Open Access Article
Open Access ArticleA MoS2 based silver-doped ZnO nanocomposite and its antibacterial activity against β-lactamase expressing Escherichia coli†
Atanu Naskar ,
Joonho Shin
,
Joonho Shin and
Kwang-sun Kim
and
Kwang-sun Kim *
*
Department of Chemistry and Chemistry Institute for Functional Materials, Pusan National University, Busan, 46241, South Korea. E-mail: kwangsun.kim@pusan.ac.kr; Fax: +82-51-516-7421; Tel: +82-51-510-2241
First published on 2nd March 2022
Abstract
Multidrug-resistant (MDR) Gram-negative bacteria including Escherichia coli are increasingly resistant to current antibiotics. Among the strategies implemented to eradicate such MDR pathogens, approaches based on two-dimensional (2D) nanomaterials have received considerable attention. In particular, the excellent physicochemical properties of 2D molybdenum disulfide (MoS2) nanosheets, including a high surface area, good conductivity, and good surface retention, are advantageous for their use as bactericidal agents. Herein, we report the fabrication of a MoS2-based nanocomposite conjugated with silver-doped zinc oxide (AZM) as an effective antibacterial agent against E. coli species. The properties of AZM were characterized, and its antibacterial activity against MDR E. coli strains with different resistance types was evaluated. MoS2 was found to activate the antibacterial activity of AZM and provide enhanced selectivity against MDR E. coli strains expressing β-lactamases. We proposed that membrane disruption of bacterial cell walls was the major cell death mechanism for MDR E. coli. Furthermore, surface charge perturbation could explain the differences in AZM activity against MDR E. coli strains expressing a β-lactamase and a mobilized colistin resistance (mcr-1) gene product. Thus, a MoS2-based nanocomposite with a functional conjugation strategy could be a selective nano-antibacterial platform against infections caused by MDR E. coli with resistance against β-lactam antibiotics.
1. Introduction
Multidrug-resistant (MDR) bacterial infections are “the silent tsunami facing modern medicine”,1 posing a serious threat to the global medical field. MDR bacteria, which have acquired resistance to nearly all current antibiotics, can be classified as either Gram-positive or Gram-negative bacteria. Gram-negative bacteria have a distinctive membrane structure, including an envelope and an outer membrane. These structural features prevent certain antibiotics from penetrating the cell,2 resulting in Gram-negative bacteria being more resistant than Gram-positive bacteria.3 Among Gram-negative bacteria, Escherichia coli is well known to human ecology, as it is the first bacterial species to colonize after birth.4 Notably, E. coli is also a main cause of disease in nearly every part of the human body, such as gastrointestinal infections, urinary tract infections, appendicitis, meningitis, pneumonia, skin abscesses, intra-amniotic and puerperal infection in pregnant women, and endocarditis.5 Moreover, owing to its steadily increasing resistance to antibiotics and the consequent impact on human health, E. coli is now included, along with the rest of the Enterobacteriaceae family, in the ‘priority pathogens’ list published by the World Health Organization (WHO).6 Therefore, urgent scientific efforts are required to manage MDR E. coli and related infections.Nanocomposites have been proven in many studies to have the potential to address the above problem by functioning as antibacterial agents.7–13 The intrinsic properties of nanomaterials, such as a large surface-area-to-volume ratio, are advantageous for bacterial growth inhibition, providing a multitarget approach as compared to the single target approach of antibiotics. In particular, owing to their distinctive physiochemical properties, two-dimensional (2D) layered materials such as graphene14,15 and black phosphorus16,17 have emerged as promising materials for applications in areas such as biomedicine, energy, and electronics.18,19 Molybdenum disulfide (MoS2), another 2D nanomaterial, has also shown potential for biomedical applications, especially in terms of antibacterial activity.15 The applicability of MoS2 nanosheets can be attributed to their high surface area, good conductivity, and unique properties. Moreover, as MoS2 nanosheets can induce physical damage resulting in bacterial inactivation, they are a potential antibacterial agent against Gram-negative bacterial infections.
