Open Access Article
Open Access ArticleSelenium nanoparticles inhibited H1N1 influenza virus-induced apoptosis by ROS-mediated signaling pathways
Xia Liu†
,
Danyang Chen†,
Jingyao Su†,
Ruilin Zheng,
Zhihui Ning,
Mingqi Zhao,
Bing Zhu* and
Yinghua Li *
*
Center Laboratory, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, No. 318 Renminzhong Road, Yuexiu District, Guangzhou, 510120, People's Republic of China. E-mail: liyinghua@gzhmu.edu.cn; zhubing@gzhmu.edu.cn
First published on 31st January 2022
Abstract
Influenza A (H1N1) viruses are distributed around the world and pose a threat to public health. Vaccination is the main treatment strategy to prevent influenza infection, but antiviral drugs also play an important role in controlling seasonal and pandemic influenza. Currently, as influenza viruses may be developing antiviral resistance, new agents with different modes of action are being investigated. Recently, selenium nanoparticles (SeNPs), which have antiviral effects, have attracted increasing attention in biomedical interventions. The appearance of nanotechnology has attracted great attention in the field of nanomedicine. SeNPs constitute an attractive vector platform for delivering a variety of drugs to action targets. SeNPs are being explored for potential therapeutic efficacy in a variety of oxidative stress and inflammation-mediated diseases, such as cancer, arthritis, diabetes, and kidney disease. SeNPs could inhibit infection of Madin–Darby canine kidney (MDCK) cells with H1N1 and prevent chromatin condensation and DNA fragmentation. ROS play a key role in physiological processes for apoptosis. SeNPs significantly inhibited the production of reactive oxygen species (ROS) in MDCK cells. Mechanistic investigation revealed that SeNPs inhibited the apoptosis induced by H1N1 virus infection in MDCK cells by improving the level of GPx1. Our results suggest that SeNPs are an effective selenium source and a promising H1N1 influenza antiviral candidate.
1 Introduction
Influenza A (H1N1) viruses are distributed globally and pose a threat to public health.1 Currently, H1N1 continues to spread around the world. The great pandemic that occurred in 2009 caused great social panic and economic losses. It is important to strengthen and coordinate preventive activities at a global level to control the spread of the virus.2 Swine play a key role in the transmission of this virus to humans as a natural host and mixing carrier of influenza A (H1N1) viruses. Furthermore, swine influenza A (H1N1) viruses have provided all eight genes or some genes to the genomes of influenza strains that have historically caused human pandemics. Therefore, continuous surveillance of influenza A (H1N1) viruses in swine herds helps prevent and control this virus.3 Influenza A virus infections occur in different species, including pigs, causing mild to severe respiratory symptoms that lead to a significant burden of disease. Hence, pigs are thought to be a genetic mixing vessel for human and avian influenza viruses. Classical H1N1 swine influenza viruses (SIVs) cause sporadic zoonotic infections, while recombinant swine-origin influenza A H1N1 virus caused a human influenza pandemic in 2009.4So far, vaccination is still considered the best way to control infection. Fortunately, in August and September 2009, a vaccine against H1N1 was reported. However, it is important to note that this vaccine is not suitable for children and people who are allergic to eggs.2 In addition, between the rapid evolution of the virus and vaccine development, restraining the spread of influenza infections is a long endeavour.5 Synthetic drugs have led to the emergence of drug-resistant strains, which cause trouble to humans.6 The research and development of antiviral drugs is very important. Selenium (Se) is an essential trace element in the human body.7–9 It is very important in physiological processes with many physiological functions and its biological functions are mainly manifested in low molecular weight Se compounds and Se-containing proteins.10,11 It is incorporated into selenoproteins as selenocysteine (Sec), representing the most important part of the active center of their enzymatic activities.12 Se controls several key biological processes, such as the elimination of reactive oxygen species (ROS) and the regulation of specific enzymes.13–15 Selenium deficiency can lead to susceptibility to infections, including respiratory viruses.16
