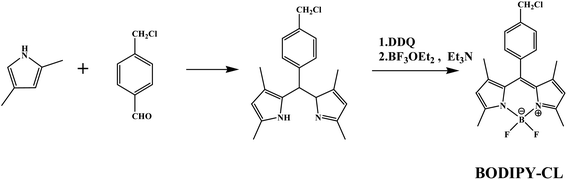
Open Access Article
Open Access ArticleSynthesis and bioimaging of a BODIPY-based fluorescence quenching probe for Fe3+†
Junqiang Leng,
Xinyu Lan,
Shuang Liu,
Wenxuan Jia,
Wenshuai Cheng,
Jianbo Cheng and
Zhenbo Liu *
*
School of Chemistry and Chemical Engineering, Yantai University, Yantai 264005, P. R. China. E-mail: zhenboliu@foxmail.com
First published on 2nd August 2022
Abstract
Iron is the main substance for maintaining life. Real-time determination of ferric ion (Fe3+) in living cells is of great significance for understanding the relationship of Fe3+ concentration changes with various physiological and pathological processes. Fluorescent probes are suitable for the detection of trace metal ions in cells due to their low toxicity and high sensitivity. In this work, a boron-dipyrromethene-based fluorescent probe (BODIPY-CL) for selective detection of Fe3+ was synthesized. The fluorescence emission of BODIPY-CL was determined at 516 nm. In a pH range of 1 to 10, the probe BODIPY-CL exhibits a quenching response to Fe3+. Meanwhile, BODIPY-CL showed a highly selective response to Fe3+ compared with 16 kinds of metal ions. The stoichiometry ratio of BODIPY-CL bound to Fe3+ was nearly 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The fluorescence quenching response obtained by the sensor was linear with the Fe3+ concentration in the range of 0–400 μM, and the detection limit was 2.9 μM. BODIPY-CL was successfully applied to image Fe3+ in cells. This study provides a promising fluorescent imaging probe for further research on the physiological and pathological effects of Fe3+.
1. The fluorescence quenching response obtained by the sensor was linear with the Fe3+ concentration in the range of 0–400 μM, and the detection limit was 2.9 μM. BODIPY-CL was successfully applied to image Fe3+ in cells. This study provides a promising fluorescent imaging probe for further research on the physiological and pathological effects of Fe3+.
1. Introduction
Iron is essential to many physiological processes in living systems,1 including oxygen transport,2 energy metabolism,3 synthesis of vitamins and steroid hormones,4 metabolism of drugs and exogenous substances,5 nucleotide synthesis and cell growth.6 However, abnormal iron concentrations can cause severe diseases, such as abdominal pain and Alzheimer's and Parkinson's disease, in humans.7,8 In addition, high levels of Fe3+ in algal cells can lead to variations in their morphology.9 The specific mechanism of Fe3+ participating in life activities in organisms is not clear. Therefore, it is of great significance to accurately determine the concentration of Fe3+ in living cells.For monitoring metal ions in living cellular organisms, fluorescent probes, with the advantages of sensitivity, simplicity, high selectivity and real-time monitoring, are an effective tool. Fluorescent probes have been widely used in the detection of trace iron ions10–19 and the imaging of Fe3+ in living cells, which provides a powerful means to study the role of Fe3+ in life activities.20
For fluorescent probes, in order to better recognize Fe3+, the design of recognition groups is the key.21–24 Previously reported25–30 probes used to detect Fe3+ mainly have two recognition modes. The first mode is the probe design based on the participation of Fe3+ in many oxidation and hydrolysis reactions, including oxidation of catechol,31 ferrocene,32 hydroxylamine33 and hydrolysis of acetal,34 Schiff base,35 and chelation of Fe3+ with fluorophores.36 And the other mode is the probe chelated with iron ions, which mostly introduce groups containing N, S and O elements into the probe. The chelation of iron ions with the introduced elements can cause changes in fluorescence properties and detect iron ions.37–39 While, can the introduction of other heteroatoms into probe molecules also interact with iron ions to cause fluorescence changes?
