Open Access Article
Open Access ArticleRapid tryptic peptide mapping of human serum albumin using DI-MS/MSALL†
Ke Zhang‡
,
Xingcheng Gong‡,
Qian Wang,
Pengfei Tu,
Jun Li and
Yuelin Song
and
Yuelin Song *
*
Modern Research Center for Traditional Chinese Medicine, School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing 100029, China. E-mail: syltwc2005@163.com
First published on 29th March 2022
Abstract
In recent decades, proteinic drugs, in particular monoclonal antibodies, are taking the leading role of small molecule drugs, and peptide mapping relying on liquid chromatography-tandem mass spectrometry (LC-MS/MS) is an emerging approach to substitute the role of a ligand-binding assay for the quality control of the proteinic drugs. However, such LC-MS/MS approaches extensively suffer from time-intensive measurements, leading to a limited throughput. To achieve accelerated measurements, here, the potential of DI-MS/MSALL towards tryptic peptide mapping was evaluated through comparing with well-defined LC-MS/MS means, and human serum albumin (HSA) was employed as the representative protein for applicability illustration. Among the 55 tryptic peptides theoretically suggested by Skyline software, 47 were successfully captured by DI-MS/MSALL through acquiring the desired MS2 spectra, in comparison to 51 detected by LC-MS/MS. DI-MS/MSALL measurements merely took 5 min, which was dramatically superior to the LC-MS/MS assay. Noteworthily, different from fruitful multi-charged MS1 signals for LC-MS/MS, most quasi-molecular ions received lower charged states. DI-MS/MSALL also possessed advantages such as lower solvent consumption and facile instrumentation; however, more sample was consumed. In conclusion, DI-MS/MSALL is eligible to act as an alternative analytical tool for LC-MS/MS towards the peptide mapping of proteinic drugs, particularly when a heavy measurement workload.
1. Introduction
In recent decades, proteinic drugs, such as monoclonal antibodies (mAbs), immunoglobulin, interferon, insulin, etc., account for an increasing proportion in the drug market.1–4 Although many analytical tools can fully address the quality control requirements of small molecular drugs, it is still a challenging task to assess the quality of so-called macromolecules.5,6 Previously, molecular weight measurements and ligand-binding assays (LBA) served as determinant roles for the quality control of protonic drugs.7 Recently, owing to the rapid development of mass spectrometric technologies, particularly Qtof-MS and Orbitrap-MS, the quality control level of protonic drugs has been significantly improved. As a widely popular technique, MALDI-TOF-MS is able to measure the exact molecular weight of a given protein without proteolysis;8,9 however, this technique fails to provide the desired amino acid sequence information. After proteolysis, such as tryptic digestion, the entire sequence is segmented into a set of peptides. When employing collision-induced dissociation (CID), their mass fragmentation pathways, similar to those small molecules, enable amino acid sequencing of each peptide, resulting in a promising strategy namely peptide mapping for in-depth quality control of proteinic drugs.10,11 Technically, peptide mapping can be regarded as a bottom-up proteomic approach.5,12,13Liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) is always a fit-for-purpose analytical tool in response to a peptide mapping strategy.14–16 LC is responsible for chromatographing the proteolytic peptide pool into pure fractions and subsequently transmitting to MS/MS for the acquisition of both MS1 and MS2 spectra for each peptide. Afterwards, those quasi-molecular ions, e.g., [M + 2H]2+ and [M + 3H]3+, together with the featured fragment ion species, such as a+, b+, c+, x+, y+, and z+, resulting from the dissociations around the amido bond,17,18 facilitate the configuration of each peptide, usually through searching relevant databases. Actually, the core-concept of peptide mapping is the construction of a well-aligned MS1–MS2–amino acid sequence dataset. Because of the unique ability to segment complicated ion populations into sequential ion fractions with relative narrow m/z windows prior to the entrance into the collision cell, the gas phase ion fractionation (GPF) theory has been demonstrated as a versatile approach to convert the selectivity advantage of MS to the so-called mass spectrometric separation ability,19–21 leading to a possibility for the removal of the LC domain when analyzing complicated matrices. In theory, narrower GPF windows theoretically facilitate greater mass spectrometric separation potential, even resulting in homogeneous ion clusters, however, a dramatic increment of MS2 spectral acquisition time. Although being able to decrease the measurement time, a wider GPF bin significantly compromises the spectral recording capacity, and even worse, generates a great technical challenge to correlate fragment ion species with their precursor ions. The SWATH program,22–24 usually bearing a GPF window as 25 Da is actually generated by balancing the MS2 spectral scan rate with peak width. If it is allowed by the great peak width, the GPF window can be defined as narrow as 1 Da and the exact program is available as the MS/MSALL technique,25–27 leading to the MS2 spectral acquisition potential for each nominal mass. Direct infusion (DI), fortunately, offers a great apparent peak width which is equal to the infusion time, to each analyte, thus allowing in theory, the acquisition of the MS2 spectrum for each 1 Da mass window. Consequently, DI-MS/MSALL should be a viable choice to build the MS1–MS2–amino acid sequence dataset within only a couple of minutes, which is attributed to the removal of the LC domain.
Previous studies have steadily proved that DI-MS/MSALL is an eligible analytical tool for the comprehensive characterization of small molecules in complicated matrices, such as herbal medicines.28–30 In the current study, we attempted to assess the potential of DI-MS/MSALL towards the universal acquisition of MS1 and MS2 spectra for each peptide in the tryptic digestion solution through comparing with the conventional LC-MS/MS approach. As a proof-of-concept, the MS1–MS2–amino acid sequence dataset construction was conducted for HSA, the entire sequence of which is available in relevant databases, e.g., PDB (http://www.wwpdb.org/) and UniProt (https://www.uniprot.org/). The obtained findings are envisioned to demonstrate DI-MS/MSALL as an alternative choice for LC-MS/MS towards peptide mapping, in particular when there is a tremendous quality assessment workload for those protonic drugs.
2. Materials and methods
2.1 Chemicals and reagents
HSA (molecular weight: 66.5 kDa) was purchased from Yuanye Biotech Co. Ltd (Shanghai, China). Tri-(β-chloroethyl)-phosphate (TCEP) and iodoacetamide (IAA) were commercially obtained from Sigma-Aldrich (St. Louis, MO). Trypsin was obtained from Promega (Fitchburg, WI). Ultrafiltration units with a 10 kDa molecular weight cut-off (MWCO) were supplied by Sartorius (Goettingen, Germany). Ultra-pure water was prepared in-house on a Milli-Q Integral water purification system (Millipore, Bedford, MA). All other chemicals and reagents were of the highest grade available and purchased from Beijing Chemical Works (Beijing, China).2.2 Sample preparation
The HSA tryptic peptide pool preparation was adapted from the well-defined Filter Aided Sample Prep (FASP) protocols with minor modifications.31 In brief, HSA was denatured at 95 °C for 5 min and subsequently reduced through incubation with 10 mmol L−1 TCEP at 67 °C for 10 min. The incubate was then transferred into the ultrafiltration unit and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. A 100 μL aliquot of 100 mmol L−1 IAA was utilized for thiol group carbamidomethylation via lucifugal incubation for 30 min at room temperature (23 ± 1 °C). Following centrifugation at 10
000 rpm for 30 min. A 100 μL aliquot of 100 mmol L−1 IAA was utilized for thiol group carbamidomethylation via lucifugal incubation for 30 min at room temperature (23 ± 1 °C). Following centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min, the residues were successively washed with 100 μL of 8 mol L−1 aqueous urea and 200 μL of 50 mmol L−1 NH4HCO3, for twice. The residues in the ultrafiltration unit then underwent tryptic digestion (trypsin/protein: 1/50) for 16 h at 37 °C. Afterwards, the ultrafiltration unit containing tryptic peptides was inserted into another clear tube to receive centrifugation at 10
000 rpm for 30 min, the residues were successively washed with 100 μL of 8 mol L−1 aqueous urea and 200 μL of 50 mmol L−1 NH4HCO3, for twice. The residues in the ultrafiltration unit then underwent tryptic digestion (trypsin/protein: 1/50) for 16 h at 37 °C. Afterwards, the ultrafiltration unit containing tryptic peptides was inserted into another clear tube to receive centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min, and the resultant filtrates were subjected for DI-MS/MSALL as well as LC-MS/MS analysis.
000 rpm for 30 min, and the resultant filtrates were subjected for DI-MS/MSALL as well as LC-MS/MS analysis.