Nanotechnology has been used to study the mechanisms and treatment options of diseases in the past three decades.17,18 Many kinds of nanostructures, including polymers, dendrimers, liposomes, metal nanoparticles (Ag, Au, Ce, Cu, Eu, Fe, Se, Ti, Y, etc.), silicon and carbon-based nanomaterials have been used as successful therapeutic agents and drug delivery carriers.18 Nanoparticles have unique features such as small size, large surface area, surface charge, surface chemistry, solubility and versatility.19 Among these nanoparticles, selenium nanoparticles (SeNPs) are one of the most extensively studied. Nanoparticles are considered to be potential materials that can alleviate the formation of biofilms, the production of ROS, and low redox activity; thus, we think that SeNPs in the biomedical field have development potential and are worthy of further exploration. SeNPs can inhibit calcium oxalate urinary calculi, inhibit the growth of Staphylococcus aureus, have a protective effect on the cardiovascular system by preventing modification of lipid oxidation and reducing platelet aggregation, inhibit tumor growth by reducing ROS levels in hypoxic tumor cells, prevent DNA oxidative damage by combination with ROS producing sites, be used for gene and drug carriers and so on.20 In this paper, SeNPs have been combined with the application of anti-influenza virus. The hope is to find a new breakthrough in the development of anti-influenza viruses. Because Se is toxic and has a narrow margin of safety, the use of Se in the form of nanoparticles has essentially solved the toxicological concerns associated with Se. SeNPs have attractive anticancer activity and reduced toxicity.18 Among the various types of nanoparticles, there are not many nanoparticles currently used for antiviral research. Selenium has superior adsorption capacity as well as a trend of improving selenium enzyme action, avoiding cell damage in the body caused by free radicals and has a defensive effect towards the oxidation of DNA. In addition, selenium nanoparticles play a central role in improving immune responses to various bacterial and viral antigens due to their low toxicity and parameterized particle size and surface characteristics.21 Free radicals from oxygen are known as reactive oxygen species (ROS), and increased ROS production leads to peroxidation of DNA, proteins, and lipids, which changes their structure, activity, and physical properties, leading to cell death and tissue damage.22 Reactive oxygen scavenging enzymes include superoxide dismutase (SOD), glutathione peroxidase (Gpx) and catalase (CAT). Gpx1, as an important antioxidant enzyme, is an enzyme necessary for antioxidant activity in organisms, and plays a positive role in preventing the accumulation of intracellular hydrogen peroxide.23 It uses glutathione, the most abundant free radical scavenger, to reduce hydrogen peroxide, organic hydroperoxides, and lipid hydroperoxides.24 As demonstrated by a mechanism investigation, SeNPs inhibited H1N1 influenza virus from infecting MDCK cells by inducing apoptosis via the suppression of AKT and p53 signaling pathways.25 This study aims to verify that selenium nanoparticles inhibit H1N1 influenza virus-induced apoptosis by improving the level of Gpx1.
2 Experimental
2.1 Materials
Madin–Darby canine kidney cells (MDCK) were used in the current study. All cells were purchased from ATCC and cultured according to the ATCC recommendations. They were cultured with serum-free minimum essential medium (MEM; Gibcol, USA) in a 37 °C incubator.26 Na2SeO3, vitamin C, MTT, DAPI, and DCF-DA were purchased from Sigma-Aldrich. Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco (Life Technologies, Carlsbad, CA, USA). H1N1 influenza virus was provided by Guangzhou Women and Children's Medical Center of Guangzhou Medical University.2.2 Preparation and characterization of the SeNPs