To date, only a few studies have investigated the antibacterial activity of MoS2 nanosheets.20–22 Recently, MoS2 nanosheets were shown to have rapid disinfection capabilities toward MDR E. coli strains and methicillin-resistant Staphylococcus aureus (MRSA) via a solar disinfection method.20 In this study, the MoS2 nanosheets were shown to have better antibacterial activity against E. coli than MRSA, but no selectivity toward specific types of MDR E. coli was observed. In another study by Ali et al.,23 thiolated ligand functionalized MoS2 nanosheets was utilized against E. coli and S. aureus, but no MDR Gram-negative bacterial strains were studied. Similar activity was observed in the work of Kasinathan et al.24 where cyclodextrin functionalized multi-layered MoS2 nanosheets were utilized against type strains of E. coli and S. aureus. Therefore, a novel approach is necessary to improve the specificity of MoS2-based nanocomposites against MDR E. coli strains resistant to different antibiotics.
Doping nanoparticles can alter their physiochemical properties, such as particle size and surface area, which can play key roles in enhancing antibacterial activity.25 For instance, a recent report showed that antibacterial activity is inversely associated with particle size.25 Moreover, several reports showed that the properties of ZnO could be altered for various applications such as photocatalysis, sensors, and antibacterial activity by doping with metal ions.26–29 Since Ag has been utilized as an antibacterial agent on its own, Ag doped ZnO appears to have significant potential as a new type of drug in the eradication of bacteria.27,28 In particular, ZO and Ag are well-known for their ability to kill E. coli, including MDR strains.16,25 Therefore, if MoS2 nanosheets are decorated with ZO or Ag-doped ZO (AZO), synergistic antibacterial activity may be acquired.
Inspired by above advantageous properties and recent studies, in the present study, we successfully synthesized AZO and immobilized it on MoS2 nanosheets (AZM) to realize the full potential of this nanocomposite material against MDR E. coli strains. The AZM nanocomposite was characterized using X-ray diffraction (XRD), transmission electron microscopy (TEM), and X-ray photoelectron spectroscopy (XPS). Moreover, the antibacterial activity of AZM was evaluated against MDR E. coli strains with different antibiotic resistance profiles. Notably, AZM was more active than ZO and AZO. Furthermore, AZM was generally more active against E. coli strains resistant to cell-wall-active antibiotics than to colistin-resistant. Scanning electron microscopy (SEM) and zeta potential measurements suggested that the major mode of action of AZM against MDR E. coli strains involved cell disruption and selective pressure strains that express cell-wall-active antibiotic-degrading enzymes (e.g., β-lactamase) through surface charge perturbation. These results suggest that MoS2 decorated with antibacterial metal oxide NPs has the potential to be used as a platform for fabricating nanoantibiotics capable of efficiently and selectively eliminating cell-wall-degrading enzyme producing MDR E. coli strains.
2. Materials and methods
2.1. Synthesis of AZO nanoparticles
Initially, 50 mM zinc chloride (ZnCl2, 98%; Sigma-Aldrich, St. Louis, MO, USA) and 5 at% (with respect to Zn2+) silver nitrate (AgNO3, ≥99.9%; Sigma-Aldrich) were uniformly dispersed in a beaker containing 200 mL of deionized water with continuous stirring for 30 min at 40 °C. Subsequently, NH4OH solution was added dropwise to the reaction mixture under continuous stirring until the pH reached 11. After stirring the mixture continuously at 90 °C for 3 h, the beaker was quickly transferred to an ice bath to stop the reaction. The precipitate was collected by centrifugation and then washed with deionized water and ethanol several times. Finally, the product was dried overnight in an air oven at 60 °C to obtain AZO nanoparticles.2.2. Syntheses of AZM nanocomposite
MoS2 nanosheets were prepared by the ultrasonication-assisted aqueous exfoliation of MoS2 powder (<2 μm, 99%; Sigma-Aldrich) using a previously developed method.30 First, 100 mg of MoS2 was added to 20 mL of 1-methyl-2-pyrrolidinone (NMP, 99%; Sigma-Aldrich), and the resulting suspension was ultrasonicated in an ice bath ultrasonicator (to prevent overheating) for 6 h to achieve direct exfoliation. Unexfoliated MoS2 were removed by centrifugation at 2000 rpm for 10 min, and then the supernatant was centrifuged again at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. After discarding the sediment, the directly exfoliated MoS2 nanosheets (3.5 mg mL−1) were stored in water for further use.
000 rpm for 10 min. After discarding the sediment, the directly exfoliated MoS2 nanosheets (3.5 mg mL−1) were stored in water for further use.