2.3 DI-MS/MSALL measurements
The peptide mixture was directly infused into the ESI interface of a SCIEX TripleTOF 6600+ mass spectrometer (FOSTER City, CA) by applying the Flow Injection Analysis (FIA) program.25 A SIL-20ACXR auto-sampler (Shimadzu, Kyoto, Japan) was responsible for delivering 50 μL sample at a rate of 10 μL min−1. The MS/MSALL program was deployed for spectral acquisition. At the first 10 seconds, all ions amongst m/z 100–2000 formed in the ion source domain were recorded to generate the MS1 spectrum. Afterwards, the ion population was fragmented into consecutive ion fractions with a 1 Da width by applying the GPF algorithm to the first quadrupole cell (Q1). Each ion cluster subsequently entered the collision cell to receive collision-induced dissociation, and all fragment ion species were transmitted into the TOF chamber to yield high-resolution MS2 spectra. Primary settings for the MS/MS spectral acquisition were defined as follows: ionization polarity, positive; curtain gas, 25 MPa; GS1, 25 MPa; GS2, 15 MPa; spray voltage, 5500 V; temperature, 400 °C; scan range of either MS1 or MS2, m/z 100–2000; accumulation time of survey scan, 10 s; collision energy (CE), 35 eV; collision energy spread (CES), 15 eV; and accumulation time of each MS2 spectral recording, 100 ms. Data processing, mainly correlating the MS2 spectrum to each signal in the MS1 spectrum, was conducted with SCIEX MasterView software.2.4 LC-MS/MS measurements
LC-MS/MS measurements were undertaken on LC (LC-20AD modular equipment, Shimadzu) coupled with the SCIEX TripleTOF 6600+ mass spectrometer, and the chromatographic separation was carried out on a Waters Acquity UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm, Milford, CT). The mobile phase composed of 0.1% aqueous formic acid (A) and ACN containing 0.1% formic acid (B) was delivered by following the gradient program as follows: 0–3 min, 10% B – 15% B; 3–13 min, 15% B – 20% B; 13–19 min, 20% B – 30% B; 19–23 min, 30% B – 95% B; 23–25 min, 95% B; 25–25.1 min, 95% B – 10% B; 25.1–30 min, 10% B; and flow rate, 0.2 mL min−1. The column was maintained at 40 °C and the injection volume was set as 2.0 μL. Except the employment of data-dependent acquisition (DDA) to automatically trigger MS2 spectral acquisition for top-3 MS1 spectral signals, the other settings were exactly identical with those of the DI-MS/MSALL program.2.5 Software-assisted peptide annotation
The canonical sequence of HSA (ID: P02768) was obtained from UniProt. The FASTA file containing the information, such as protein ID and sequence, was imported into Skyline software32 to generate the tryptic peptide list as well as the quasi-molecular ions and theoretical computational fragment ion species of each peptide. Peptide settings were defined as below: enzyme, trypsin [KR|P]; missed cleavage, 0; sequence length, 4–40 amino acids; and fixed modification, carbamidomethyl (C). Regarding the ion transition settings, the charge-states of the precursor ions were defined as 1–6, and all a+, x+, b+, y+, c+ and z+ were involved as the fragment ion types.When matching the acquired MS/MS spectral data from either DI-MS/MSALL or LC-MS/MS with the accurate m/z information suggested by Skyline software, a mass tolerance of ±10 ppm was allowed to assign the fragment ion species.
3. Results and discussion
3.1 Peptide characterization with DI-MS/MSALL
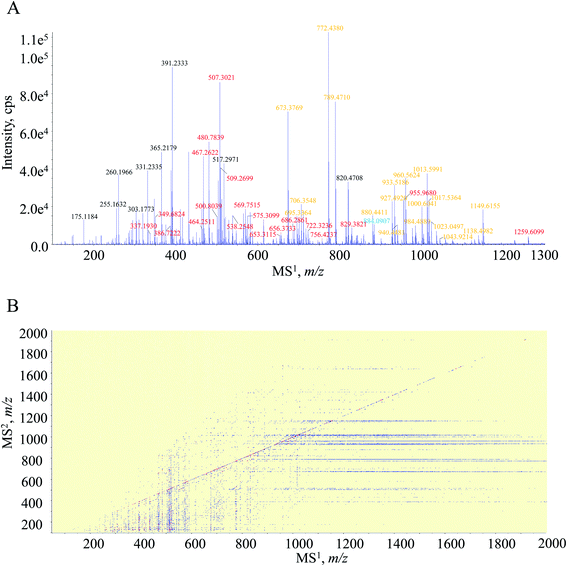
The entire MS/MSALL measurement actually contained the MS1 full scan experiment. To ensure the MS1 spectrum quality, the initial ten seconds were scheduled for the MS1 spectral recording. As shown in MS1 spectrum (Fig. 1A), the primary signals occurred at m/z 467.2622, 480.7839, 507.3021, 569.7515, 673.3769, 772.4380, 789.4710 and 1013.5991. Attention was paid afterwards to clarify the charge-state of each MS1 spectral signal, which was of great importance for the molecular formula calculation and the subsequent amino acid sequencing. Because of the significant natural abundance (approximately 1.1%) and the determinant role for peptides, the 13C-isotopic signal was employed to indicate the charge-state. If the distance between the concerned signal and its 13C-isotopic signal was 1.000 Da ± 5 mDa, the ion should be singly charged. Moreover, the 0.500 Da ± 5 mDa difference corresponded to doubly charged and so forth. Taking m/z 1013.5991 for instance, the isotopic signal was observed at m/z 1014.6024, demonstrating that it should be a singly charged ion, and the molecular formula was thereby calculated as C45H80O14N12. Meanwhile, because of the occurrence of the isotopic signal at m/z 481.2850, m/z 480.7839 was defined as the doubly charged ion, and hence, the elemental composition was generated as C44H73O11N13. Overall, regarding those primary signals, 36 were singly charged ions and 37 were doubly charged ions, whilst triply charged ions, even higher charged states, rarely occurred in the MS1 spectrum. Thereafter, we found that several MS1 spectral signals corresponded to a single peptide, and for instance, m/z 1000.6041 and 500.8039 were the singly and doubly charged molecular ions, respectively, of the peptide bearing a molecular formula as C45H81O14N11. Following careful data processing, those obvious MS1 spectral signals are assigned as Table 1.| No. | Peptide | LC-MS/MS | MS/MSALL | |||
|---|---|---|---|---|---|---|
| tR | MS1 | MS2 | MS1 | MS2 | ||
| 1 | SEVAHR | 1.18 | 698.3598(1) | — | 698.3569(1) | 524.2386, 496.2567, 482.2795, 465.2551, 366.1822, 316.1464, 295.1463, 217.0823, 175.1164, 158.0921 |
| 349.6838(2) | 611.3245, 482.2845, 465.2558, 383.2155, 366.1883, 316.1497, 312.1781, 295.1520, 234.1324, 217.0826, 201.1243, 189.0876, 175.1191, 158.0926, 105.0660 | 349.6824(2) | 611.3216, 482.2819, 465.2561, 383.2123, 366.1877, 316.1513, 312.1742, 295.1537, 288.1457, 234.1313, 217.0817, 201.1244, 189.0867, 175.1188, 158.0904, 105.0660 | |||