Selenium nanoparticles (SeNPs), a new form of selenium, have attracted worldwide attention because of their biological activity and low toxicity. The main factors determining their efficiency are the size, stability and surface properties of the nanomaterial. SeNPs were prepared according to the method reported in the literature.27 Briefly, 0.1 M Na2SeO3 was freshly prepared with ultrapure water. 0.25 ml stock solution of Na2SeO3 (0.1 M) was gradually added into a 2 ml stock solution (50 mM) of vitamin C. Finally, the solution was dialyzed overnight using a dialysis bag. Its purpose was to remove superfluous vitamin C and Na2SeO3. 5 μl of the samples were dripped onto a copper grid that was coated with a holey carbon film to dry and observed using transmission electron microscopy. The TEM images were captured on an H-7650 (Hitachi, Tokyo, Japan) performed at 80 kV. A Nova Nano scanning electron microscope 430 (FEI, Hillsboro, OR, USA) equipped with EDX was used. SEM micrographs and elemental composition of the SeNPs was obtained using the Oxford system.28 The particle size distribution, mean diameter and zeta potential of the SeNPs were measured using a Zetasizer Nano ZS90 particle analyzer (Malvern, Worcestershire, UK).29–342.3 Determination of cell viability
As described in the literature, the cytotoxicity of the SeNPs was assessed by cell viability. The anti-proliferative activity of the SeNPs in vitro was explored by the MTT reduction method.35 MDCK cells were inoculated on a 96-well plate for 24 h in an incubator at 37 °C, 5% CO2. MDCK cells were added with H1N1 influenza virus for 2 h, and the indicated concentrations of SeNPs were then added to MDCK cells for 24 h. 20 μl per well MTT in serum-free medium was added to each well and left to stand for 5 h. Formazan crystals were dissolved by adding 150 μl per well dimethyl sulfoxide. Absorbance values were recorded at 570 nm using a VersaMax ELISA microplate reader (Molecular Devices, San Jose, CA, USA).36 Cell viability was measured using the MTT reduction method; we detected the cell proliferation and investigated the antiviral activity of the SeNPs.2.4 Detection of mitochondrial membrane potential (ΔΨm)
JC-1 is an ideal fluorescent probe to detect the mitochondrial membrane potential (ΔΨm). The fluorescence intensity of JC-1 was used to estimate the state of ΔΨm in the MDCK cells exposed to SeNPs.37 As described above, plasma membrane changes in the SeNP-treated MDCK cells were detected. The cells were inoculated on 6-well plates, infected with virus according to the above experimental steps, and detected after SeNP treatment. Firstly, we sucked the culture medium and washed them once with PBS. After adding another 1 ml of cell culture solution, 1 ml of JC-1 staining solution was added to each well. MDCK cells were incubated at 37 °C for 30 min. Centrifugation was performed at 600 g for 4 min, and the supernatant was removed. The cells were washed twice with 10 μg ml−1 JC-1 PBS buffer, and the cells were then suspended. The MDCK cells were collected by 600 g centrifugation for 4 min, and the cells were suspended with JC-1 containing PBS buffer.38 JC-1 fluorescence was detected by flow cytometry under excitation (485 nm) and dual emission (from green to red at 530 nm).392.5 TUNEL-DAPI co-staining assay
The effect of SeNPs on DNA fragmentation was detected with fluorescence staining using the TUNEL apoptosis kit, as described above. MDCK cells were confirmed with TUNEL for 1 h and incubated with DAPI for 15 min, and nuclear staining was performed at 37 °C.2.6 Caspase-3 activity
Caspase-3 activity was determined by a colorimetric assay kit. In summary, MDCK cells were treated with SeNPs for 48 h, and the cell culture medium was absorbed into a 15 ml centrifuge tube for use. The cells were digested with trypsin and collected in a spare cell culture medium. They were centrifuged at 1500 rpm for 5 min. Then, we removed the supernatant and washed them with PBS once. Then, cell lysate was added and lysed for 15 min. We centrifuged this mixture at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 15 min. After centrifugation, we mixed 50 μl cell lysis solution with 50 μl reaction buffer and added it to a 96-well plate. Then, 10 μl caspase substrate was added to each plate and cultured at 37 °C in the dark for 1 h. Absorbance values were measured at 380 nm (excitation) and 460 nm (emission) and caspase activity was expressed as pleated changes compared to the control.40