To prepare the AZM sample, 0.5 mL (3.5 mg mL−1) of as-prepared MoS2 nanosheets was mixed with 50 mg of as-synthesized AZO nanoparticles in 40 mL of deionized water. After ultrasonication for 10 min, the mixture was stirred continuously for 6 h. Finally, the AZM nanocomposites were collected after centrifugation and dried at 60 °C for 6 h.
2.3. Characterization
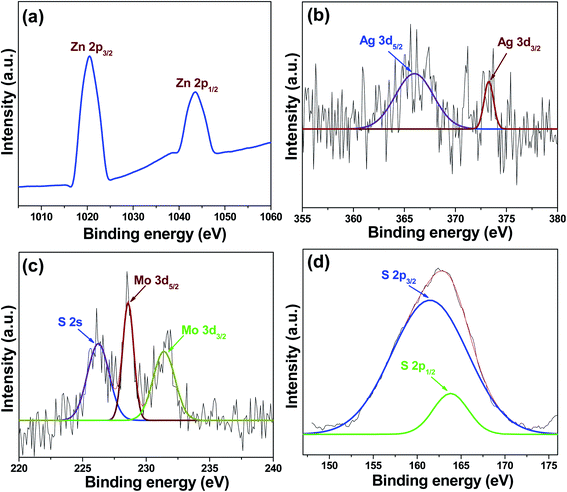
3. Results and discussion
3.1. Material properties
D = kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, θ,
| (1) |
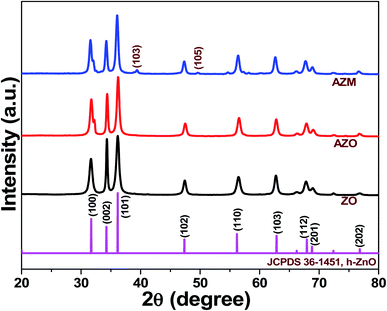 | ||
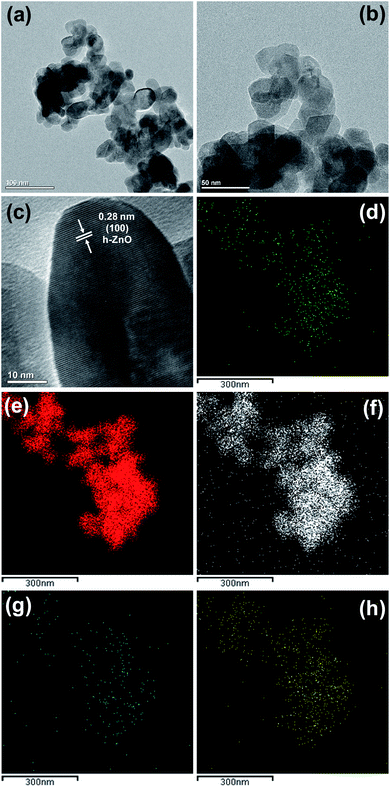
| Fig. 1 X-ray diffraction (XRD) patterns of ZnO (ZO), Ag-doped ZnO (AZO), and Ag-doped ZnO/MoS2 (AZM) samples. | ||
The D values calculated for ZO, AZO, and AZM were approximately 25, 21, and 21 nm, respectively. The reduction of the ZO crystallite size in AZO indicated that Ag doping occurred. This reduction in crystallite size may also have implications for enhancing the antibacterial activity, as discussed later. Overall, the XRD results confirmed the successful preparation of the ZO, AZO, and AZM samples.