| 2 | DLGEENFK | 5.62 | 951.4476(1) | 836.4223, 723.3375 | 951.4425(1) | 537.2641, 408.2357 |
| 476.2238(2) | 723.3299, 706.3057, 666.3093, 537.2672, 520.2390, 408.2236, 391.1960, 294.1816, 286.1400, 229.1184, 201.1240, 147.1125, 130.0856 | 476.2230(2) | 723.3306, 537.2475, 432.2603, 415.2388, 286.1494, 229.1356, 201.1228, 173.1231, 147.1110, 133.0473, 130.0906 | |||
| 3 | ALVLIAFAQYLQQCPFEDHVK | — | — | — | — | — |
| 4 | LVNEVTEFAK | 10.24 | 1149.6180(1) | — | 1149.6155(1) | — |
| 575.3099(2) | 1036.5326, 1019.5066, 937.4629, 920.4387, 823.4207, 694.3782, 656.3597, 595.3097, 555.2431, 494.2614, 477.2357, 456.2458, 391.1618, 365.2185, 327.2029, 244.0932, 218.1503, 213.1601, 201.1230, 185.1654, 147.1131, 130.0860 | 575.3099(2) | 1036.5237, 937.4616, 920.4336, 823.4157, 694.3746, 595.3064, 494.2579, 456.2419, 365.2159, 244.0924, 218.1483, 213.1588, 185.1644, 130.0862 | |||
| 383.8747(3) | — | — | — | |||
| 5 | TCVADESAENCDK | 1.73 | 1498.5834(1) | — | — | — |
| 749.7927(2) | 1237.4992, 1138.4326, 1067.3959, 952.3675, 823.3261, 736.2928, 676.2595, 665.2544, 547.2183, 536.2133, 432.1919, 422.1706, 361.1550, 262.0865, 234.0908, 119.0815 | 749.7915(2) | 1237.5019, 1138.4293, 1121.4071, 1067.3852, 952.3578, 823.3382, 763.2907, 736.2967, 547.2214, 536.2098, 432.1851, 422.1710, 404.1968, 361.1507, 262.0837, 234.0927, 147.1125, 130.0797, 119.0797 | |||
| 500.1971(3) | — | — | — | |||
| 6 | SLHTLFGDK | 8.70 | 1017.5374(1) | — | 1017.5364(1) | 871.4240, 756.3999, 699.3757, 579.3075, 524.3187, 466.2261, 439.2260, 338.1783, 147.1117 |
| 509.2713(2) | — | 509.2699(2) | 871.4292, 817.4184, 680.3581, 671.3100, 579.3110, 492.2258, 466.2262, 439.2288, 345.1568, 338.1817, 319.1596, 262.1392, 201.1213, 173.1280, 147.1121 | |||
| 339.8508 (3) | 817.4207, 680.3633, 671.3200, 579.3137, 552.3129, 524.3175, 466.2309, 439.2313, 411.2337, 338.1830, 319.1624, 310.1858, 262.1419, 201.1239, 173.1287, 147.1132, 130.0869 | |||||
| 7 | LCTVATLR | 6.38 | 933.5207(1) | — | 933.5186(1) | — |
| 467.2633(2) | 820.4347, 660.4058, 559.3561, 460.2875, 460.2875, 443.2611, 389.2512, 389.2512, 372.2243, 288.2031, 274.1227, 271.1767, 234.0907, 201.1213, 175.1190, 158.0922, 131.1182 | 467.2622(2) | 820.4316, 660.4020, 559.3545, 460.2865, 443.2576, 389.2501, 375.1674, 372.2218, 274.1205, 271.1745, 246.1266, 201.1220, 175.1184, 131.1179 | |||
| 8 | ETYGEMADCCAK | 4.42 | 1434.5359(1) | — | 1434.5366(1) | — |
| 717.7710(2) | 1204.4447, 1041.3801, 984.3584, 855.3156, 724.2756, 711.2642, 653.2384, 580.2252, 538.2114, 378.1796, 231.0975, 218.1498, 203.1023, 147.1121, 102.0556 | 717.7695(2) | 1204.4326, 1041.3765, 984.3524, 855.3069, 724.2694, 653.2303, 538.2096, 378.1797, 231.0955 | |||
| 478.8487(3) | 1041.4015, 855.3085, 724.2765, 653.2377, 580.2249, 538.2118, 451.1784, 378.1812, 231.0954, 218.1480, 147.1114, 102.0556 | — | — | |||
| 9 | QEPER | 1.20 | 658.3172(1) | 484.2093; 401.2146; 384.1886; 355.1591; 258.1103; 175.1191 | 658.3143(1) | 401.2118; 384.1795; 175.1181 |
| 329.6621(2) | 530.2355; 401.2151; 384.1859; 304.1607; 230.1133; 175.1201; 129.0649 | 329.6608(2) | 401.2140; 304.1573; 175.1176 | |||
| 10 | NECFLQHK | 4.03 | 1075.5022(1) | — | 1075.5020(1) | — |
| 538.2531(2) | — | 538.2548(2) | 961.4519, 901.4185, 832.4090, 672.3751, 664.2945, 655.3508, 636.2792, 551.1681, 525.3079, 508.2800, 421.1879, 412.2298, 404.1213, 395.2019, 284.1696, 244.0938, 216.0958, 147.1109, 130.0939, 115.0884 | |||
| 359.1712(3) | 832.4132, 672.3837, 525.3133, 508.2867, 412.2315, 395.2054, 284.1723, 261.0506, 244.0819, 216.0976, 147.1129, 132.0803, 130.0857 | — | — | |||
| 11 | DDNPNLPR | 3.64 | 940.4505(1) | 825.4200, 710.3957, 693.3673, 596.3488, 579.3354, 482.2694, 345.1005, 272.1674, 255.1430 | 940.4481(1) | 825.4157, 710.3957, 669.2960, 596.3538, 255.1492, 175.1208 |
| 470.7283(2) | 710.3941, 693.3702, 596.3507, 579.3222, 499.2989, 482.2715, 385.2559, 368.2293, 345.1039, 272.1725, 255.1455, 231.0614, 203.0664, 175.1193, 158.0917 | 470.7271(2) | 710.3928, 693.3638, 596.3509, 579.3180, 499.2963, 482.2701, 385.2529, 368.2284, 345.1022, 272.1720, 255.1463, 231.0601, 203.0665, 175.1182, 158.0919, 116.0682 | |||
| 12 | LVRPEVDVMCTAFHDNEETFLK | 18.28 | 1325.6376(2) | 1279.6002, 995.4704, 880.4579, 809.4365, 508.2960 | — | — |
| YLYEIAR | 9.31 | 884.0922(3) | 1611.7058, 1518.7510, 1451.6917, 1371.6873, 1350.6299, 1300.6491, 1279.5971, 1199.5927, 1132.5303, 1039.5654, 995.4666, 908.5204, 880.4415, 809.4528, 766.3946, 694.4218, 637.3533, 595.3585, 508.3144, 407.2649, 369.2601, 260.1970, 213.1594, 185.1645, 147.1129 | 884.4256 (3) | 1655.8134, 1611.6562, 1518.7436, 1490.7492, 1451.6591, 1371.6761, 1350.6283, 1343.6808, 1279.5740, 1132.5275, 1039.5580, 995.4681, 908.5144, 880.4309, 809.4469, 766.3838, 694.4204, 637.3443, 508.3152, 407.2743, 390.2358, 369.2548, 260.1975, 185.1651, 147.1119, 130.0867 | |
| 663.3205(4) | 1279.5974, 1132.5277, 1039.5616, 995.4682, 908.5196, 880.4491, 809.4519, 694.4242, 663.3179, 595.3568, 567.3621, 508.3112, 407.2651, 369.2603, 260.1971, 213.1596, 185.1650, 147.1130, 130.0864 | — | — | |||
| 13 | 927.4958(1) | — | 927.4920(1) | 682.3449, 654.3515, 569.2554, 541.2525, 440.2183, 412.2254, 359.2407, 175.1186 | ||
| 464.2506(2) | 764.4310, 651.3467, 634.3193, 488.2837, 359.2409, 342.2141, 277.1558, 249.1608, 246.1564, 229.1300, 175.1196, 158.0928, 136.0766 | 464.2511(2) | 764.4249, 651.3413, 488.2785, 359.2351, 277.1525, 249.1594, 229.1012, 175.1185, 158.0900, 136.0746 | |||
| 14 | HPYFYAPELLFFAK | 21.42 | 1742.8997(1) | — | — | — |
| 871.9500(2) | — | — | — | |||
| 581.6349(3) | 1231.6172, 1118.5403, 1035.5770, 1005.4517, 964.5522, 867.5023, 779.3545, 751.3624, 738.4587, 708.3173, 680.3227, 625.3757, 545.2563, 517.2589, 512.2915, 398.1863, 365.2229, 235.1217, 218.1518, 147.1144, 130.0876, 110.0737 | — | — | |||
| 476.4785(4) | — | — | — | |||
| 15 | AAFTECCQAADK | 3.75 | 1371.5716(1) | — | 1371.5686(1) | — |