000 g for 15 min. After centrifugation, we mixed 50 μl cell lysis solution with 50 μl reaction buffer and added it to a 96-well plate. Then, 10 μl caspase substrate was added to each plate and cultured at 37 °C in the dark for 1 h. Absorbance values were measured at 380 nm (excitation) and 460 nm (emission) and caspase activity was expressed as pleated changes compared to the control.40
2.7 Determination of Gpx1
We used a glutathione peroxidase kit. The cells were first washed with PBS, then digested with trypsin and collected into appropriate centrifuge tubes. We centrifuged them at 1500 rpm for 5 min. We removed the supernatant and added cell lysate. We centrifuged them at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 10 min, and the supernatant was taken for detection. We added the corresponding liquid to the 96-well plate according to the instructions of the Gpx1 test kit. At the corresponding time of incubation at 37 °C, the absorbance of each well was measured immediately at 340 nm with an enzyme standard instrument.41
000 g for 10 min, and the supernatant was taken for detection. We added the corresponding liquid to the 96-well plate according to the instructions of the Gpx1 test kit. At the corresponding time of incubation at 37 °C, the absorbance of each well was measured immediately at 340 nm with an enzyme standard instrument.41
2.8 Determination of reactive oxygen species
The mitochondrion is a significant component of intracellular ROS production. The overexpression of ROS causes damage to mitochondrial ATP synthesis and leads to mitochondrial dysfunction, which further induces cell apoptosis.42 ROS play a key role in physiological processes for apoptosis.43–46 Cells were inoculated on 12-well plates, infected with virus according to the previous experimental steps, and detected after SeNP treatment. We adopted a kit for reactive oxygen species detection, which used the fluorescent probe DCFH-DA (2,7-dichlorodihydrofluorescein diacetate). DCFH-DA does not fluoresce itself and can pass through the cell membrane freely. After entering the cell, DCFH-DA can be hydrolyzed by intracellular esterase to generate DCFH. Since DCFH cannot penetrate the cell membrane, the probe can be easily loaded into the cell. Intracellular reactive oxygen species can oxidize non-fluorescent DCFH to produce fluorescent DCF. The level of intracellular reactive oxygen species can be determined by measuring the fluorescence of DCF. Firstly, DCFH-DA was diluted with serum-free medium to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000. After the original cell culture medium was removed, an appropriate volume of diluted DCFH-DA was added. Then, they were incubated in a 37 °C cell culture chamber for 30 min. The cells were washed with serum-free medium thrice to fully remove the DCFH-DA that did not enter the cells, and fluorescence intensity analysis of DCF was then performed with 488 nm excitation and 525 nm emission to obtain the mitochondrial ROS levels.47
1000. After the original cell culture medium was removed, an appropriate volume of diluted DCFH-DA was added. Then, they were incubated in a 37 °C cell culture chamber for 30 min. The cells were washed with serum-free medium thrice to fully remove the DCFH-DA that did not enter the cells, and fluorescence intensity analysis of DCF was then performed with 488 nm excitation and 525 nm emission to obtain the mitochondrial ROS levels.47
2.9 Statistical analysis
All data are presented as mean ± SD. One-way analysis of variance (ANOVA) was used in multiple group comparisons. Differences with P <0.05 (*) or P <0.01 (**) were considered statistically significant.3 Results and discussion
3.1 Preparation and characterization of the SeNPs
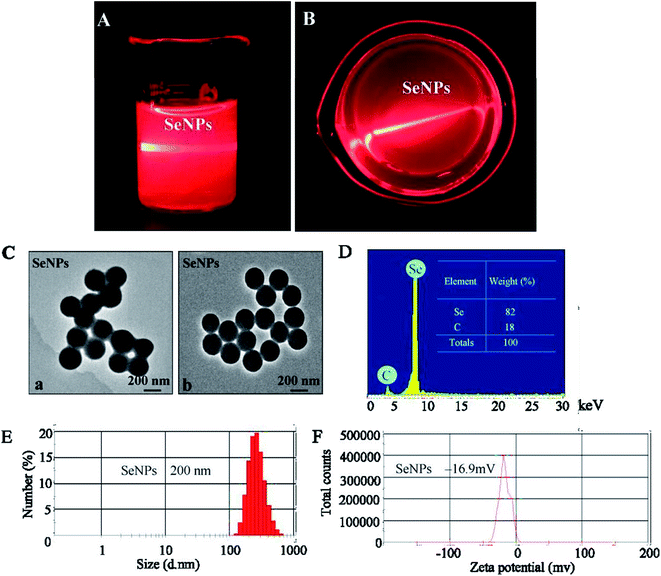
The optical images and Tyndall effect of the SeNPs are shown in Fig. 1A and B; the results indicate that the SeNP nanoparticles were composited. The morphology of SeNPs presented spherical particles, as shown in Fig. 1C. As shown in Fig. 1D, energy-dispersive X-ray spectroscopy indicated a signal of C (18%); the percentage of Se atoms was 82%. As shown in Fig. 1E and F, the size of the SeNPs was 200 nm and the zeta potential of the SeNPs was −16.9 mv.3.2 Antiviral activity of the SeNPs