3.2. Antibacterial activity
The antibacterial activities of ZO, AZO, and AZM were determined as MIC values (Table 1) against two different types of MDR E. coli strains expressing (1) cell-wall-active antibiotic-degrading β-lactamases encoded by blaAmpC, blaTEM-1/blaCTX-M-27, blaESBL, and blaNDM-1, and (2) a mobilized colistin resistance (mcr-1) gene product, which has recently become problematic. The MIC values of AZM against all the investigated E. coli cells were in the range of 15.63–125 μg mL−1, indicating the AZM is 2- to 8-fold more active than AZO. AlamarBlue™ Cell Viability Reagent (Invitrogen, CA, USA), a ready-to-use resazurin (7-hydroxy-3H-phenoxazin-3-one 10-oxide)-based solution has been utilized in respect of determine the MIC clearly. As discussed in method section, when the reagent color is changed from blue to pink (Fig. S1†) it indicates the bacterial growth occurred, while no color change means bacterial growth was inhibited. The bactericidal activity of AZM was further confirmed by the non-detection of viable cells from the fraction of ATCC BAA-2471 cells treated with samples at the MIC level on the agar plate (Fig. S2†). Among different types of antibiotic-resistant E. coli strains, those expressing β-lactamases had lower MIC values (6 of 8 tested strains: 75%) than MDR strains containing mcr-1 gene. To determine whether the presence of β-lactamase encoding gene is the determinant for AZM activity, the MIC value was determined against a strain containing both blaNDM-1 and mcr-1 genes (NCCP 16284). The presence of NDM-1 did not increase AZM activity, indicatng that the presence of the β-lactamase encoding gene itself is not a determinant for AZM activity, rather the mcr-1 gene is likely a strong inhibitor of AZM activity. Therefore, the AZM nanocomposite is a promising nano-antibacterial agent for the selective disinfection of MDR E. coli strains producing β-lactamase.| Strain identification numbera | Resistant antibioticsb | Genetic Featuresc | MIC (μg mL−1) | ||
|---|---|---|---|---|---|
| ZO | AZO | AZM | |||
a ATCC, CCARM, and NCCP represent American Type Culture Collection (https://www.atcc.org/), Culture Collection of Antimicrobial Resistant Microbes (HYPERLINK "http://knrrb.ccarm-bio.or.kr/index.jsp?rrb=ccarm" \o "http://knrrb.ccarm-bio.or.kr/index.jsp?rrb=ccarm"http://knrrb.ccarm-bio.or.kr/index.jsp?rrb = ccarm), and National Culture Collection for Pathogens (https://nccp.kdca.go.kr/), respectively.b Acronyms: AMC, Amoxicillin/Clavulanate (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1); AMP, Ampicillin; CAZ, Ceftazidime; CEP, Cephalothin; CHL, Chloramphenicol; CIP, Ciprofloxacin; COL, Colistin; CTX, Cefotaxime; DOR, Doripenem; ETP, Ertapenem; FEP, Cefepime; FOX, Cefoxitin; GEN, Gentamicin; IMP, Imipenem; MEM, Meropenem; NAL, Nalidixic acid; NOR, Norfloxacin; TET, Tetracycline; SXT, Sulfamethoxazole-Trimethoprim.c bla, β-lactamase expressing gene; CTX-M-27: CTX-M β-lactamase detected in E. coli 27; ESBL: extended-spectrum β-lactamase; mcr-1: mobilized colistin resistance gene; NDM-1: New Delhi metallo-β-lactamase; TEM-1: a broad-spectrum β-lactamase. 1); AMP, Ampicillin; CAZ, Ceftazidime; CEP, Cephalothin; CHL, Chloramphenicol; CIP, Ciprofloxacin; COL, Colistin; CTX, Cefotaxime; DOR, Doripenem; ETP, Ertapenem; FEP, Cefepime; FOX, Cefoxitin; GEN, Gentamicin; IMP, Imipenem; MEM, Meropenem; NAL, Nalidixic acid; NOR, Norfloxacin; TET, Tetracycline; SXT, Sulfamethoxazole-Trimethoprim.c bla, β-lactamase expressing gene; CTX-M-27: CTX-M β-lactamase detected in E. coli 27; ESBL: extended-spectrum β-lactamase; mcr-1: mobilized colistin resistance gene; NDM-1: New Delhi metallo-β-lactamase; TEM-1: a broad-spectrum β-lactamase. |