| 686.2874(2) | 1229.4235, 1082.4235, 1082.4235, 981.3755, 852.3338, 692.3016, 532.2714, 520.2390, 515.2436, 404.2131, 391.1967, 333.1762, 290.1503, 262.1397, 262.1397, 245.1293, 147.1127, 143.0813, 115.0864 | 686.2861(2) | 1229.4778, 1225.4405, 1082.4189, 1082.4189, 1039.3828, 981.3688, 968.3539, 852.3296, 835.2966, 692.2943, 532.2681, 520.2386, 515.2366, 404.2141, 391.1935, 333.1740, 290.1472, 262.1396, 245.1228, 147.1108, 143.0806, 130.0860, 115.0847 | |||
| 457.8596(3) | — | — | — | |||
| 16 | AACLLPK | 6.22 | 772.4403(1) | 630.3612, 529.2766, 501.2852, 416.1946, 357.2448, 303.1117, 244.1636, 147.1130 | 772.4380(1) | 630.3598, 529.2782, 501.2865, 470.3312, 416.1957, 388.2002, 357.2490, 303.1126, 244.1657, 147.1121 |
| 386.7234(2) | 701.4005, 630.3639, 470.3340, 416.1963, 357.2497, 303.1129, 244.1661, 147.1133, 143.0813, 130.0862, 115.0870 | 386.7222(2) | 701.3975, 630.3604, 613.3555, 470.3281, 416.1956, 357.2483, 303.1087, 244.1647, 160.1072, 147.1120, 143.0797, 115.0867 | |||
| 17 | LDELR | 3.67 | 645.3572(1) | — | 645.3558(1) | 417.2424; 358.1562; 288.2007; 229.1195; 175.1175 |
| 323.1828(2) | 532.2747; 471.2575; 417.2465; 358.1595; 288.2046; 229.1194; 201.1242; 175.1192 | — | — | |||
| 18 | DEGK | 1.11 | 448.2033(1) | 333.1731; 302.0981; 245.0705; 217.0792; 204.1337; 147.1107 | 448.2037(1) | 333.1778; 217.0792; 204.1314; 147.1118 |
| 19 | ASSAK | 1.05 | 463.2518(1) | 392.2136; 314.1464; 305.1823; 246.1087; 218.1506; 159.0766; 147.1134 | 463.2501(1) | 392.2127; 305.1821; 246.1063; 218.1496; 159.0755; 147.1121 |
| 232.1292(2) | — | — | — | |||
| 20 | CASLQK | 1.72 | 706.3556(1) | — | 706.3548(1) | 560.2492, 546.3191, 475.2844, 432.1887, 404.1893, 388.2523, 319.1068, 275.1696, 232.0739, 204.0792, 147.1112, 133.0415 |
| 353.6822(2) | 560.2476, 546.3231, 529.2936, 475.2859, 458.2619, 449.2114, 404.1945, 388.2554, 319.1069, 291.1115, 275.1722, 258.1454, 232.0744, 204.0804, 161.0370, 147.1128, 133.0431, 130.0868 | 353.6812(2) | 546.3336, 529.2870, 475.2884, 388.2510, 319.1084, 275.1700, 258.1419, 232.0726, 204.0787, 161.0395, 147.1109, 133.0417, 130.0864 | |||
| 21 | FGER | 1.80 | 508.2520(1) | 361.1852; 334.1382; 205.0971; 175.1192; 120.0811 | — | — |
| 254.6302(2) | 361.1841; 175.1199; 120.0827 | — | — | |||
| 22 | AWAVAR | 5.24 | 673.3794(1) | 585.3137, 499.2674, 471.2702, 428.2324, 416.2609, 399.2333, 345.2234, 329.1629, 328.1997, 258.1236, 246.1551, 229.1302, 175.1196, 158.0916 | 673.3769(1) | 499.2717, 428.2254, 400.2292, 399.2332, 329.1599, 328.1964, 258.1228, 175.1195 |
| 337.1939(2) | 602.3408, 471.2669, 416.2625, 399.2348, 345.2246, 328.1975, 258.1240, 246.1499, 230.1291, 229.1298, 175.1193, 158.0899 | 337.1930(2) | 602.3306, 416.2603, 399.2359, 345.2185, 328.1892, 258.1188, 246.1504, 230.1289, 229.1281, 175.1180, 158.0894 | |||
| 23 | LSQR | 1.18 | 503.2945(1) | 390.2106; 303.1790; 175.1197 | 503.2925(1) | 303.1766; 175.1188 |
| — | — | 252.1505(2) | 390.2003; 175.1205 | |||
| 24 | AEFAEVSK | 4.37 | 880.4428(1) | — | 880.4411(1) | 734.3247, 680.3574, 647.3019, 548.2344, 533.2896, 520.2334, 462.2505, 419.1902, 348.1560, 333.2121, 234.1418, 201.0840 |
| 440.7243(2) | 809.4044, 680.3619, 533.2934, 462.2557, 419.2262, 348.1552, 333.2133, 320.1609, 234.1452, 217.1180, 201.088, 173.0923, 173.0923, 147.1129, 130.0861 | 440.7232(2) | 809.4115, 680.3579, 533.2867, 462.2541, 419.2357, 348.1554, 333.2098, 234.1439, 201.0846, 173.0911, 173.0911, 147.1130, 130.0846 | |||
| 25 | LVTDLTK | 5.38 | 789.4722(1) | — | 789.4710(1) | 676.3794, 643.3671, 577.3193, 559.3115, 542.3169, 514.3209, 476.2678, 429.2334, 361.2417, 248.1598, 230.1500, 213.1586, 185.1642, 147.1107 |
| 395.2394(2) | 676.3873, 577.3213, 559.3088, 476.2714, 401.2395, 361.2449, 248.1610, 230.1501, 213.1594, 185.1651, 147.1134, 130.0861 | 395.2389(2) | 676.3848, 577.3157, 559.3016, 476.2608, 401.2475, 361.2391, 248.1598, 230.1498, 185.1642, 147.1132, 130.0856 | |||
| 26 | VHTECCHGDLLECADDR | 4.99 | 1043.9234(2) | 1850.6635, 1749.6969, 1620.6358, 1300.5453, 1163.4928, 991.4630, 765.2914, 636.2446, 290.1440 | — | — |
| 696.2853(3) | 1460.5873, 1300.5592, 1163.4987, 1106.4764, 991.4545, 878.3672, 787.2908, 765.2827, 759.2194, 636.2398, 476.2096, 405.1708, 388.1452, 338.1807, 310.1133, 290.1460, 237.1348, 209.1397, 175.1187, 158.0917 | — | — | |||
| 522.4647(4) | 1209.4845, 1096.3983, 924.3481, 765.2844, 636.2407, 476.2107, 405.1737, 338.1826, 290.1458, 237.1354, 209.1403, 175.1193 | — | — | |||
| 27 | ADLAK | 1.69 | — | — | 517.2971(1) | 371.1911; 331.2319; 300.1543; 218.1496; 147.1126 |
| 259.1529(2) | 446.2590; 331.2315; 218.1501; 147.1128 | — | — | |||
| 28 | YICENQDSISSK | 3.85 | 1443.6476(1) | — | 1443.6457(1) | — |
| 722.3249(2) | 1280.5795, 1167.4930, 1150.4703, 1123.4735, 1007.4649, 923.3578, 878.4219, 861.3944, 808.3293, 764.3787, 747.3523, 680.2705, 636.3207, 566.2298, 521.2939, 437.1854, 434.2607, 409.1880, 321.1772, 277.1551, 249.1606, 234.1449, 147.1124, 136.0761, 130.0862 | 722.3206(2) | 1167.4952, 1150.4563, 1007.4564, 878.4153, 636.3211, 521.2912, 437.1825, 434.2712, 321.1747, 277.1547, 249.1585, 136.0742 | |||
| 481.8849(3) | — | — | — | |||
| 29 | ECCEKPLLEK | 3.92 | 1305.6239(1) | — | — | — |
| 653.3133(2) | 917.3810, 856.5104, 727.4617, 599.3729, 389.2331, 290.0785, 276.1525, 147.1128 | 653.3115(2) | 1159.4666, 1016.5355, 856.5168, 727.4792, 599.3723, 389.2356, 290.0756, 276.1549, 147.1099, 147.1099, 102.0516 | |||
| 435.8778(3) | 1016.5269, 856.5163, 727.4704, 599.3771, 502.3265, 450.1096, 389.2409, 290.0810, 276.1560, 262.0854, 173.0916, 147.1125, 130.0866, 102.0554 | — | — | |||
| 30 | SHCIAEVENDEMPADLPSLAADFVESK | — | — | — | — | — |
| 31 | DVCK | 1.21 | 521.2406(1) | 406.2118; 375.1335; 347.1399; 307.1441; 232.1280; 215.1026; 187.1084; 147.1132 | 521.2376(1) | 406.2088; 375.1301; 347.1353; 307.1417; 232.1235; 215.1010; 187.1066; 147.1114; 133.0420 |
| 32 | NYAEAK | 1.67 | 695.3376(1) | 695.3364(1) | 581.2833, 549.2288, 521.2286, 478.1906, 450.1956, 418.2296, 349.1499, 347.1923, 278.1125, 250.1175, 218.1499, 201.0852, 147.1123, 130.0849 | |