As for the composition of fluorescent probes, coumarin,40–42 rhodamine,43–45 cyanine,46–48 boron-dipyrromethene (BODIPY)49–51 and 1,8-naphthalimide52,53 are the most common fluorophores. Among them, BODIPY have the characteristics of high fluorescence quantum yield and excellent optical stability.54–57 In addition, its chemical stability, insensitivity to the polarity and pH, and low cytotoxicity make it ideal for in bioimaging.58–60
Recently, we found that the introduction of p-phenylmethyl chloride on the BODIPY fluorophore can react with Fe3+, resulting in a change in fluorescence properties. Taking advantage of this, we synthesized a fluorescent probe not been reported and established a method for Fe3+ detection. This work may provide a new strategy for the determination of iron ions.
2. Experimental
2.1 Reagents and instrumentation
All reagents were analytical pure and were used directly without further purification. The salts of FeCl3·6H2O, NaCl, KCl, LiCl, MgCl2·6H2O, AlCl3·9H2O, BaCl2·2H2O, CaCl2, ZnCl2, HgCl2, CdCl2, NiCl2·6H2O, CoCl2·6H2O, CrCl3·6H2O, CuCl2·2H2O, and Pb(NO3)2 were dissolved in ultra-pure water, respectively, to prepare the concentration of 100.0 mM solution. The fluorescent probe BODIPY-CL was dissolved in CH3CN to prepare the stock solution with a concentration of 1.0 mM. The stock solution was further diluted in buffers to obtain working solutions. All buffers were phosphate-buffered saline solution (PBS, pH = 7.4) containing CH3CN, (PBS/CH3CN = 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v). The column chromatography used 200–300 mesh silica gel (Qingdao Haiyang Chemical Co., China). Thin-layer chromatography (TLC) was performed on silica gel 60 F254 plates (20 cm × 20 cm, 0.25 mm thickness) (Merck, Germany). The 1H and 13C nuclear magnetic resonance (NMR) absorption spectra were measured by using a 400 MHz Bruker spectrometer. The ultra-violet (UV) spectrum was measured with PerkinElmer LAMBDA 365 spectrophotometer using a quartz cuvette with a path length of 1.0 cm. Fluorescence spectra were recorded by Varian Cary Eclipse fluorescence spectrophotometer using 1 cm quartz cuvette with excitation wavelength of 371 nm and temperature of 25 °C. Cell images were taken with a fluorescence microscope (Olympus, IX73). The wavelength of the filter is 505 nm. The pH value was measured using a pH meter (Ohaus, ST3100).
2, v/v). The column chromatography used 200–300 mesh silica gel (Qingdao Haiyang Chemical Co., China). Thin-layer chromatography (TLC) was performed on silica gel 60 F254 plates (20 cm × 20 cm, 0.25 mm thickness) (Merck, Germany). The 1H and 13C nuclear magnetic resonance (NMR) absorption spectra were measured by using a 400 MHz Bruker spectrometer. The ultra-violet (UV) spectrum was measured with PerkinElmer LAMBDA 365 spectrophotometer using a quartz cuvette with a path length of 1.0 cm. Fluorescence spectra were recorded by Varian Cary Eclipse fluorescence spectrophotometer using 1 cm quartz cuvette with excitation wavelength of 371 nm and temperature of 25 °C. Cell images were taken with a fluorescence microscope (Olympus, IX73). The wavelength of the filter is 505 nm. The pH value was measured using a pH meter (Ohaus, ST3100).