|||||
| ATCC 25922 | None | Type strain | 500 | 62.5 | 62.5 |
| CCARM 1011 | AMP, CEP, GEN, NOR | blaAmpC | 250 | 62.5 | 31.25 |
| CCARM 1013 | AMP, CEP, GEN, NOR | blaAmpC | 250 | 62.5 | 31.25 |
| CCARM 1368 | AMP, CEP, CTX, GEN, NOR | blaESBL | 250 | 250 | 125 |
| CCARM1381 | AMP, CEP, CTX, GEN, NOR | blaESBL | 250 | 250 | 125 |
| ATCC BAA-2452 | ETP, IMP | blaNDM-1 | >250 | 125 | 31.25 |
| ATCC BAA-2469 | ETP, IMP | blaNDM-1 | >250 | 125 | 15.63 |
| ATCC BAA-2471 | ETP, IMP | blaNDM-1 | 250 | 125 | 31.25 |
| NCCP 16283 | AMP, CAZ, CHL, CIP, COL, FEP, FOX, GEN, NAL, SXT, TET | mcr-1 | 250 | 250 | 125 |
| NCCP 16284 | AMC, AMP, CAZ, CHL, CIP, COL, DOR, ETP, FEP, FOX, IMP, MEM, NAL, SXT, TET | mcr-1; blaNDM-1; blaTEM-1, blaCTX-M-27 | 250 | 250 | 125 |
3.3. Proposed antibacterial action mechanism of AZM
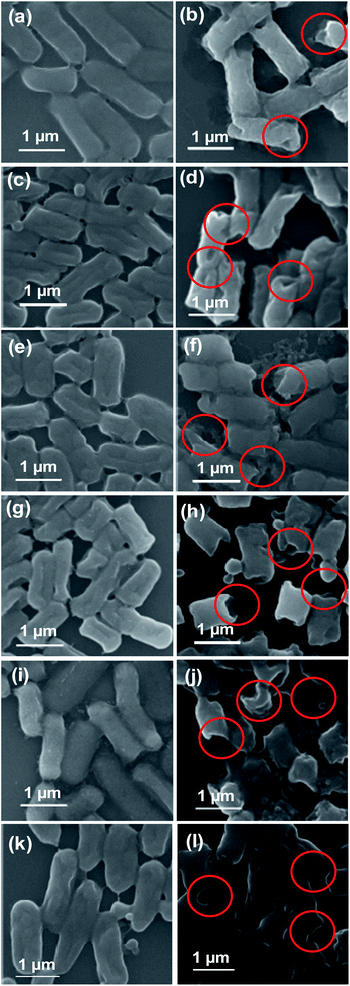
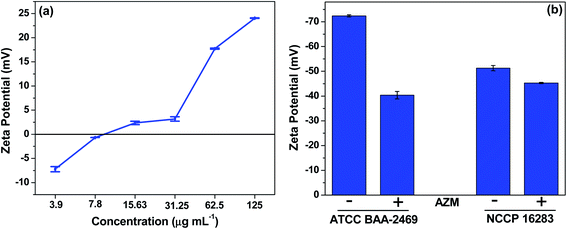
It has been reported that the antibacterial activity of NPs is primarily due to membrane damage caused by the adsorption of NPs on the cell walls, followed by penetration into the cell. To identify the antibacterial action mechanism of AZM nanocomposite, the morphologies of E. coli cells were analyzed using SEM with or without treatment by AZM nanocomposite at the sub-MIC (15.63 μg mL−1) level, at which good antibacterial activity was observed. As shown in Fig. 4, the untreated bacteria cells have smooth surfaces without ruptures, and all cells in the population have similar sizes. However, treatment with the AZM nanocomposite caused the bacterial cells to rupture, resulting in morphological defects, such as smaller sizes than the nontreated cells and wrinkled membranes (as indicated by red circles in Fig. 4). Following AZM treatment, an increase in the population of smaller ruptured cells was observed, particularly in β-lactamase expressing MDR strains (Fig. 4h and j), suggesting that AZM preferentially targets such strains. Therefore, membrane damage is confirmed as a plausible mechanism for the antibacterial activity of AZM against cell-wall-active antibiotic-degrading MDR E. coli strains.Further, we investigated whether any surface modification by AZM is additional mechanism of action. To this end, the role of surface charge of AZM, which might perturb the membrane structure, in the antibacterial activity was assessed by zeta-potential measurements at a range of concentrations corresponding to the MICs for different resistance type of MDR E. coli strains expressing either Mcr-1 or β-lactamase. As shown in Fig. 5a, the charge of AZM increased in a concentration-dependent manner, becoming positive at the MIC level (15.63 μg mL−1: 2.35 mV to 125 μg mL−1: 24.05 mV). E. coli is generally known to have a negative surface charge owing to the presence of negatively charged lipid A on the LPS component,16 whereas the strain expressing Mcr-1 has a more positive surface charge owing to LPS remodeling by ArnT and EptA proteins.33 Therefore, the positively charged AZM nanocomposite could neutralize negatively charged E. coli, ultimately paving the way for excellent antibacterial activity by charge neutralization mechanism. From the MIC results (Table 1), we hypothesized that cells with more negatively charged surfaces, such as β-lactamase expressing MDR strains of E. coli, will be more prone to interactions with positively charged AZM, which will aid in bacterial cell killing. In contrast, the positively charged strains expressing Mcr-1 will repulse AZM from the cell surface.