| 348.1718(2) | 581.2964, 418.2313, 347.1928, 278.1227, 250.1197, 218.1508, 173.0925, 147.1130, 130.0868 | — | — | |||
| 33 | DVFLGMFLYEYAR | 21.67 | 1623.7919(1) | — | — | — |
| 812.3975(2) | 1409.6962, 1262.6298, 1215.5815, 1149.5434, 1092.5209, 1086.5336, 961.4794, 923.4712, 814.4092, 810.3871, 701.3276, 663.3192, 538.2638, 532.2781, 504.2808, 475.2559, 447.2602, 409.2196, 362.1722, 246.1575, 215.1030, 187.1079, 175.1191 | 812.3986(2) | 1378.6618, 1262.6046, 1149.5330, 1092.4912, 961.4964, 940.4506, 701.3266, 538.2550, 475.2480, 409.2311, 362.1734, 187.1098, 175.1170 | |||
| 541.9329(3) | 1262.6022, 1149.5416, 1092.5186, 961.4781, 814.4095, 810.3821, 701.3288, 663.3143, 538.2615, 532.2784, 475.2527, 409.2197, 392.1912, 362.1679, 246.1568, 229.1292, 215.1021, 187.1076, 175.1181 | — | — | |||
| 34 | HPDYSVVLLLR | 18.51 | 1311.7501(1) | — | 1311.7430(1) | — |
| 656.3746(2) | — | 656.3733(2) | 1174.6775, 1077.6293, 1060.6117, 1024.5528, 962.5924, 799.5280, 798.3687, 712.4986, 613.4525, 514.3685, 513.2237, 384.2651, 350.1406, 235.1166, 175.1177, 158.0900, 110.0714 | |||
| 437.9192(3) | 798.3763, 699.3082, 671.3120, 613.4398, 600.2419, 572.2412, 514.3723, 513.2084, 497.3393, 401.2891, 384.2623, 350.1461, 288.2029, 271.1764, 235.1185, 175.1188, 158.0917, 110.0710 | — | — | |||
| 35 | TYETTLEK | 4.08 | 984.4909(1) | — | 984.4889(1) | 591.3265, 490.2805, 389.2293, 147.1125 |
| 492.7483(2) | 883.4399, 720.3763, 591.3341, 490.2869, 394.1597, 389.2398, 276.1552, 265.1185, 237.1241, 147.1126, 130.0861, 102.0552 | 492.7478(2) | 883.4323, 720.3710, 703.3700, 591.3315, 389.2360, 276.1515, 265.1202, 237.1219, 147.1105 | |||
| 36 | CCAAADPHECYAK | 1.81 | 1552.6050(1) | — | — | — |
| 776.8027(2) | — | 776.8010(2) | 1406.4691, 1090.4576, 1019.4181, 1012.3537, 904.3963, 807.3317, 670.2848, 649.2004, 541.1721, 534.1721, 506.1735, 463.1412, 392.1050, 321.0678, 218.1501, 178.0642, 147.1134, 133.0429 | |||
| 518.2037(3) | 1090.4624, 1019.4351, 904.3976, 807.3457, 670.2868, 649.2085, 541.2430, 534.1802, 506.1837, 463.1430, 435.1402, 409.1298, 392.1051, 381.2099, 293.0744, 218.1500, 201.1217, 178.0646, 161.0387, 147.1126, 133.0433, 130.0864 | — | — | |||
| 37 | VFDEFKPLVEEPQNLIK | 19.42 | 1023.0530(2) | 1279.7284, 766.3710, 712.4221, 491.2062 | 1023.0483(2) | 1898.9958, 1785.8923, 1407.8146, 1333.665, 1279.7279, 1204.6113, 1075.5932, 1069.5734, 976.5194, 970.5191, 841.4679, 766.3785, 712.4308, 638.2785, 610.2786, 508.2487, 491.2145, 463.2258, 362.1697, 260.1909, 219.1520 |
| 682.3703(3) | 1333.6722, 1279.7282, 1262.7056, 1204.6275, 1182.6767, 1075.5844, 1069.5875, 976.5163, 970.5231, 841.4795, 766.3767, 712.4356, 695.4035, 638.2830, 615.3809, 610.2893, 598.3559, 491.2142, 487.3251, 463.2175, 373.2810, 362.1724, 260.1974, 247.1448, 219.1502, 147.1133, 130.0863 | — | — | |||
| 512.0283(4) | 1204.6269, 976.5154, 948.5137, 863.4322, 712.4335, 638.2852, 615.3758, 487.3257, 362.1690, 260.1962, 247.1412, 219.1480, 147.1119 | — | — | |||
| 38 | QNCELFEQLGEYK | 17.47 | 1657.7569(1) | — | 1657.7595(1) | — |
| 829.3804(2) | 1415.6538, 1328.5837, 1255.6217, 1162.5183, 1126.5778, 1013.4929, 921.3764, 866.4258, 792.3326, 737.3833, 720.3538, 645.2659, 609.3240, 532.1823, 496.2400, 439.2188, 403.1473, 310.1767, 243.1088, 147.1126, 101.0706 | 829.3821(2) | 1415.6514, 1255.6221, 1162.5076, 1126.5761, 1049.4437, 1013.4950, 921.3805, 866.4200, 792.3456, 737.3794, 645.2968, 617.2533, 609.3184, 532.2712, 496.2365, 403.1626, 310.1746, 243.1085, 215.1051, 130.0815 | |||
| 553.2549(3) | — | — | — | |||
| 39 | FQNALLVR | 10.92 | 960.5653(1) | — | 960.5624(1) | — |
| 480.7844(2) | 813.4956, 796.4692, 685.4366, 668.4108, 571.3930, 500.3562, 483.3293, 461.2154, 390.1792, 387.2727, 370.2452, 276.1352, 274.1879, 257.1611, 248.1396, 175.1196, 165.1031, 158.0930, 120.0817 | 480.7839(2) | 813.4908, 796.4663, 685.4335, 668.4070, 574.2914, 571.3925, 554.3533, 546.2971, 500.3548, 483.3238, 461.2093, 433.2182, 390.1752, 387.2700, 370.2441, 276.1335, 257.1601, 175.1188, 165.1020, 158.0924, 120.0811 | |||
| 40 | VPQVSTPTLVEVSR | 13.50 | 756.4262(2) | 1315.7213, 1187.6683, 1088.5972, 1001.5618, 900.5131, 612.3332, 589.3301, 511.2868, 490.2599, 424.2539, 396.2607, 325.1878, 197.1277, 175.1153, 169.1325 | 756.4237(2) | 1315.7190, 1298.6990, 1187.6609, 1088.5940, 1001.5701, 940.4374, 900.5110, 612.3214, 589.3220, 572.2987, 490.2652, 424.2563, 396.2541, 361.2138, 325.1845, 297.1873, 197.1256, 175.1162, 169.1345 |
| 504.6181(3) | 702.4016, 589.3265, 490.2573, 424.2572, 325.1872, 262.1595 | — | — | |||
| 41 | NLGK | 1.40 | 431.2627(1) | 317.2197; 285.1560; 228.1350; 204.1350; 200.1403; 147.1132 | 431.2603(1) | 317.2177; 285.1536; 228.1341; 204.1337; 200.1383; 147.1123 |
| 42 | VGSK | 1.79 | 390.2312(1) | 291.1674; 244.1291; 234.1459; 216.1339; 157.0974; 147.1133; 129.1028 | 390.2339(1) | 291.1652; 244.1273; 234.1444; 216.1329; 157.0968; 147.1122; 129.1016 |
| 43 | HPEAK | 1.01 | 581.3041(1) | 444.2439; 435.1973; 364.1611; 235.1187; 218.1492; 147.1128; 110.0714 | 581.3029(1) | 444.2442; 435.1950; 364.1592; 235.1178; 218.1490; 147.1114; 110.0702 |
| 291.1566(2) | — | 291.1563(2) | 444.2498; 435.1900; 364.1687; 235.1171; 218.1485; 147.1116; 110.0712 | |||
| 44 | MPCAEDYLSVVLNQLCVLHEK | 21.50 | 1259.6129(2) | — | 1259.6099(2) | — |
| 840.0758(3) | 1538.8403, 1451.8046, 1378.6395, 1352.7366, 1335.7076, 1265.5597, 1253.6670, 1236.6439, 1166.4859, 1140.5833, 1123.5567, 1067.4168, 1026.5399, 1009.5147, 980.3858, 898.4829, 867.3019, 785.3982, 704.2380, 625.3666, 589.2113, 526.2990, 460.1659, 413.2139, 389.1310, 276.1551, 229.1015, 201.1051, 147.1125, 104.0535 | 840.0747 (3) | 1538.8354, 1352.7322, 1265.5512, 1253.6692, 1166.4802, 1140.5795, 1067.3993, 1026.5395, 980.3891, 898.4737, 867.2965, 785.3961, 704.2385, 526.2951, 460.1624, 413.2119, 389.1286, 276.1510, 229.0958, 104.0529 | |||