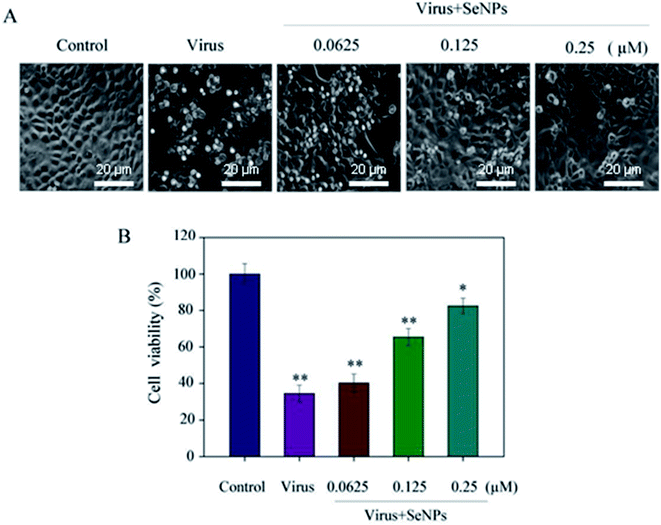
MTT assay was used to detect the effects of virus and SeNPs on MDCK cells. Fig. 2A shows the morphology of the cells in the corresponding dose-adding group; the morphology of the cells infected with the virus alone significantly changed. When the cells were treated with H1N1, they showed cytoplasmic shrinkage, loss of cell contract, and reduction in cell numbers. With the increase of the concentration of SeNPs, the virus-infected cells were effectively inhibited, and appeared healthy, with regularity in shape. Fig. 2B shows that after virus infection, the cell survival rate was 34.5%, which decreased significantly compared with the control group. Compared with the control group, when the concentrations of SeNPs were 0.0625 μM and 0.125 μM, the cell survival rates were 40.2% and 65.4%, respectively. When the SeNPs concentration was 0.25 μM, the survival rate increased to 82.5%. The results showed that SeNPs effectively inhibited H1N1 influenza virus replication.3.3 Depletion of mitochondrial membrane potential (ΔΨm) induced by SeNPs
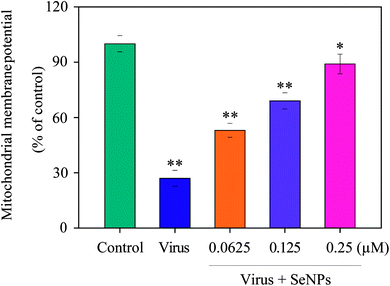
Increase in intracellular and mitochondrial ROS can cause oxidative stress and lead to the loss of mitochondrial membrane potential. As shown in Fig. 3, MDCK cells treated with H1N1 virus showed increased mitochondrial depolarization and dysfunction. Compared to the virus-infected group, when MDCK cells were exposed to SeNPs, the percentage of mitochondrial membrane potential increased significantly. The mitochondrial membrane potential of the virus group was 27%, and those of the SeNP-treated groups markedly dropped to 53% (0.0625 μM), 69% (0.125 μM) and 89% (0.25 μM), compared with the virus group. It could be concluded that SeNPs inhibited H1N1 influenza viruses via apoptosis by inducting mitochondrial dysfunction in MDCK cells.3.4 Inhibition of H1N1 infection in MDCK cells by SeNPs
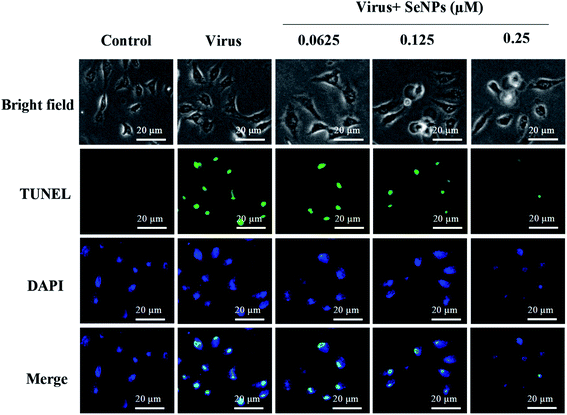
TUNEL staining uses a terminal transferase marker, which can accurately reflect the biochemical and morphological characteristics of apoptotic cells. As shown in Fig. 4, MDCK cells exhibited apoptotic characteristics; DNA fragmentation and nuclear condensation were observed under the action of H1N1 influenza virus. However, treatment with SeNPs significantly prevented the H1N1-induced nuclear morphology changes. It can be concluded from the figure that SeNPs rescue the apoptosis of MDCK cells induced by H1N1.3.5 Detection of caspase-3 activity
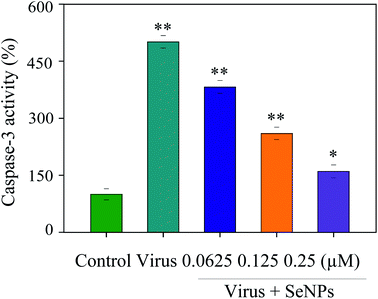
Apoptosis was induced during the H1N1 virus infection. Apoptosis was detected by inhibiting caspase-3 activation. In this paper, MDCK cells were treated with different concentrations of SeNPs after H1N1 virus infection. Then, the activity of caspase-3 was detected by an enzyme standard instrument. As reflected in Fig. 5, caspase-3 activity of the control group in which cells were uninfected was 100%; however, caspase-3 activity in untreated H1N1 virus-infected cells was 501%. MDCK cell activity remarkably increased after treatment with H1N1 influenza virus, while that of the SeNP-treated groups markedly dropped to 382% (0.0625 μM), 260% (0.125 μM) and 160% (0.25 μM), compared with the virus group. These results suggest that H1N1 virus may induce apoptosis through caspase-3, which is effectively suppressed by SeNPs.3.6 Inhibition of apoptosis signaling pathways by SeNPs
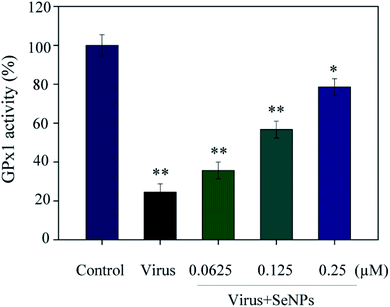
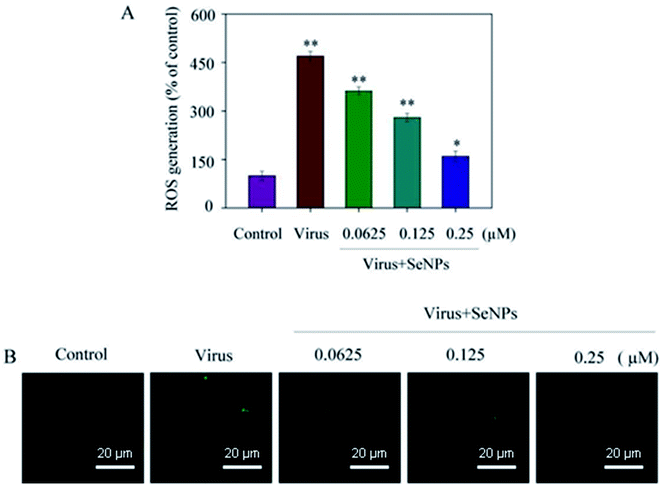
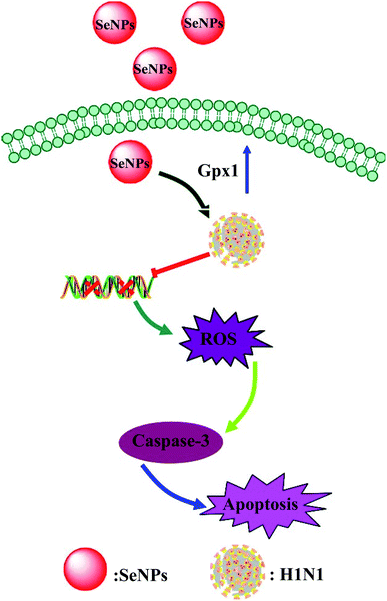
