and materials science. In addition, improvement of the prediction accuracy was achieved by imaging the missing values in data.23 New concepts and modeling methods are required for utilization of small data, such as experimental and literature data in laboratories. Our group has studied SpM coupled with chemical perspectives for small data, namely SpM-S, in chemistry and materials science.24–30 The method facilitated the controlled synthesis of 2D materials and exploration of new organic anodes using our own small experimental data. In the present work, the advantages of SpM-S were studied in a data-scientific manner to enable comparison with other ML methods (Fig. 1).SpM is a general concept for explanation of whole data using a limited number of significant descriptors.31,32 The method has been widely used for data compression in image analyses,33–35 such as diagnosis using magnetic resonance imaging. In SpM, the dimension of data is reduced by ML. However, only the automatic selection of descriptors causes rejection of the significant descriptors and/or adoption of insignificant descriptors, particularly in small data. In addition, noise, errors, and outliers in small data have negative effects on the extraction of the descriptors and prediction accuracy of the constructed models. Our group has studied coupling our experience, perspective, and intuition with SpM in all the processes including preparation of the dataset and selection of the descriptors toward development of small-data-driven materials science and chemistry.24–30 In the initial stage, a small yet balanced dataset is prepared using experimental, calculational, and literature data. The descriptors are extracted by ML, such as exhaustive search with linear regression (ES-LiR) and minimax concave penalty and penalized linear unbiased selection algorithm (MC+).36–38 Then, the significant descriptors are selected from the results of ML on the basis of our chemical perspectives as the prior knowledge. This process facilitates finding the significant descriptors with the chemically reasonable correlation to the targets and avoiding overtraining with adoption of insignificant descriptors. The chemical perspectives and implicit knowledge can be included in the predictors. The linear-regression models are constructed using a limited number of the selected descriptors. The straightforward linear regression models are applied to the exploration of optimized conditions and new materials. In the present work, SpM-S was compared with the other ML methods on the same datasets to elucidate the advantages. Three real experimental datasets about the yield, size, and size distribution of exfoliated transition-metal-oxide nanosheets were used for the training and validation (Fig. 1a–c). Linear regression was performed by least absolute shrinkage and selection operator (LASSO) with variable selection and multiple linear regression (ML-R) without variable selection (Fig. 1e). Nonlinear-regression models were constructed by support vector regression (SV-R), random forest regression (RF-R), and neural network regression (NN-R) (Fig. 1f). The results indicate that SpM-S provided more accurate, generalizable, and interpretable prediction models. Moreover, combination of ML and researchers can enable the construction of better predictors even on small data.
Nanosheets, such as monolayers and few-layers, have attracted much interest as 2D materials with characteristic properties.39–41 Although a typical route to obtain 2D materials is exfoliation of layered materials, the processes still have challenges. In general, exfoliation of precursor layered materials is not easily controlled by experimental parameters because of the unpredictable down-sizing processes in the liquid phase. Time- and effort-consuming microscopy analyses are required for the characterization of the resultant nanosheets. Therefore, new approaches are needed to achieve efficient control of the exfoliation behavior toward tailored synthesis of nanosheets. Application of ML to 2D materials has been studied in recent years. ML has been applied to the optimization of processes for bottom-up synthesis,42 exploration of precursor layered compounds,43 estimation of the thickness combined with image analyses on the microscopy images,44,45 and improvement of the properties.46,47 In these previous studies, ML-assisted approaches were not applied to control the exfoliation processes providing 2D materials. Our group has applied ML, namely SpM-S, to achieve high-yield and size-controlled synthesis of nanosheets through controlled exfoliation.24–29 However, the data-scientific validity for construction of the predictors was not studied in our previous reports. In the present work, the validity and advantages of our SpM-S were studied using the small datasets about the yield, lateral size, and lateral-size distribution of nanosheets in comparison with other ML methods.