| 630.3088(4) | — | — | — | |||
| 45 | TPVSDR | 1.55 | 674.3480(1) | — | — | — |
| 337.6777(2) | 573.3006, 476.2475, 459.2192, 377.1787, 360.1523, 290.1468, 273.1197, 199.1083, 175.1181, 171.1131, 158.0903 | 337.6802(2) | 573.2887, 556.2792, 476.2456, 377.1744, 360.1518, 273.1170, 175.1183, 171.1122, 158.0914 | |||
| 46 | CCTESLVNR | 4.00 | 1138.5005(1) | — | 1138.4982(1) | — |
| 569.7519(2) | 978.4716, 961.4460, 818.4379, 717.3911, 638.1893, 588.3476, 551.1593, 501.3155, 422.1175, 388.2315, 371.2040, 321.0697, 293.0749, 289.1631, 272.1361, 178.0652, 175.1197, 161.0381, 158.0926, 133.0435 | 569.7515(2) | 978.4629, 961.4601, 850.3317, 818.4313, 801.4100, 743.3786, 717.3866, 700.3718, 638.1851, 588.3421, 501.3112, 484.2740, 422.1135, 388.2296, 371.2011, 321.0660, 289.5950, 272.1331, 178.0645, 175.1175, 161.0362, 158.0905, 133.0422 | |||
| 380.1700(3) | — | — | — | |||
| 47 | RPCFSALEVDETYVPK | 15.89 | 955.9689(2) | 1667.7745, 1568.7009, 1405.6319, 1304.5837, 1175.5469, 1060.5170, 961.4535, 832.4091, 736.3781, 607.3368, 343.2319, 244.1652, 226.1549, 147.1122, 129.1014 | 955.9680(2) | — |
| 637.6487(3) | 1405.6409, 1304.5938, 1192.6170, 1175.5538, 1079.5268, 1060.5261, 1032.5270, 961.4562, 950.4837, 933.4608, 851.4148, 834.3870, 832.4132, 804.4192, 719.3278, 691.3337, 648.2933, 607.3453, 561.2590, 533.2641, 506.2974, 414.1900, 343.2336, 254.1602, 244.1661, 227.1371, 226.155, 157.1339, 147.1134, 130.0861, 129.1018 | 637.6471 (3) | 1192.6132, 1175.5350, 1060.5220, 961.4492, 950.4834, 933.4458, 851.4079, 832.4068, 804.4239, 736.3764, 719.3251, 648.2835, 607.3404, 414.1905, 343.2375, 254.1588, 244.1654, 147.1130, 130.0858 | |||
| 478.4871(4) | 832.4120, 506.2937, 254.1612, 244.1626, 157.1087, 147.1165 | — | — | |||
| 48 | EFNAETFTFHADICTLSEK | — | — | — | — | — |
| 49 | QTALVELVK | 13.45 | 1000.6073(1) | 854.4979, 771.5044, 755.4255, 642.3453, 587.3739, 513.3045, 488.3048, 414.2323, 359.2633, 301.1493, 230.1132 | 1000.6041(1) | — |
| 500.8058(2) | 872.5454, 771.4984, 700.4616, 587.3781, 488.3093, 359.2665, 246.1816, 147.1131, 130.0866, 101.0716 | 500.8039(2) | 872.5383, 771.4908, 700.4577, 587.3735, 488.3031, 471.2894, 385.2454, 359.2628, 342.2008, 301.1470, 246.1813, 229.1172, 202.1175, 147.1120, 130.0858, 101.0691 | |||
| 50 | HKPK | 0.91 | 509.3203(1) | 372.2598; 363.2139; 266.1659; 244.1659; 147.1127; 138.0654; 110.0720 | — | — |
| 255.1642(2) | 372.2624; 355.2336; 266.1607; 244.1651; 227.1386; 147.1128; 110.0721 | 255.1632(2) | 266.1587; 244.1666; 147.1115; 138.0641; 110.0717 | |||
| 51 | EQLK | 1.56 | 517.2985(1) | 388.2541; 371.2246; 260.1978; 258.1095; 147.1117; 102.0556 | 517.2917(1) | 371.1911; 260.1951; 258.1084; 147.1126 |
| 259.1529(2) | — | — | — | |||
| 52 | AVMDDFAAFVEK | 19.74 | 1342.6398(1) | — | 1342.6383(1) | — |
| 671.8229(2) | 1172.5323, 1041.4895, 926.4637, 821.3499, 811.4363, 750.3139, 679.2764, 664.3678, 593.3313, 532.1062, 522.2932, 417.1812, 375.2247, 302.1541, 276.1561, 171.1136, 147.1127, 143.1186 | 671.8201(2) | 1172.5279, 1067.4721, 1067.4721, 1041.4834, 952.4236, 926.4624, 837.3588, 811.4316, 664.3631, 651.2413, 593.3261, 548.2222, 532.1979, 522.2893, 417.1756, 375.2229, 302.1581, 302.1581, 276.1528, 171.1125, 147.1138, 143.1171 | |||
| 448.2164(3) | — | — | — | |||
| 53 | ADDK | — | — | — | — | — |
| 54 | ETCFAEEGK | 3.36 | 535.7264(2) | 941.4031, 840.3469, 680.3217, 533.2541, 462.2152, 333.1727, 231.0967, 204.1328, 203.1034, 130.0847, 102.0542 | — | — |
| 55 | LVAASQAALGL | 16.95 | 1013.6008(1) | — | 1013.5991(1) | 882.4960, 825.4799, 712.3986, 641.3573, 613.3274, 570.3192 |
| 507.3024(2) | 882.5043, 825.4831, 801.4458, 712.3996, 641.3613, 613.3346, 570.3246, 542.2963, 444.2803, 355.2328, 327.2243, 302.2084, 284.1964, 213.1602, 189.1241, 185.1633, 132.1022 | 507.3021(2) | 882.5017, 825.4781, 801.4553, 712.3988, 684.3937, 659.3703, 641.3565, 570.3225, 459.2871, 444.2797, 442.2656, 373.2455, 355.2326, 327.2361, 302.2039, 284.1958, 256.1647, 213.1588, 189.1230, 185.1646, 132.1014 | |||
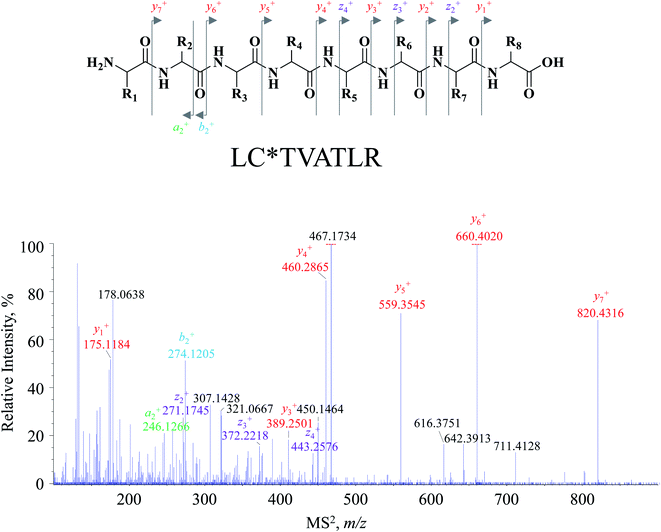
Attention was turned to the MS2 spectral profile acquired by the DI-MS/MSALL program. All fragment ion species are summarized as a scattering plot as Fig. 1B and actually, the plot should be a three-dimensional plot. As shown in Fig. 1B, most fragment ions are distributed under the line of y = x, whilst those dots above the line are resulted from the generation of singly charged fragment ions from multiply charged precursor ions. The MS2 spectrum of each primary MS1 signal could be extracted by the line of x = b where b was the m/z value of the MS1 signal-of-choice. After applying this rule, the correlation of the fragment ion species to the precursor ions was achieved, and peptide annotation was subsequently conducted by inquiring each MS1–MS2 item to Skyline software. Taking m/z 467.2622 for instance, all the fragment ion species are distributed on the line of x = 467.2622, and the extracted MS2 spectrum is shown in Fig. 2. Those primary MS2 spectral signals together with the precursor ion were inquired to Skyline software to assign each signal-of-interest. The peptide was outputted as LC*TVATLR. Following the well-defined ion nomenclature rule,33 m/z were assigned as y7+, y6+, y5+, y4+, z4+, y3+, b2+, and y1+, accordingly (Fig. 2). Obviously, y-type ions usually received greater responses (Fig. 2), whereas any x-type ions were absent in the MS2 spectrum.
A total of 47 peptides were characterized through matching the aligned MS1–MS2 items (Table 1) with the information calculated by Skyline software. Actually, Skyline software suggested that 55 peptides (Fig. S1, ESI†) could be given by the tryptic digestion of HSA, whilst the information of the other eight theoretical peptides, such as ALVLIAFAQYLQQC*PFEDHV, HPYFYAPELLFFAK, LSQR, VHTEC*C*HGDLLEC*ADDR, SHC*IAEVENDEMPADLPSLAADFVESK, EFNAETFTFHADIC*TLSEK, ETC*FAEEGK and ADDK, failed to exist in the DI-MS/MSALL data profile.