As shown in Fig. 6, the MDCK cells were treated with SeNPs and Gpx1 activity was detected. The concentrations of SeNPs were 0.0625 μM, 0.125 μM and 0.25 μM. In the virus group alone, the activity of Gpx1 was 24.5%. With the addition of SeNPs, the activity of Gpx1 gradually increases to 35.6%, 56.7% and 78.5%, respectively. Compared with the virus-infected group, the activity of Gpx1 in MDCK cells treated with SeNPs increased and was correlated with the concentration of SeNPs. It can be seen from the figure that H1N1 influenza virus can significantly reduce the activity of Gpx1, while SeNPs can increase the activity of Gpx1. It can be concluded from the figure that SeNPs may inhibit the proliferation of H1N1 influenza virus by increasing the activity of Gpx1. This ultimately leads to DNA damage by regulating the apoptosis signaling pathway. A few biological metal and metal oxide nanoparticles, including CuONPs and AgNPs, have been studied in HeLa cells, showing effective anticancer activity by increasing intracellular ROS in a concentration-dependent manner. Similarly, in the lung cancer cell line A549, MgONPs can also lead to apoptosis of cancer cells by increasing production of ROS. ROS production was detected by DCF-DA to indicate the action mechanism of the SeNPs. After treatment with H1N1, intracellular ROS production increased to 470%, as shown in Fig. 7A. MDCK cells were infected with H1N1, treated with SeNPs and then were preincubated with 10 μM DCFH-DA for 20 min. The concentration of SeNPs was 0.0625 μM, 0.125 μM and 0.25 μM. The corresponding intracellular ROS generation was 362% (0.0625 μM), 280% (0.125 μM) and 160% (0.25 μM). The higher the concentration of SeNPs, the more significant the reduction rate of ROS genes. As shown in Fig. 7B, the fluorescence intensity of DCF was found by treatment with H1N1 influenza virus. The fluorescence intensity of the H1N1 influenza virus-treated group was much stronger than that of the SeNP-treated groups. These results show that SeNPs can effectively inhibit the production of ROS caused by the H1N1 virus. Generally, as shown in Fig. 8, H1N1 infection induced intracellular apoptosis in MDCK cells and SeNPs inhibited apoptosis via upregulation of Gpx1 levels in this study.4 Conclusions
In conclusion, in this study, SeNPs have low toxicity and show superior antiviral ability to inhibit H1N1 influenza virus infection. Their antiviral mechanism suggested that SeNPs inhibited the caspase-3-mediated apoptosis of MDCK cells by increasing and regulating Gpx1 levels. Particularly, selenium is an essential nutrient, and small amounts of SeNPs are distributed in healthy tissues throughout the body and could even be beneficial to provide anti-cancer and antioxidant effects at low concentrations. In addition, the slow degradation of SeNPs in the body will lead to sustained long-term supplementation of selenium, which may contribute to the prevention of cancer; therefore, appropriate concentrations of SeNPs are beneficial to human health. In summary, this study illustrates that SeNPs have been widely used in the functional study of antiviral drugs, and using SeNPs may be a highly effective strategy to control the transmission of H1N1, whether for drug delivery or other therapeutic purposes, which is crucial for the development of new antiviral drugs. In the future, there is still a lot to explore and develop in the antiviral aspect of nanomaterials, and experimental research on the surface functionalization of SeNPs and drugs is needed, which will open up a new path for biomedicine. It is important to explore the potential of SeNPs as delivery vectors for targeted antiviral therapy.Author contributions
Xia Liu, Danyang Chen and Jingyao Su designed the study, analyzed the experimental data and drafted the manuscript. Ruilin Zheng and Zhihui Ning carried out the experiments. Mingqi Zhao analyzed the data. Bing Zhu and Yinghua Li refined the manuscript and coordinated the study. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the fund from the Open Project of Guangdong Key Laboratory of Marine Materia (LMM2020-7), the Technology Planning Projects of Guangzhou (202102010202), the Guangdong Natural Science Foundation (2020A1515110648), the Open Fund of Guangdong Provincial Key Laboratory of Functional Supramolecular Coordination Materials and Applications (2020A03) and the Guangzhou Medical University Students' Science and Technology Innovation Project (2021AEK119, 2021AEK122, 2021AEK125 and 2021AEK128).Notes and references
- J. Y. Song, J. Y. Noh, W. S. Choi, H. J. Cheong and W. J. Kim, Expert Rev. Anti-Infect. Ther., 2015, 13, 1361–1372 CrossRef CAS PubMed.