Results and discussion
Training datasets based on experimental data
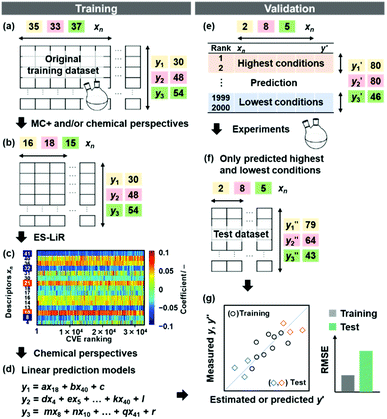
The small datasets for training were prepared with the experimental data in our group.24–27 The soft layered composites of transition-metal oxides and organic guests were exfoliated into nanosheets in organic dispersion media (Fig. 1a and b). The exfoliation behavior was different with changes in the combinations of the host layers, intercalated guests, and dispersion media. The yields (y1), size (y2), and size distribution (y3) of the exfoliated nanosheets as the objective variables (y) were measured in the different host–guest–medium combinations (Fig. 1a, b and 2a).24–27 The precursor layered materials were exfoliated in dispersion media under 60 °C for 5 days. Then, the dispersion liquids containing the exfoliated nanosheets were filtered using filters with pore size larger than 2 μm to remove the unexfoliated bulky materials. Then, the nanosheets in the colloidal liquids were collected using a membrane filter with a pore size of 0.1 μm. The yield (y1) was defined as the weight percentage of the collected exfoliated nanosheets (W) to the initial weight of the precursor layered materials (W0), i.e. 100 × W/W0. The lateral size (y2) was represented by the size-reduction rate (RL = Lave/L0) calculated from the average lateral sizes of the precursor layered materials (L0) and exfoliated nanosheets (Lave). The coefficient of variation of the lateral size (LCV = σ/Lave) was calculated from the average lateral size (Lave) and its standard deviation (σ) to define the size distribution (y3). The Lave and σ values were measured by dynamic light scattering (DLS) of the dispersion liquids containing the nanosheets to achieve high-throughput analysis without time-consuming microscopy techniques.26,27 Although the precise size was not measured by DLS analysis on the dispersion liquid, the lateral size and its distribution were roughly estimated without microscopy analyses. The particle size measured by DLS analysis had a rough correlation with the lateral size measured on the images of transmission electron microscopy (TEM).26,27,48On the basis of our chemical perspectives, the explanatory variables (xn: n = 1–41) were prepared by the calculation and literature data of physicochemical parameters related to the objective variables (Table 1). The original training datasets contained the following data: 30 y1 and 35 xn (n = 1–26, 28–32, 34, 36, 39, 40) for the yield, 48 y2 and 33 xn (n = 1, 3–24, 26, 28–32, 34, 36, 39, 40) for the size, and 54 y3 and 37 xn (n = 1, 3–24, 26, 27, 30–41) for the size distribution (Fig. 2a and Tables S1–S4 in the ESI†).24–27 The mean, standard deviation (SD), and sample number (n) of the training and test data are summarized in Table S1 in the ESI.† The predictors were constructed by SpM-S and the other ML methods using these original datasets about the yield, size, and size distribution of the nanosheets.
| n/— | Parameters | x n for |
|---|---|---|
| a Literature data. b Calculation data. c Experimental data. | ||
| Dispersion media | ||
| 1 | Molecular weight | y 1, y2, y3 |
| 2 | Molecular lengthb | y 1 |
| 3 | Melting pointa | y 1, y2, y3 |
| 4 | Boiling pointa | y 1, y2, y3 |
| 5 | Densitya | y 1, y2, y3 |
| 6 | Relative permittivitya | y 1, y2, y3 |
| 7 | Vapor pressurea | y 1, y2, y3 |
| 8 | Viscositya | y 1, y2, y3 |
| 9 | Refractive indexa | y 1, y2, y3 |
| 10 | Surface tensiona | y 1, y2, y3 |
| 11 | Heat capacityb | y 1, y2, y3 |
| 12 | Entropyb | y 1, y2, y3 |
| 13 | Enthalpyb | y 1, y2, y3 |
| 14 | Dipole momentb | y 1, y2, y3 |
| 15 | Polarizabilityb | y 1, y2, y3 |
| 16 | HSP-dispersion termb | y 1, y2, y3 |
| 17 | HSP-polarity termb | y 1, y2, y3 |
| 18 | HSP-hydrogen bonding termb | y 1, y2, y3 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Guest molecules | ||
| 19 | Molecular weight | y 1, y2, y3 |
| 20 | Polarizabilityb | y 1, y2, y3 |
| 21 | Dipole momentb | y 1, y2, y3 |
| 22 | Heat capacityb | y 1, y2, y3 |
| 23 | Entropyb | y 1, y2, y3 |
| 24 | Enthalpyb | y 1, y2, y3 |
| 25 | Molecular lengthb | y 1 |
| 26 | Layer distancec | y 1, y2, y3 |
| 27 | Layer distance expansionc | y 3 |
| 28 | Composition (x)c | y 1, y2 |
| 29 | Interlayer densityc | y 1, y2 |
| 30 | HSP-dispersion termb | y 1, y2, y3 |
| 31 | HSP-polarity termb | y 1, y2, y3 |
| 32 | HSP-hydrogen bonding termb | y 1, y2, y3 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Guest–medium combinations | ||
| 33 | Δ Polarizability (=x14 − x19)b | y 3 |
| 34 | Δ Polarizability (=|x29|)b | y 1, y2, y3 |
| 35 | Δ Dipole moment (=x13 − x20)b | y 3 |
| 36 | Δ Dipole moment (=|x31|)b | y 1, y2, y3 |
| 37 | Product of dipole moment (=x13 × x20)b | y 3 |
| 38 | Δ Heat capacity (=x10 − x21)b | y 3 |
| 39 | Δ Heat capacity (=|x35|)b | y 1, y2, y3 |
| 40 | HSP distanceb | y 1, y2, y3 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Host layers | ||
| 41 | Bulk sizec | y 3 |
Construction of predictors based on SpM-S
The number of xn was reduced to lower the calculation cost for the following ES-LiR. In the initial screening, ten to twenty descriptors were extracted by ML and/or our chemical perspective. For example, the descriptors were extracted using MC+ and our prior knowledge.27 The number of xn (N) was reduced to N = 16 for y1, N = 18 for y2, and N = 15 for y3 (Fig. 2b). Then, linear-regression models were exhaustively prepared by ES-LiR for all the possible combinations of the descriptors, namely 2N − 1 combinations, on the screened datasets. For example, a total of 3.3 × 104 to 2.6× 105 patterns of the linear-regression models were prepared in the range of N = 15–18. The models were sorted in the ascending order of cross-validation error (CVE) values. The coefficients of each descriptor with positive and negative values are summarized in the weight diagram using specific colors (Fig. 2c). The weight diagram indicates that the