3.2 Comparison between DI-MS/MSALL and LC-MS/MS
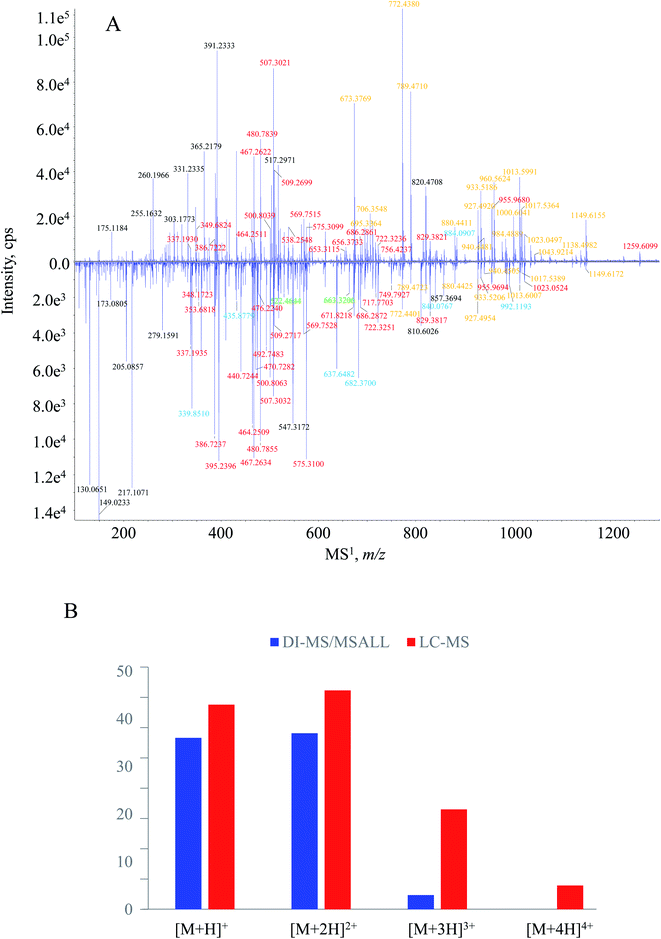
The tryptic peptide pool of HSA was also subjected to peptide mapping by LC-MS/MS. MS1 spectra were acquired by fully scanning the ions generated in the ion source, and MS2 spectral recording was automatically triggered by MS1 experiment via DDA manner. Because of the optimized chromatographic program, e.g., column, mobile phase and elution procedure, those tryptic peptides received satisfactory chromatographic behaviors in terms of retention time and resolution (Fig. S2, ESI†). Hence, it was quite convenient to build the MS1–MS2 dataset. After the inquiry of each MS1–MS2 item to Skyline software, a total of 51 tryptic peptides were captured, 4 additional ones (i.e., HPYFYAPELLFFAK, LSQR, VHTEC*C*HGDLLEC*ADDR and ETC*FAEEGK) in comparison to the DI-MS/MSALL measurements.MS1 spectral comparison between the two measurement pipelines is illustrated in Fig. 3A. It was worthy to mention that the downer MS1 spectrum corresponded to the average spectrum amongst the entire measurement time (0–25 min). Except for possessing most signals of DI-MS/MSALL, more signals were generated by LC-MS/MS. Overall, signals with greater m/z values gained greater responses from DI-MS/MSALL whereas those low m/z signals, in particular m/z 130.0651, 149.0233, 205.0857, and 217.1071, were quite abundant in the MS1 spectrum in the presence of LC. When concern was paid to the charge-state profile, we found that the quasi-molecular ion bearing high charged levels obtained significant responses from LC-MS/MS rather than DI-MS/MSALL. Regarding the downer MS1 spectrum, 21 and 5 signals were assigned as triply and quadruply charged quasi-molecular ions, respectively (Fig. 3B), whereas only three triply ions and no quadruply charged ions were captured by the DI-MS/MSALL program (Fig. 3B). For those singly and doubly charged quasi-molecular ions, comparable performances occurred between DI-MS/MSALL (36 singly charged ions and 37 doubly charged ions) and LC-MS/MS (43 singly charged ions and 46 doubly charged ions). The different charge-state features between these measurement approaches could be attributed to the greater ionization competition for DI-MS/MSALL because all peptides as well as some substrates arrived at the ion source at the same time.34,35 On the other hand, those signals distributed within the low m/z region, such as m/z 130.0651, 149.0233, 205.0857 and 217.1071, were assigned as the fragment ion species resulting from the extensive in-source dissociation when undergoing LC-MS/MS measurement.
In comparison of the significant difference occurring for the MS1 spectrum, a greater similarity was observed for the MS2 spectral pattern. Taking LC*TVATLR for instance (Fig. S3, ESI†), most fragment ion species, e.g., m/z 820.4347, 660.4043, 559.3561, 460.2875, 389.2515, 274.1227 and 175.1190 detected by DI-MS/MSALL were also captured by LC-MS/MS. Because those MS2 spectral signals played the determinant role for amino acid sequencing, DI-MS/MSALL therefore showed comparable ability with the well-recommended LC-MS/MS means for peptide annotation. Combining the performances on both the MS1 and MS2 spectrum acquisition, overall, these two methods were comparable in peptide coverage and sequence annotation.
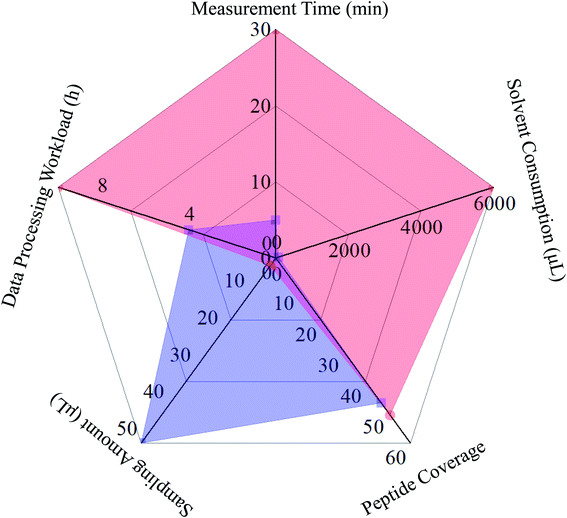
Owing to the robust chromatographic separation ability, LC was able to transmit even pure analytes into the ion source of the mass spectrometer, and a given MS2 spectrum usually corresponded exactly to the single precursor ion. Moreover, DDA manner would further facilitate correlating MS2 spectra to their precursor ions. Consequently, LC-MS/MS in combination with the DDA algorithm served as the most favored pipeline for peptide mapping. In the case of DI-MS/MSALL, the separation potential towards the ion population generated from all analytes was totally resulted from the gas phase ion fractionation algorithm, and the ion current consisting of ions with identical nominal mass entered, fortunately, the collision cell to yield a high-quality MS2 spectrum. Because those tryptic peptides were oriented from a single protein, their quasi-molecular were usually distributed in separated mass windows with a 1 Da width, and the ion fraction frequently comprised of a single ion species. Although MS/MSALL inherently corresponded to the data-independent acquisition (DIA) manner, one-to-one correspondence could be achieved between the MS2 spectrum and precursor ion.29 On the basis of our experience, the data processing workload (mainly correlating MS2 spectra to their precursor ions), rather than the analytical measurement course, of DI-MS/MSALL and LC-MS/MS would cost 4 hours and 10 hours, respectively. Further, the comparison of measurement time, solvent consumption, and sampling amount were also taken into account for comparison. Obviously, merely a 2 μL sample was consumed by LC-MS/MS, whereas 25 volumes were mandatory for the entire DI-MS/MSALL program because the 10 μL min−1 sampling rate lasted for five minutes. On the other side, the whole DI-MS/MSALL measurement could be finished within five minutes. Only 50 μL solvent was consumed for washing the tubing, whereas a total of 6000 μL solvent was costed by LC-MS/MS, suggesting DI-MS/MSALL should be an eco-friendly approach. Systematic comparison between these two measurement ways is illustrated in Fig. 4. Above all, either analytical tool possessed inherent pros and cons, with the prerequisite that either could fully address the analytical requirements of peptide mapping for a single protein. However, when analyzing the peptidome of complicated matrices, LC-MS/MS instead of DI-MS/MSALL should be the more suitable choice, because the defaulted 1 Da binning could not avoid the co-occurrence of more than two peptides in a single mass window, hence, it might be a challenging task to construct a MS1–MS2 dataset.