To confirm this hypothesis, we measured the zeta potential values of the representative MDR E. coli strains (BAA-2469 and NCCP 16283), which showed the highest and lowest activity against AZM. The zeta potential values were measured with and without treatment of AZM at the same concentration (7.8 μg mL−1), which is the sublethal concentration of AZM against BAA-2469. As shown in Fig. 5b, both strains showed negative surface charges (−72.40 and −51.27 mV for BAA-2469 and NCCP 16283, respectively), confirming that the surface charge of the strain containing mcr-1 gene product is less negative than that of the β-lactamase expressing strain. Correspondingly, in the presence of AZM, the net negative surface charge of BAA-2469 was remarkably reduced, indicating that the cells interacted with positively charged AZM. In contrast, the change in the zeta potential of NCCP 16283 was minimal, as its increased positive charge resulting from lipid modification inhibited its ability to interact with positively charged AZM. From these results, we conclude that membrane disruption through cell surface charge perturbation is the major mechanism of action for AZM in killing MDR E. coli strains, particularly those expressing β-lactamase.
4. Conclusions
In conclusion, an AZM nanocomposite was synthesized by immobilizing AZO NPs on MoS2 nanosheets. The AZM nanocomposite outperformed the AZO nanoparticles as an antibacterial material against MDR E. coli strains by 2- to 8-fold with selectivity for those expressing β-lactamase. SEM analysis showed that AZM acted by disrupting cell membranes. Furthermore, zeta potential analysis revealed that the observed selectivity toward β-lactamase expressing MDR E. coli strains originated from enhanced interactions between this negatively charged strain and positively charged AZM at the MIC. This work shows that the combination of MoS2 nanosheets with antibacterial metal oxide NPs provides a potential nanoantibiotic platform for fabricating antibacterial materials that can efficiently target β-lactamase expressing MDR E. coli strains.Author contributions
Conceptualization, A. N. and K.-s. K.; Investigation, A. N. and J. S.; Formal analysis, A. N.; Writing, A. N. and K.-s. K.; Supervision, K.-s. K.; funding acquisition, K.-s. K. All authors have read and agreed to the published version of the manuscript.Funding
This research was funded by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (grant number NRF-2021R1A2C1007413).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The pathogen resources (NCCP No. 16283 and 16284) for this study were provided by the National Culture Collection for Pathogens (NCCP).References
- D. Cox, Antibiotic resistance: the race to stop the silent tsunami facing modern medicine, The Guardian. 2015 Aug 21, available from: http://www.theguardian.com/society/blog/2015/aug/21/antibiotic-resistance-the-race-to-stop-the-silent-tsunami-facing-modern-medicine Search PubMed.
- H. I. Zgurskaya, C. A. Löpez and S. Gnanakaran, ACS Infect. Dis., 2015, 1(11), 512–522 CrossRef CAS PubMed.
- Z. Breijyeh, B. Jubeh and R. Karaman, Molecules, 2020, 25(6), 1340 CrossRef CAS PubMed.
- J. H. Hewitt and J. Rigby, J. Hyg., 1976, 77(1), 129–139 CrossRef CAS PubMed.
- L. Poirel, J. Y. Madec, A. Lupo, A. K. Schink, N. Kieffer, P. Nordmann and S. Schwarz, Microbiol. Spectrum, 2018, 6(4), ARBA-0026-2017 CrossRef PubMed.
- Antibiotic Resistance Threats in the United States, 2019, available online: https://www.cdc.gov/drugresistance/biggest-threats.html, (accessed on 28 February 2022) Search PubMed.
- A. Naskar, S. Lee, Y. Lee, S. Kim and K. S. Kim, Pharmaceutics, 2020, 12(9), 841 CrossRef CAS PubMed.
- A. Naskar, S. Lee and K. S. Kim, Front. Bioeng. Biotechnol., 2020, 8, 216 CrossRef PubMed.
- M. Bhushan, S. Muthukamalam, S. Sudharani and A. K. Viswanath, RSC Adv., 2015, 5, 32006–32014 RSC.
- K. V. P. Kumar, O. S. N. Ghosh, G. Balakrishnan, P. Thirugnanasambantham, S. K. Raghavan and A. K. Viswanath, RSC Adv., 2015, 5, 16815–16820 RSC.