more densely colored and darker descriptors have the stronger correlation. Therefore, the significant descriptors were selected by our chemical perspectives on the weight diagram. The exfoliation of the layered composites was initiated by the intercalation of the organic dispersion media in the interlayer space based on the guests.29 The guest–medium combinations and their affinity have effects on the exfoliation behavior in the vertical direction and fracture behavior in the lateral direction. In addition, the flexibility of the layers is different in the host layered materials. These chemical insights were applied to the selection of the descriptors.24–28 A detailed discussion regarding the contribution of each descriptor to the yield, lateral size, and size distribution is provided in our previous studies.25–27 In particular, such prior knowledge is important for selection of the descriptors using SpM on small data. In this manner, the linear-regression models were constructed by the selected descriptors for estimation of y1, y2, and y3 (Fig. 2d). The estimated values, namely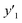 ,
, 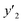 , and
, and 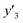 , are represented by the following equations (eqn (1)–(3)).25–27
, are represented by the following equations (eqn (1)–(3)).25–27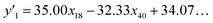 | (1) |
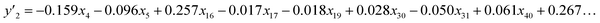 | (2) |
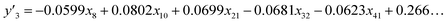 | (3) |
As xn was converted to the normalized frequency distribution such that the mean is 0 and standard deviation is 1, the coefficients indicate the weight of the descriptors. The correlation between the estimated (y′) and measured (y) values was represented by a root-mean-square error (RMSE) of 17.9% for 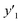 , 0.091 for
, 0.091 for  , and 0.116 for
, and 0.116 for  (the black circles in Fig. 3a).
(the black circles in Fig. 3a).
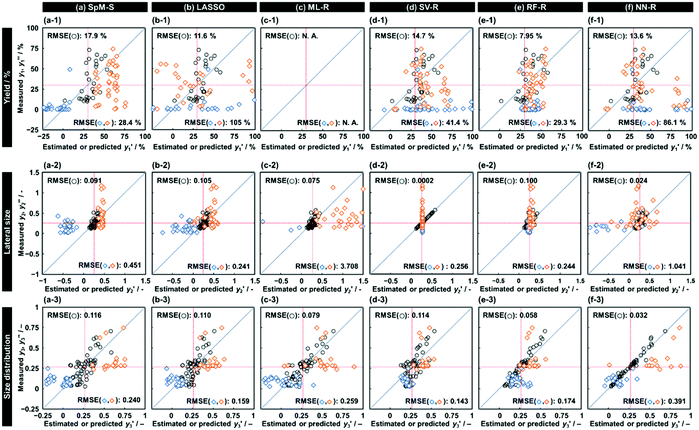 | ||
| Fig. 3 Relationship between the estimated y′ and measured y for the training datasets (black circles) and relationship between the predicted y′ and measured y′′ for the test datasets (colored diamonds) for the yield (top), lateral size (middle), and size distribution (bottom) using the different predictors constructed by SpM-S (a), LASSO (b), ML-R (c), SV-R (d), RF-R (e), and NN-R (f). The red lines indicate the threshold to evaluate the prediction accuracy based on the rate of successful experiments in Table 2. | ||
The validation of these prediction models was experimentally performed to synthesize the 2D materials under the predicted new conditions (Fig. 2e). These prediction models recommended the host–guest–medium combinations for achieving high-yield and size-controlled synthesis of the exfoliated nanosheets. Prior to the experiments, the predicted  ,
, 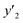 , and
, and 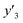 values were virtually calculated in unknown 200–2500 host–guest–medium combinations (Fig. 2e). The exfoliation experiments were only performed under the conditions providing the highest and lowest
values were virtually calculated in unknown 200–2500 host–guest–medium combinations (Fig. 2e). The exfoliation experiments were only performed under the conditions providing the highest and lowest 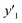 ,
, 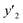 , and
, and 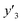 , namely 80, 80, and 46 conditions, respectively. When the precursor layered materials were not synthesized because of experimental reasons, such conditions were excluded from the list. The experimentally measured values (
, namely 80, 80, and 46 conditions, respectively. When the precursor layered materials were not synthesized because of experimental reasons, such conditions were excluded from the list. The experimentally measured values (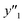 ,
, 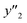 , and
, and 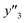 ) are summarized in the test data (Fig. 2f). The test dataset included 79
) are summarized in the test data (Fig. 2f). The test dataset included 79 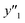 and 2 xn (n = 18, 40) for the yields, 64
and 2 xn (n = 18, 40) for the yields, 64 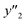 and 8 xn (n = 4, 5, 16, 17, 19, 30, 31, 40) for the size, and 43
and 8 xn (n = 4, 5, 16, 17, 19, 30, 31, 40) for the size, and 43 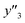 and 5 xn (n = 8, 10, 21, 32, 41) for the size distribution (Tables S5–S7 in the ESI†). The relationship between the predicted (y′) and measured (y′′) values was represented by RMSE of 28.4% for yield, 0.451 for size, and 0.240 for size distribution (the colored diamonds in Fig. 3a). Although the accuracy of the prediction models was not very precise, the averages of the higher and lower
and 5 xn (n = 8, 10, 21, 32, 41) for the size distribution (Tables S5–S7 in the ESI†). The relationship between the predicted (y′) and measured (y′′) values was represented by RMSE of 28.4% for yield, 0.451 for size, and 0.240 for size distribution (the colored diamonds in Fig. 3a). Although the accuracy of the prediction models was not very precise, the averages of the higher and lower 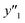 ,
, 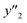 and
and 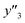 showed significant differences. Therefore, the higher and lower
showed significant differences. Therefore, the higher and lower 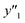 ,
, 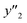 , and
, and 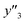 values were selectively achieved in a limited number of experiments using the predictors.
values were selectively achieved in a limited number of experiments using the predictors.