- L. Zhang, X. Zhang, Q. Ma, F. Ma and H. Zhou, Genomics, Proteomics Bioinf., 2010, 8, 139–144 CrossRef CAS.
- Z. Yu, K. Cheng, H. He and J. Wu, Transboundary Emerging Dis., 2020, 67, 450–454 CrossRef PubMed.
- X. Li, L. Guo, C. Liu, Y. Cheng, M. Kong, L. Yang, Z. Zhuang, J. Liu, M. Zou, X. Dong, X. Su and Q. Gu, Emerging Microbes Infect., 2019, 8, 1535–1545 CrossRef PubMed.
- D. Xiang, Y. Zheng, W. Duan, X. Li, J. Yin, S. Shigdar, M. L. O'Connor, M. Marappan, X. Zhao, Y. Miao, B. Xiang and C. Zheng, Int. J. Nanomed., 2013, 8, 4103–4113 CrossRef PubMed.
- J. T. Nguyen, D. F. Smee, D. L. Barnard, J. G. Julander, M. Gross, M. D. de Jong and G. T. Went, PLoS One, 2012, 7, e31006 CrossRef CAS PubMed.
- P. Erkekoglu, A. Asci, M. Ceyhan, M. Kizilgun, U. Schweizer, C. Atas, A. Kara and G. B. Kocer, Turk. J. Pediatr., 2013, 55, 271–282 Search PubMed.
- Y. Li, X. Li, Y. S. Wong, T. Chen, H. Zhang, C. Liu and W. Zheng, Biomaterials, 2011, 32, 9068–9076 CrossRef CAS PubMed.
- K. Bai, B. Hong, J. He, Z. Hong and R. Tan, Int. J. Nanomed., 2017, 12, 4527–4539 CrossRef CAS PubMed.
- Y. Li, Z. Lin, G. Gong, M. Guo, T. Xu, C. Wang, M. Zhao, Y. Xia, Y. Tang, J. Zhong, Y. Chen, L. Hua, Y. Huang, F. Zeng and B. Zhu, J. Mater. Chem. B, 2019, 7, 4252–4262 RSC.
- Z. Lin, Y. Li, M. Guo, M. Xiao, C. Wang, M. Zhao, T. Xu, Y. Xia and B. Zhu, RSC Adv., 2017, 7, 35290–35296 RSC.
- C. S. Lee, H. Kim, J. Yu, S. H. Yu, S. Ban, S. Oh, D. Jeong, J. Im, M. J. Baek and T. H. Kim, Eur. J. Med. Chem., 2017, 142, 416–423 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, H. Xia, H. Wang and B. Zhu, Int. J. Nanomed., 2016, 11, 3065–3076 CrossRef CAS PubMed.
- Y. Li, X. Li, W. Zheng, C. Fan, Y. Zhang and T. Chen, J. Mater. Chem. B, 2013, 1, 6365–6372 RSC.
- F. Yang, Q. Tang, X. Zhong, Y. Bai, T. Chen, Y. Zhang, Y. Li and W. Zheng, Int. J. Nanomed., 2012, 7, 835–844 CrossRef CAS PubMed.
- L. Yu, L. Sun, Y. Nan and L. Y. Zhu, Biol. Trace Elem. Res., 2011, 141, 254–261 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, T. Xu, C. Wang, H. Xia, H. Wang and B. Zhu, Int. J. Nanomed., 2016, 11, 3065–3076 CrossRef CAS PubMed.
- A. Khurana, S. Tekula, M. A. Saifi, P. Venkatesh and C. Godugu, Biomed. Pharmacother., 2019, 111, 802–812 CrossRef CAS PubMed.
- M. F. Dias, B. Figueiredo, J. Teixeira-Neto, M. Guerra, S. L. Fialho and C. A. Silva, Biomed. Pharmacother., 2018, 103, 1107–1114 CrossRef CAS PubMed.
- V. Nayak, K. R. Singh, A. K. Singh and R. P. Singh, New J. Chem., 2021, 45, 2849–2878 RSC.
- S. Chaudhary, A. Umar and S. K. Mehta, J. Biomed. Nanotechnol., 2014, 10, 3004–3042 CrossRef CAS PubMed.
- P. F. Surai, I. I. Kochish, V. I. Fisinin and D. T. Juniper, Animals, 2019, 9, 1–25 CrossRef PubMed.
- Y. Zhuang, L. Li, L. Feng, S. Wang, H. Su, H. Liu, H. Liu and Y. Wu, Nanoscale, 2020, 12, 1389–1396 RSC.