- M. Bhushan, Y. Kumar, L. Periyasamy and A. K. Viswanath, Appl. Nanosci., 2018, 8, 137–153 CrossRef CAS.
- M. Bhushan, Y. Kumar, L. Periyasamy and A. K. Viswanath, Mater. Sci. Eng. C, 2019, 96, 66–76 CrossRef CAS PubMed.
- M. Bhushan, D. Mohapatra, Y. Kumar and A. K. Viswanath, Mater. Sci. Eng. B, 2021, 268, 115119 CrossRef CAS.
- K. Tadyszak, J. K. Wychowaniec and J. Litowczenko, Nanomaterials, 2018, 8(11), 944 CrossRef PubMed.
- A. Naskar, H. Khan and S. Jana, J. Sol-Gel Sci. Technol., 2018, 86, 599–609 CrossRef CAS.
- H. Cho, A. Naskar, S. Lee, S. Kim and K. S. Kim, Pharmaceutics, 2021, 13(2), 250 CrossRef CAS PubMed.
- A. Naskar and K. S. Kim, Microb. Pathog., 2019, 137, 103800 CrossRef CAS PubMed.
- Z. Wu, J. Qi, W. Wang, Z. Zeng and Q. He, J. Mater. Chem. A, 2021, 9, 18793–18817 RSC.
- A. Rai, H. C. P. Movva, A. Roy, D. Taneja, S. Chowdhury and S. K. Banerjee, Crystals, 2018, 8(8), 316 CrossRef.
- Y. Zhao, Y. Jia, J. Xu, L. Han, F. He and X. Jiang, Chem. Commun., 2021, 57(24), 2998–3001 RSC.
- J. Li, J. Zheng, Y. Yu, Z. Su, L. Zhang and X. Chen, Nanotechnology, 2020, 31(12), 125101 CrossRef CAS PubMed.
- S. Roy, A. Mondal, V. Yadav, A. Sarkar, R. Banerjee, P. Sanpui and A. Jaiswal, ACS Appl. Bio Mater., 2019, 2(7), 2738–2755 CrossRef CAS PubMed.
- S. R. Ali and M. De, ACS Appl. Nano Mater., 2021, 4(11), 12682–12689 CrossRef CAS.
- K. Kasinathan, K. Marimuthu, B. Murugesan, N. Pandiyan, B. Pandi, S. Mahalingam and B. Selvaraj, J. Mol. Liq., 2021, 323, 114631 CrossRef CAS.
- A. Naskar, S. Lee and K.-s. Kim, RSC Adv., 2020, 10, 1232–1242 RSC.
- R. S. Kumar, S. H. S. Dananjaya, M. D. Zoysa and M. Yang, RSC Adv., 2016, 6, 108468–108476 RSC.
- H. B. Dias, M. I. B. Bernardi, V. S. Marangoni, A. C. de Abreu Bernardi, A. N. de Souza Rastelli and A. C. Hernandes, Mater. Sci. Eng., C, 2019, 96, 391–401 CrossRef CAS PubMed.
- C. Karunakaran, V. Rajeswari and P. Gomathisankar, Mater. Sci. Semicond. Process., 2011, 14, 133–138 CrossRef CAS.
- G. Iqbal, S. Faisal, S. Khan, D. F. Shams and A. Nadhman, J. Photochem. Photobiol., B, 2019, 192, 141–146 CrossRef CAS PubMed.
- T. I. Kim, B. Kwon, J. Yoon, I. J. Park, G. S. Bang, Y. Park, Y. S. Seo and S. Y. Choi, ACS Appl. Mater. Interfaces, 2017, 9(9), 7908–7917 CrossRef CAS PubMed.
- Y. Qi, N. Wang, Q. Xu, H. Li, P. Zhou, X. Lu and G. Zhao, Chem. Commun., 2015, 51, 6726–6729 RSC.
- A. Kaur, A. O. Ibhadon and S. K. Kansal, J. Mater. Sci., 2017, 52, 5256–5267 CrossRef CAS.
- S. M. Zimmerman, A.-A. J. Lafontaine, C. M. Herrera, A. B. Mclean and M. S. Tren, Antimicrob. Agents Chemother., 2020, 64, e01677-19 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00163b |
| This journal is © The Royal Society of Chemistry 2022 |