In general, the exfoliation of layered materials is not easily controlled by the experimental parameters based on the chemical insights of senior researchers because of the unpredictable down-sizing processes in the liquid phase. The prediction models facilitated the high-yield and size-controlled synthesis of the nanosheets in the limited number of time-consuming exfoliation experiments.25–27 For example, the number of experiments was reduced to 89% for the high-yield synthesis,25 98% for the lateral-size control,26 and 98% for the control of the size distribution.27 Moreover, the elucidation of the structural and chemical factors facilitates further understanding and control of the exfoliation processes.
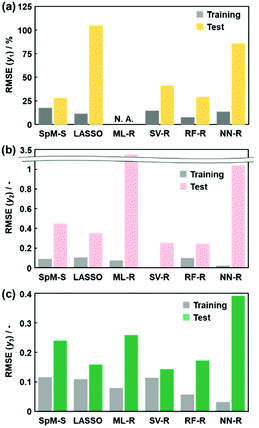
Construction of predictors based on the other ML methods
The other ML methods including two linear and three nonlinear regressions were applied to the same training and test datasets to study the suitability for the small data (Fig. 1e and f). LASSO, ML-R, SV-R, RF-R, and NN-R were performed on the original training dataset containing 30 y1 and 35 xn for the yield, 48 y2 and 33 xn for the size, and 54 y3 and 37 xn for the size distribution (Fig. 2a). The detailed methods, such as the execution environments and model constructions, are described in the ESI.† The relationship between the estimated y′ and measured y was summarized with the RMSE values (the black circles in Fig. 3b–f). In addition, the test datasets including 79 y1, 64 y2, and 43 y3 were applied to validate the constructed predictors using the same descriptors on the training dataset (Fig. 2f and the colored diamonds in Fig. 3b–f). Fig. 4 summarizes the RMSE values for each predictor to the training (black) and test (colored) datasets.LASSO, a typical algorithm for sparse modeling, extracts the descriptors using an L1-regularization term.49 The number of descriptors was reduced to 16 for 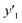 , 16 for
, 16 for 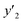 , and 13 for
, and 13 for 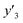 . ML-R uses all the descriptors, i.e. 35 for
. ML-R uses all the descriptors, i.e. 35 for 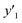 , 33 for
, 33 for 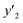 , and 37 for
, and 37 for 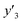 , without extraction of the descriptors. The linear-regression models were constructed using these descriptors and then validated in the training and test datasets. In the three linear-regression models, the relationship between the measured (y) and estimated (y′) values on the training datasets was not significantly different (the black circles in Fig. 3a–c), even though the number of descriptors and their coefficients were different. On the test datasets, SpM-S showed the better correlation between y′ and y′′ compared with the other two linear regression models (the colored diamonds in Fig. 3c–e).
, without extraction of the descriptors. The linear-regression models were constructed using these descriptors and then validated in the training and test datasets. In the three linear-regression models, the relationship between the measured (y) and estimated (y′) values on the training datasets was not significantly different (the black circles in Fig. 3a–c), even though the number of descriptors and their coefficients were different. On the test datasets, SpM-S showed the better correlation between y′ and y′′ compared with the other two linear regression models (the colored diamonds in Fig. 3c–e).
Support vector machine (SVM) solves binary classification problems by formulating them as convex optimization problems.50 The optimization problem entails finding the maximum margin separating the hyperplane, while correctly classifying as many training points as possible. SVM represents this optimal hyperplane with support vectors. The sparse solution and good generalization of the SVM lend themselves to adaptation to regression problems.51 SV-R constructs a nonlinear model based on classification of data including y and y′ with the maximized margins between support vectors.52 RF-R provides a nonlinear model based on multiple decision trees and their ensemble to prevent overfitting.53 NN-R constructs a nonlinear model by graph architectures consisting of input, hidden, and output layers. While these nonlinear prediction models generally had smaller RMSE values for the training data (the black circles in Fig. 3d–f), larger RMSE values were recorded for the test datasets (the colored diamonds in Fig. 3d–f).