- R. Tian, Y. Geng, Y. Yang, I. Seim and G. Yang, Integrative Zoology, 2021, 16, 696–711 Search PubMed.
- Y. Li, Z. Lin, M. Guo, Y. Xia, M. Zhao, C. Wang, T. Xu, T. Chen and B. Zhu, Int. J. Nanomed., 2017, 12, 5733–5743 Search PubMed.
- M. Graham, B. Liang, G. Van Domselaar, N. Bastien, C. Beaudoin, S. Tyler, B. Kaplen, E. Landry and Y. Li, PLoS One, 2011, 6, e16087 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Guo, M. Zhao, Y. Xia, C. Wang, T. Xu and B. Zhu, Int. J. Nanomed., 2018, 13, 2005–2016 CrossRef CAS PubMed.
- Z. Lin, Y. Li, M. Guo, T. Xu, C. Wang, M. Zhao, H. Wang, T. Chen and B. Zhu, RSC Adv., 2017, 7, 742–750 RSC.
- J. Singh, K. R. Singh, M. Kumar, R. Verma, R. Verma, P. Malik, S. Srivastava, R. P. Singh and D. Kumar, Mater. Adv., 2021, 2, 6665–6675 RSC.
- P. Singh, K. R. Singh, J. Singh, P. Prasad and R. P. Singh, RSC Adv., 2021, 11, 25752–25763 RSC.
- M. Fernandes, K. R. Singh, T. Sarkar, P. Singh and R. P. Singh, Adv. Mater. Lett., 2020, 11, 081543 CrossRef.
- K. R. Singh, V. Nayak, J. Singh, A. K. Singh and R. P. Singh, RSC Adv., 2021, 11, 24722–24746 RSC.
- P. Singh, K. R. Singh, R. Verma, J. Singh and R. P. Singh, Mater. Lett., 2022, 307, 131053 CrossRef CAS.
- S. Mallick, K. R. Singh, V. Nayak, J. Singh and R. P. Singh, Mater. Lett., 2022, 306, 130912 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- W. Liu, P. Stachura, H. C. Xu, G. N. Umesh, F. Cox, R. Wang, K. S. Lang, J. Gopalakrishnan, D. Haussinger, B. Homey, P. A. Lang and A. A. Pandyra, J. Exp. Clin. Cancer Res., 2020, 39, 38 CrossRef PubMed.
- Y. Zhang, X. Li, Z. Huang, W. Zheng, C. Fan and T. Chen, Nanomedicine, 2013, 9, 74–84 CrossRef CAS PubMed.
- Y. Li, Z. Lin, G. Gong, M. Guo, T. Xu, C. Wang, M. Zhao, Y. Xia, Y. Tang, J. Zhong, Y. Chen, L. Hua, Y. Huang, F. Zeng and B. Zhu, J. Mater. Chem. B, 2019, 7, 4252–4262 RSC.
- H. Wu, X. Li and W. Liu, J. Mater. Chem., 2012, 22, 9602–9610 RSC.
- X. Gao, X. Li, J. Mu, C. T. Ho, J. Su, Y. Zhang, X. Lin, Z. Chen, B. Li and Y. Xie, Int. J. Biol. Macromol., 2020, 152, 605–615 CrossRef CAS PubMed.
- Z. Cui, S. Ren, J. Lu, F. Wang, W. Xu, Y. Sun, M. Wei, J. Chen, X. Gao, C. Xu, J. H. Mao and Y. Sun, Urol. Oncol., 2013, 31, 1117–1123 CrossRef CAS PubMed.
- M. A. Moosavi, A. Haghi, M. Rahmati, H. Taniguchi, A. Mocan, J. Echeverria, V. K. Gupta, N. T. Tzvetkov and A. G. Atanasov, Cancer Lett., 2018, 424, 46–69 CrossRef CAS PubMed.
- X. Li, L. Ma, W. Zheng and T. Chen, Biomaterials, 2014, 35, 8596–8604 CrossRef CAS PubMed.
- Y. Li, Z. Lin, M. Zhao, M. Guo, T. Xu, C. Wang, H. Xia and B. Zhu, RSC Adv., 2016, 6, 89679–89686 RSC.
- J. Zhang, X. Wang, V. Vikash, Q. Ye, D. Wu, Y. Liu and W. Dong, Oxid. Med. Cell. Longevity, 2016, 2016, 1–18 CAS.
- W. Liu, X. Li, Y. S. Wong, W. Zheng, Y. Zhang, W. Cao and T. Chen, ACS Nano, 2012, 6, 6578–6591 CrossRef CAS PubMed.
- Y. You, H. Hu, L. He and T. Chen, Chem.–Asian J., 2015, 10, 2744–2754 CrossRef CAS PubMed.
Footnote |
| † Xia Liu, Danyang Chen and Jingyao Su contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2022 |