4. Conclusions
Tryptic peptide mapping serves as a workhorse for the quality control of proteinic drugs, and this versatile strategy currently relies heavily on LC-MS/MS, resulting in a time-intensive workload. In the current study, we aimed to pursue a high-throughput analytical approach enabling rapid peptide mapping. Because the GPF theory led to the so-called MS separation potential, the rate-limiting LC separation was removed. Instead, the DI-MS/MSALL program that enabled the acquisition of the MS2 spectrum for each ion fraction with 1 Da width was evaluated regarding the tryptic peptide mapping potential of HSA. After inquiring the acquired MS2 spectrum to Skyline software, 47 out of 55 theoretical peptides were captured by DI-MS/MSALL, which was comparable with the 51 detected by LC-MS/MS. Except for the comparable peptide coverage and sequence annotation, DI-MS/MSALL was superior to LC-MS/MS in regards of time-saving, solvent-saving, and facile instrumentation as well. However, DI-MS/MSALL was inferior in terms of sampling amount. Therefore, DI-MS/MSALL should be eligible to serve as an integral part of an analyst's toolbox for tryptic peptide mapping of proteinic drugs, in particular when there is a dramatic workload of quality assessment.Author contributions
K Zhang: methodology, investigation, writing – original draft. XC Gong: methodology, investigation, writing – original draft. Q Wang: formal analysis, writing – original draft. PF Tu: supervision, writing – review & editing. J Li: funding acquisition, writing – review & editing. YL Song (corresponding author): conceptualization, supervision, funding acquisition, writing – review & editing. All authors contributed to data interpretation and preparation of the manuscript for publication and they approved the final version.Conflicts of interest
This work has no conflicts of interest that might be relevant to the contents of this manuscript.Acknowledgements
This study was financially supported by the National Key Research and Development Project (No. 2018YFC1707300) and the National Natural Science Foundation of China (No. 81973444 and 81773875).References
- T. T. Li, X. J. Han, C. J. Gu, H. T. Guo, H. J. Zhang, Y. M. Wang, C. Hu, K. Wang, F. J. Liu, F. Y. Luo, Y. N. Zhang, J. Hu, W. Wang, S. L. Li, Y. N. Hao, M. Y. Shen, J. J. Huang, Y. Y. Long, S. Y. Song, R. X. Wu, S. Mu, Q. Chen, F. X. Gao, J. W. Wang, S. H. Long, L. Li, Y. Wu, Y. Gao, W. Xu, X. Cai, D. Qu, Z. R. Zhang, H. Q. Zhang, N. Li, Q. Z. Gao, G. J. Zhang, C. L. He, W. Wang, X. Y. Ji, N. Tang, Z. H. Yuan, Y. H. Xie, H. T. Yang, B. Zhang and A. L. Huang, Nat. Commun., 2021, 12, 6304 CrossRef CAS PubMed.
- M. Fassler, M. S. Rappaport, C. B. Cuño and J. George, Journal of Neuroinflammation, 2021, 18, 19 CrossRef CAS PubMed.
- Z. Zajkowska, A. Borsini, N. Nikkheslat, A. Russell, G. F. Romano, S. Tomassi, N. Hepgul, D. Forton, K. Agarwal, M. Hotopf, V. Mondelli, P. Zunszain and C. M. Pariante, Brain Behav. Immun., 2020, 90, 248–258 CrossRef CAS PubMed.
- L. van den Boom and K. Kostev, Diabetes Obes. Metab., 2022, 24(2), 296–301 CrossRef CAS PubMed.
- W. J. Liu, J. Yu, W. Li, Z. Z. Jiang, T. Li, L. B. Cao, P. F. Tu, J. Li and Y. L. Song, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2021, 1171, 122624 CrossRef CAS PubMed.
- M. Gülfen, Y. Canbaz and A. Özdemir, J. Anal. Test., 2020, 4, 45–53 CrossRef.
- P. Saravanan, A. Sen, V. Balamurugan, S. K. Bandyopadhyay and R. K. Singh, Biologicals, 2008, 36, 1–6 CrossRef CAS PubMed.
- J. Gao, K. Meyer, K. Borucki and P. M. Ueland, Anal. Chem., 2018, 90, 3366–3373 CrossRef CAS PubMed.
- A. S. Kritikou, R. Aalizadeh, D. E. Damalas, I. V. Barla, C. Baessmann and N. S. Thomaidis, Food Chem., 2021, 370, 131057 CrossRef PubMed.
- X. C. Yu, K. Joe, Y. Zhang, A. Adriano, Y. Wang, H. Gazzano-Santoro, R. G. Keck, G. Deperalta and V. Ling, Anal. Chem., 2011, 83, 5912–5919 CrossRef CAS PubMed.
- T. Mouchahoir and J. E. Schiel, Anal. Bioanal. Chem., 2018, 410, 2111–2126 CrossRef CAS PubMed.
- W. J. Liu, Q. Q. Song, Y. B. Cao, Y. N. Zhao, H. X. Huo, Y. T. Wang, Y. L. Song, J. Li and P. F. Tu, Anal. Chim. Acta, 2019, 1069, 89–97 CrossRef CAS PubMed.
- L. Zhong, L. Zhu and Z.-W. Cai, J. Anal. Test., 2021, 1–16 Search PubMed.
- M. J. Traylor, A. V. Tchoudakova, A. M. Lundquist, J. E. Gill, F. L. Boldog and B. S. Tangarone, Anal. Chem., 2016, 88, 9309–9317 CrossRef CAS PubMed.
- P. Jiang, F. M. Li and J. Ding, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2020, 1137, 121895 CrossRef CAS PubMed.
- H. H. Chiu, Y. J. Tsai, C. Lo, H. W. Liao, C. H. Lin, S. C. Tang and C. H. Kuo, Anal. Chim. Acta, 2021, 1189, 339231 CrossRef PubMed.
- A. Guthals, K. R. Clauser, A. M. Frank and N. Bandeira, J. Proteome Res., 2013, 12, 2846–2857 CrossRef CAS PubMed.
- A. Theisen, B. Yan, J. M. Brown, M. Morris, B. Bellina and P. E. Barran, Anal. Chem., 2016, 88, 9964–9971 CrossRef CAS PubMed.
- Z. X. Yan and R. Yan, Anal. Chem., 2015, 87, 2861–2868 CrossRef CAS PubMed.
- E. C. Yi, M. Marelli, H. Lee, S. O. Purvine, R. Aebersold, J. D. Aitchison and D. R. Goodlett, Electrophoresis, 2002, 23, 3205–3216 CrossRef CAS PubMed.
- A. Scherl, S. A. Shaffer, G. K. Taylor, H. D. Kulasekara, S. I. Miller and D. R. Goodlett, Anal. Chem., 2008, 80, 1182–1191 CrossRef CAS PubMed.
- M. K. Midha, D. S. Campbell, C. Kapil, U. Kusebauch, M. R. Hoopmann, S. L. Bader and R. L. Moritz, Nat. Commun., 2020, 11, 1–8 CrossRef PubMed.
- M. X. Chen, Y. Zhang, A. R. Fernie, Y. G. Liu and F. Y. Zhu, Trends Biotechnol., 2021, 39, 433–437 Search PubMed.
- L. C. Gillet, P. Navarro, S. Tate, H. Röst, N. Selevsek, L. Reiter, R. Bonner and R. Aebersold, Mol. Cell. Proteomics, 2012, 11, O111.016717 CrossRef PubMed.
- https://www.sciex.com.cn/content/dam/SCIEX/pdf/posters/asms2018/Academia-Omics/494_Tuesday_Maldini.pdf.
- B. Simons, D. Kauhanen, T. Sylvänne, K. Tarasov, E. Duchoslav and K. Ekroos, Metabolites, 2012, 2, 195–213 CrossRef CAS PubMed.
- F. Gao, J. McDaniel, E. Y. Chen, H. E. Rockwell, J. Drolet, V. K. Vishnudas, V. Tolstikov, R. Sarangarajan, N. R. Narain and M. A. Kiebish, Nutr. Metab., 2017, 14, 28 CrossRef PubMed.
- K. Zhang, W. J. Liu, Q. Q. Song, J.-B. Wan, J. Yu, X. C. Gong, L. B. Cao, D. D. Si, P. F. Tu and J. Li, Anal. Chem., 2021, 93, 2541–2550 CrossRef CAS PubMed.
- P. J. Zhang, J. Jiang, K. Zhang, W. J. Liu, P. F. Tu, J. Li, Y. L. Song, J. Zheng and L. Tang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2021, 1176, 122735 CrossRef CAS PubMed.
- X. C. Gong, W. J. Liu, L. B. Cao, J. Yu, D. D. Si, J. Li, P. F. Tu and Y. L. Song, China J. Chin. Mater. Med., 2021, 46, 2220–2228 Search PubMed.
- J. R. Wiśniewski, A. Zougman, N. Nagaraj and M. Mann, Nat. Methods, 2009, 6, 359–362 CrossRef PubMed.
- L. K. Pino, B. C. Searle, J. G. Bollinger, B. Nunn, B. MacLean and M. J. MacCoss, Mass Spectrom. Rev., 2020, 39, 229–244 CrossRef CAS PubMed.
- K. Biemann, Methods Enzymol., 1990, 193, 886–887 CAS.
- E. Chekmeneva, G. dos Santos Correia, Q. Chan, A. Wijeyesekera, A. Tin, J. H. Young, P. Elliott, J. K. Nicholson and E. Holmes, J. Proteome Res., 2017, 16, 1646–1658 CrossRef CAS PubMed.
- I. Kourtchev, P. Szeto, I. O'Connor, O. A. M. Popoola, W. Maenhaut, J. Wenger and M. Kalberer, Anal. Chem., 2020, 92, 8396–8403 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08717g |
| ‡ These two authors contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2022 |