Advantages of SpM-S to small data
Our SpM-S provides the predictors with accuracy, generalizability, and interpretability (Fig. 4). As RMSE represents the difference between the measured and estimated (or predicted) values, a smaller RMSE indicates the higher prediction accuracy of the models. Fig. 4 summarizes the RMSE of each predictor to the training (black) and test (colors) datasets. SpM-S showed smaller differences in the RMSE values between the training and test datasets compared with the other ML methods (Fig. 4). The fact indicates that SpM-S provides more generalizable models with suppression of the overtraining resulting from the small training data.SpM-S shows an appropriate prediction accuracy to both the training and test datasets compared with the other ML methods even though the simple linear regression models contain a limited number of descriptors, i.e. two for y1, eight for y2, and five for y3. In these experiments, the exfoliation proceeds with intercalation of the dispersion media in the interlayer space of the layered composites. Then, the swelling induces exfoliation into nanosheets.54–57 The selected descriptors in the models (eqn (1)–(3)) are interpretable and reasonable in the context of the real exfoliation behavior.25–27 The selection of the descriptors assisted by the experience and perspective of researchers contributes to both suppression of the overtraining and construction of a more generalizable model. The descriptors and coefficients of the linear regression models imply the important factors and their weight, respectively. The interpretable model is helpful for designing the next experiments. Although the descriptors and coefficients are clear in the models constructed by LASSO and ML-R, the number of descriptors is not appropriate for identification of the significant factors. In the nonlinear models, it is not easy to extract the positive and negative correlations of each descriptor for discussion based on our chemical insights. Recently, Hatakeyama-Sato and Oyaizu reported that a generative model was used to construct a better predictor with imaging the missing data.23 Although the prediction accuracy is improved by the new method, the interpretability is lowered by combination with the more complex prediction models. In the present work, straightforward and interpretable prediction models are constructed by SpM-S with the assistance of our chemical perspectives using a limited number of descriptors. Therefore, SpM-S is regarded as a preferred method for small data.
The RMSE values of the models based on SpM-S were not always the smallest compared with those based on RF-R and SV-R (Fig. 4). The purpose of our work is the selective synthesis of the larger and smaller y1, y2, and y3 using the predictors in a limited number of experiments. Although the performance of machine learning is generally discussed only with the prediction accuracy,58 the important factor to evaluate the prediction accuracy is not only the RMSE values but also the rate of successful experiments. The rate was calculated as another metric to evaluate the selective synthesis using the predictors in the real experiments (Table 2). If both the predicted (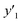 ,
, 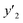 , and
, and 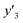 ) and measured (
) and measured (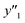 ,
, 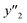 , and
, and 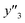 ) values are larger (or smaller) than a certain threshold, the experimental trial can be regarded as successful. After setting the thresholds, the rate of successful experiments (Rs) is defined as the number of successful experiments to the number of predicted conditions. The thresholds were set at 30% for the yield, 0.267 for the lateral size (size reduction rate, L/L0), and 0.266 for the size distribution (LCV). The threshold at 30% for the yield was used in our previous work.25 The thresholds of the lateral size and size distribution were the average values of the training datasets (Table S1 in the ESI†). These thresholds were displayed using the red lines in Fig. 3. For example, the number of conditions with the predicted yield lower and higher than 30% was 11 and 19 by SpM-S (the number of colored plots to the left and right of the red threshold line in Fig. 3), respectively (Table 2). The number of conditions with the measured yield higher and lower than 30% was 9 and 12 (the number of colored plots to the lower left and upper right of the red threshold line in Fig. 3), respectively. Therefore, the Rs values were 0.818 and 0.632 for the high- and low-yield conditions, respectively. The total Rs value including the high- and low-yield conditions was 21/30 = 0.700. In addition to the RMSE values, the accuracy of the prediction models was compared with the Rs values. In the training datasets, the total Rs of SpM-S was lower than those of SV-R and RF-R (Table 2). In contrast, in the test datasets, the total Rs of SpM-S was higher than those of SV-R and RF-R (Table 2). SV-R and RF-R show an overtraining trend compared with SpM-S. In addition to the RMSE values, Rs indicates that the models based on SpM-S have the robustness and better performances. Moreover, the selective syntheses were successfully achieved by the models based on SpM-S in the real experiments. According to the results, not only the RMSE values but also other metrics, such as the Rs values, are important for evaluation of the prediction accuracy in small data.
) values are larger (or smaller) than a certain threshold, the experimental trial can be regarded as successful. After setting the thresholds, the rate of successful experiments (Rs) is defined as the number of successful experiments to the number of predicted conditions. The thresholds were set at 30% for the yield, 0.267 for the lateral size (size reduction rate, L/L0), and 0.266 for the size distribution (LCV). The threshold at 30% for the yield was used in our previous work.25 The thresholds of the lateral size and size distribution were the average values of the training datasets (Table S1 in the ESI†). These thresholds were displayed using the red lines in Fig. 3. For example, the number of conditions with the predicted yield lower and higher than 30% was 11 and 19 by SpM-S (the number of colored plots to the left and right of the red threshold line in Fig. 3), respectively (Table 2). The number of conditions with the measured yield higher and lower than 30% was 9 and 12 (the number of colored plots to the lower left and upper right of the red threshold line in Fig. 3), respectively. Therefore, the Rs values were 0.818 and 0.632 for the high- and low-yield conditions, respectively. The total Rs value including the high- and low-yield conditions was 21/30 = 0.700. In addition to the RMSE values, the accuracy of the prediction models was compared with the Rs values. In the training datasets, the total Rs of SpM-S was lower than those of SV-R and RF-R (Table 2). In contrast, in the test datasets, the total Rs of SpM-S was higher than those of SV-R and RF-R (Table 2). SV-R and RF-R show an overtraining trend compared with SpM-S. In addition to the RMSE values, Rs indicates that the models based on SpM-S have the robustness and better performances. Moreover, the selective syntheses were successfully achieved by the models based on SpM-S in the real experiments. According to the results, not only the RMSE values but also other metrics, such as the Rs values, are important for evaluation of the prediction accuracy in small data.
| Yield | SpM-S | SV-R | RF-R | |
|---|---|---|---|---|
| a The numbers in parentheses (NS/Npred) indicate the number of successful experiments (NS) and predicted conditions (Npred). | ||||
| Training |
y
1 and 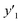 < 30% < 30% |
0.818 (9/11) | 0.929 (13/14) | 1.000 (16/16) |
y
1 and 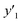 > 30% > 30% |
0.632 (12/19) | 0.813 (13/16) | 1.000 (14/14) | |
| Total | 0.700 (21/30) | 0.867 (26/30) | 1.000 (30/30) | |
| Test |
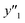 and and 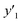 < 30% < 30% |
0.974 (38/39) | 0.875 (14/16) | 0.750 (6/8) |
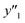 and and 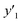 > 30% > 30% |
0.500 (20/40) | 0.302 (19/63) | 0.268 (19/71) | |
| Total | 0.734 (58/79) | 0.418 (33/79) | 0.316 (25/79) | |
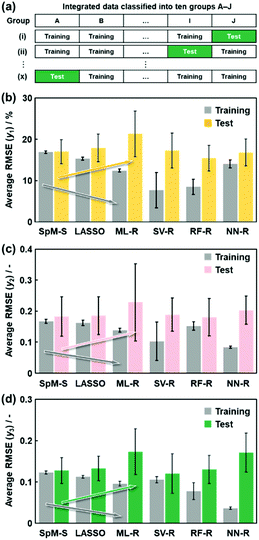
The advantage of the models constructed by SpM-S was studied with changes in the training and test data (Fig. 5). The training and test data were mixed and then divided into ten datasets (Fig. 5a). Nine of the datasets and the remaining one were used for training and validation, respectively. The ten-fold segmentalized validation of the training and test data was performed ten times with changes in the assignment of the test data. The average RMSE and its standard deviation in the ten trials were calculated for each method (Fig. 5b–d). The training and validation were performed using the same descriptors in Fig. 4. Although the RMSE values decreased with an increase in the quantity of the training data compared with that in Fig. 4, the overall trends for the accuracy and generalizability did not change much in this ten-fold segmentalized validation (Fig. 5). In the linear-regression models, while the average RMSE values of SpM-S, LASSO, and ML-R on the training datasets decreased in this order, those on the test datasets increased in the same order (arrows in Fig. 5). The results support that a limited number of significant descriptors are selected by SpM-S in small data to avoid overfitting. In contrast, the nonlinear models showed overtraining with large differences in the RMSE values between the training and test datasets. Therefore, the advantage of SpM-S is not characteristic to the original datasets including the training and test datasets in Fig. 2a and f. Moreover, SpM-S can construct generalizable and interpretable prediction models using a limited number of significant descriptors. The methodology can be applied to other small data for acceleration of research activities without wasting time, money, and effort.
Conclusions
Advantages of SpM-S in materials science and chemistry were studied using real experimental small data about the controlled synthesis of 2D materials. In SpM-S, SpM is combined with our chemical perspectives for application to small data. The new data-scientific approach was compared with other ML methods, such as LASSO and ML-R for linear regression, and SV-R, RF-R, and NN-R for nonlinear regression. A limited number of descriptors were extracted and selected by sparse modeling coupled with chemical perspectives. Straightforward and interpretable linear-regression models using the selected descriptors were constructed for the prediction of the yield, size, and size distribution of 2D materials. The test datasets for the validation were prepared by the controlled synthesis of 2D materials under the predicted conditions. The RMSE values of the prediction models to the training and test datasets were compared with the different ML methods. The more accurate, generalizable, and interpretable models were constructed by SpM-S compared with the other ML methods. Introduction of experience and chemical perspectives can suppress the overlearning typically caused by small data. The methodology can be applied to a wide variety of small data, such as experimental data in laboratories.Experimental methods
Preparation of the datasets
Both the training and test datasets were prepared from the data in our previous studies about yield,24,25 size,26 and size-distribution.27 The datasets are available in Tables S2–S7.†ML and construction of the predictors
ML was implemented using the Scikit-learn package (ver. 0.22.1) in Python (ver. 3.7.6). ES-LiR and ten-fold segmentalized validation were also mounted in Python. MC+ was performed using the ncvreg package (ver. 3.13.0) in R programing language (ver. 3.6.3). The prediction models, namely eqn (1)–(3), based on SpM-S were already constructed in our previous studies about the yield,24,25 size,26 and size-distribution.27The hyperparameter for LASSO, lambda, was determined by five-fold cross validation (CV). In the ten-fold segmentalized validation, the descriptors for ten models were fixed as those used in the training and test data while the coefficients and lambda were different. In other words, the multiple linear regressions were performed ten times using the same descriptors.
The radial basis function (RBF) kernel was used to construct the SV-R model. The hyperparameters, such as gamma, C, and epsilon, were determined by five-fold grid search CV. Grid search CV selects the hyperparameters from each parameter when the model is most accurate within a range of pre-given values. The hyperparameters were tuned for each entry of the ten-fold segmentalized validation.
In RF-R, the parameters to be tuned were maximum tree depth (max_depth) and the number of trees (n_estimators). These parameters were determined by five-fold grid search CV in the range of 1–10 for max_depth and 1–500 for n_estimators. The hyperparameters were tuned for each entry of the ten-fold segmentalized validation.
A multilayer perceptron (MLP) neural network model was used for NN-R. The number of nodes in a hidden layer and the way of updating the learning rate of the weights were tuned by five-fold grid search CV. In the present work, the number of hidden layers was set to one. The number of nodes was set in the range of 22–28. The learning rates were selected from ‘invscaling’, ‘adaptive’ and ‘constant’. In the ten-fold segmentalized validation, the appropriate hyperparameters were tuned for each entry of the ten-fold segmentalized validation.
Data availability
All the training and test data in Tables S2–S7 are available in the ESI.† The information for the program code is provided in the Experimental section.Author contributions
Y. O. conceptualized and supervised this project with funding acquisition. Y. H. analyzed the data and discussed the results with Y. I. and Y. O. Y. I. provided the program code of ES-LiR and supervised the data analysis. The original draft was prepared by Y. H. and Y. O. The manuscript was reviewed and edited by all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JST PRESTO (Y. O., JPMJPR16N2 and Y. I., JPMJPR17N2).Notes and references
- M. I. Jordan and T. M. Mitchell, Science, 2015, 349, 255 CrossRef CAS PubMed.
- T. Posner and L. Fei-Fei, Nature, 2020, 588, S118 CrossRef CAS.
- K. Rajan, Annu. Rev. Mater. Res., 2015, 45, 153 CrossRef CAS.
- B. Sun, M. Fernandez and A. S. Barnard, Nanoscale Horiz., 2016, 1, 89 RSC.
- R. Ramprasad, R. Batra, G. Pilania, A. Mannodi-Kanakkithodi and C. Kim, npj Comput. Mater., 2017, 3, 54 CrossRef.
- A. O. Oliynyk and A. Mar, Acc. Chem. Res., 2018, 51, 59 CrossRef CAS PubMed.
- J. S. Peerless, N. J. B. Milliken, T. J. Oweida, M. D. Manning and Y. G. Yingling, Adv. Theory Simul., 2019, 2, 1800129 CrossRef.
- A. C. Mater and M. L. Coote, J. Chem. Inf. Model., 2019, 59, 2545 CrossRef CAS PubMed.
- L. Himanen, A. Geurts, A. S. Foster and P. Rnke, Adv. Sci., 2019, 6, 190808 Search PubMed.
- C. Chen, Y. Zuo, W. Ye, X. Li, Z. Deng and S. P. Ong, Adv. Energy Mater., 2020, 10, 1903242 CrossRef CAS.
- G. H. Gu, C. Choi, Y. Lee, A. B. Situmorang, J. Noh, Y. H. Kim and Y. Jung, Adv. Mater., 2020, 32, 1907865 CrossRef CAS PubMed.
- K. Terayama, M. Sumita, R. Tamura and K. Tsuda, Acc. Chem. Res., 2021, 54, 1334 CrossRef CAS PubMed.
- R. Pollice, G. P. Gomes, M. Aldeghi, R. J. Hickman, M. Krenn, C. Lavigne, M. Lindner-D'Addario, A. K. Nigam, C. T. Ser, Z. Yao and A. Aspuru-Guzik, Acc. Chem. Res., 2021, 54, 849 CrossRef CAS PubMed.
- J. Kimmg, S. Zechel and U. S. Schubert, Adv. Mater., 2021, 33, 2004940 CrossRef PubMed.
- Y. Oaki and Y. Igarashi, Bull. Chem. Soc. Jpn., 2021, 94, 2410 CrossRef CAS.
- J. M. Granda, L. Donina, V. Dragone, D. L. Long and L. Cronin, Nature, 2018, 559, 377 CrossRef CAS PubMed.
- T. N. Nguyen, T. T. P. Nhat, K. Takimoto, A. Thakur, S. Nishimura, J. Ohyama, I. Miyazato, L. Takahashi, J. Fujima, K. Takahashi and T. Taniike, ACS Catal., 2020, 10, 921 CrossRef CAS.
- R. Shimizu, S. Kobayashi, Y. Watanabe, Y. Ando and T. Hitosugi, APL Mater., 2020, 8, 111110 CrossRef CAS.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai, X. Wang, X. Li, B. M. Alston, B. Li, R. Clowes, N. Rankin, B. Harris, R. S. Sprick and A. I. Cooper, Nature, 2020, 583, 237 CrossRef CAS PubMed.
- Y. Zhang and C. Ling, npj Comput. Mater., 2018, 4, 25 CrossRef.
- S. J. Pan and Q. Yang, IEEE Trans. Knowl. Data Eng., 2010, 22, 1345 Search PubMed.
- P. Chlap, H. Min, N. Vandenberg, J. Dowling, L. Holloway and A. Haworth, J. Med. Imaging Radiat. Oncol., 2021, 65, 545 CrossRef PubMed.
- K. Hatakeyama-Sato and K. Oyaizu, ACS Omega, 2021, 6, 14566 CrossRef CAS PubMed.
- G. Nakada, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2019, 2, 1800180 Search PubMed.
- K. Noda, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2020, 3, 2000084 CrossRef CAS.
- R. Mizuguchi, Y. Igarashi, H. Imai and Y. Oaki, Nanoscale, 2021, 13, 3853 RSC.
- Y. Haraguchi, Y. Igarashi, H. Imai and Y. Oaki, Adv. Theory Simul., 2021, 4, 2100158 CrossRef.
- K. Noda, Y. Igarashi, H. Imai and Y. Oaki, Chem. Commun., 2021, 57, 5921 RSC.
- Y. Oaki, Chem. Lett., 2021, 50, 305 CrossRef CAS.
- H. Numazawa, Y. Igarashi, K. Sato, H. Imai and Y. Oaki, Adv. Theory Simul., 2019, 2, 1900130 CrossRef CAS.
- R. Tibshirani, M. Wainwright and T. Hastie, Statistical Learning with Sparsity: The Lasso and Generalizations, Chapman and Hall, Philadelphia, PA, 2015 Search PubMed.
- Y. Igarashi, K. Nagata, T. Kuwatani, T. Omori, Y. Nakanishi-Ohno and M. Okada, J. Phys.: Conf. Ser., 2016, 699, 012001 CrossRef.
- E. Candès and J. Romberg, Inverse Probl., 2007, 23, 969 CrossRef.
- M. Honma, K. Akiyama, M. Uemura and S. Ikeda, Publ. Astron. Soc. Jpn., 2014, 66, 95 Search PubMed.
- T. Yamamoto, K. Fujimoto, T. Okada, Y. Fushimi, A. Stalder, Y. Natsuaki, M. Schmidt and K. Togashi, Invest. Radiol., 2016, 51, 372 CrossRef PubMed.
- Y. Igarashi, H. Takenaka, Y. Nakanishi-Ohno, M. Uemura, S. Ikeda and M. Okada, J. Phys. Soc. Jpn., 2018, 87, 044802 CrossRef.
- C. H. Zhang, Ann. Stat., 2010, 38, 894 Search PubMed.
- P. Breheny and J. Huang, Ann. Appl. Stat., 2011, 5, 232 Search PubMed.
- M. Osada and T. Sasaki, Adv. Mater., 2012, 24, 210 CrossRef CAS PubMed.
- V. Nicolosi, M. Chhowalla, M. G. Kanatzidis, M. S. Strano and J. N. Coleman, Science, 2013, 340, 1226419 CrossRef.
- M. A. Timmerman, R. Xia, R. T. P. Le, Y. Wang and J. E. Ten Elshof, Chem.–Eur. J., 2020, 27, 9084 CrossRef PubMed.
- B. Tang, Y. Lu, J. Zhou, T. Chouhan, H. Wang, P. Golani, M. Xu, Q. Xu, C. Guan and Z. Liu, Mater. Today, 2020, 41, 73 Search PubMed.
- N. C. Frey, J. Wang, G. I. V. Bellido, B. Anasori, Y. Gogotsi and V. B. Shenoy, ACS Nano, 2019, 13, 3031 CrossRef CAS PubMed.
- X. Lin, Z. Si, W. Fu, J. Yang, S. Guo, Y. Cao, J. Zhang, X. Wang, P. Liu, K. Jiang and W. Zhao, Nano Res., 2018, 11, 6316 CrossRef CAS.
- B. Han, Y. Lin, Y. Yang, N. Mao, W. Li, H. Wang, K. Yasuda, X. Wang, V. Fatemi, L. Zhou, J. I.-J. Wang, Qi. Ma, Y. Cao, D. Rodan-Legrain, Y.-Q. Bie, E. Navarro-Moratalla, D. Klein, D. MacNeill, S. Wu, H. Kitadai, X. Ling, P. Jarillo-Herrero, J. Kong, J. Yin and T. Palacios, Adv. Mater., 2020, 32, 2000953 CrossRef CAS PubMed.
- Z. Zhang, Y. Hong, B. Hou, Z. Zhang, M. Negahban and J. Zhang, Carbon, 2019, 148, 115 CrossRef CAS.
- H. Yang, Z. Zhang, J. Zhang and X. C. Zeng, Nanoscale, 2018, 10, 19092 RSC.
- M. Lotya, A. Rakovich, J. F. Donegan and J. N. Coleman, Nanotechnology, 2013, 24, 265703 CrossRef PubMed.
- R. Tibshirani, J. Roy. Stat. Soc. B Stat. Methodol., 1996, 58, 267 Search PubMed.
- H. Drucker, C. J. Burges, L. Kaufman, A. Smola and V. Vapnik, Adv. Neural Inf. Process. Syst., 1997, 9, 155 Search PubMed.
- V. Vapnik, Nonlinear modeling, Springer, Boston, MA, 1998, pp. 55–85 Search PubMed.
- A. J. Smola and B. Schölkopf, Stat. Comput., 2004, 14, 199 CrossRef.
- L. Breiman, Mach. Learn., 2001, 45, 5 CrossRef.
- R. Mizuguchi, H. Imai and Y. Oaki, Nanoscale Adv., 2020, 2, 1168 RSC.
- M. Honda, Y. Oaki and H. Imai, Chem. Mater., 2014, 26, 3579 CrossRef CAS.
- G. Nakada, H. Imai and Y. Oaki, Chem. Commun., 2018, 54, 244 RSC.
- Y. Yamamoto, H. Imai and Y. Oaki, Bull. Chem. Soc. Jpn., 2019, 92, 779 CrossRef CAS.
- D. Morgan and R. Jacobs, Annu. Rev. Mater. Res., 2020, 50, 71 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods, list of descriptors and all the datasets. See DOI: 10.1039/d1dd00010a |
| This journal is © The Royal Society of Chemistry 2022 |