2.2 Synthesis of BODIPY-CL
The synthesis procedure was modified according to the procedure described in the literature.61 0.95 g (10 mmol) of 2,4-dimethylpyrrole was added to 100 mL of dichloromethane solution containing 4-chloromethylbenzaldehyde (0.77 g, 5 mmol). The solution was stirred under N2 atmosphere at room temperature for 12 h. Add triethylamine (10 mL) and stir for 1 h. Thereafter, 1.14 g (5 mmol) of 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) and 10 mL of boron trifluoride ether complex were added and stirred for 3 h. Then the crude product obtained by removing the solvent through rotary evaporator was purified by column chromatography with petroleum ether/ethyl acetate (1/3) as eluent to obtain orange solid BODIPY-CL (0.43 g, 23.4%). 1H NMR (400 MHz, CDCl3) δ: 7.58–7.51 (m, 2H), 7.35–7.29 (m, 2H), 6.01 (s, 2H), 4.68 (s, 2H), 2.58 (s, 6H), and 1.40 (s, 6H); 13C NMR (101 MHz, DMSO-d6) δ: 155.46, 143.15, 141.92, 139.45, 134.38, 131.07, 130.13, 128.64, 121.93, 46.01, 14.69, and 14.50. HRMS (EI): calcd for C20H20BClF2N2 [M + Na]+: 395.1346, found: 395.1271.2.3 Limit of detection (LOD)
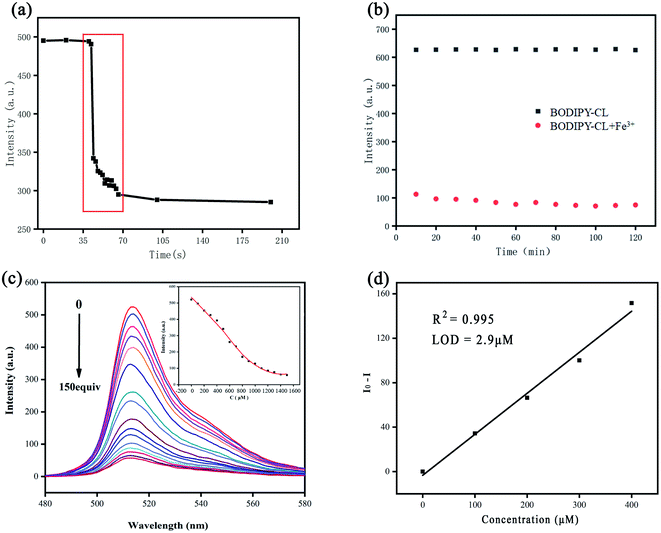
To calculate the standard deviation (σ) of blank solution, the fluorescence intensity of BODIPY-CL without Fe3+ was determined 11 times. The emission intensity of probe at 516 nm was measured with the increase of Fe3+ concentration. And the slope (k) of the curve that the emission intensity decreases with the increase of Fe3+ concentration was obtained (Fig. 2d). The LOD was based on 3σ/k according to the literature report.622.4 Computational details
Using density functional theory (DFT), the optimized structure of BODIPY-CL was obtained at B3LYP/6-31+G(d,p) level.63 The HOMO and LUMO energies of BODIPY-CL were calculated. The absorption spectrum was computed by time-dependent (TD)-DFT using B3LYP functional with the 6-31+G(d,p) basis set. The calculations were all conducted with GAUSSIAN 09 program package.642.5 Cytotoxicity assays
HeLa cells were purchased from Shanghai iCell Bioscience Inc. (Shanghai, China). Cells (4 × 103 cells per well) were seeded into 96-well plates in a 37 °C, 5% CO2 incubator for 24 h. Next, the probe BODIPY-CL at four concentrations (3.125 μM, 6.25 μM, 12.5 μM or 25.0 μM) was added and incubated for 12 h. After that, 0.5 mg mL−1 of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was added. After 4 h, remove medium carefully, and add 100.0 μL of dimethyl sulphoxide (DMSO) to the wells with the purpose of dissolving formazan crystals. Afterward, the absorbance was measured at 570 nm.2.6 Fluorescence imaging
HeLa cells were cultured in Dulbecco's Modified Eagle's Medium containing 10% fetal bovine serum (FBS) in 24-well plates at 37 °C with 5% CO2 for 24 h. Then probe BODIPY-CL dissolved in DMSO at a concentration of 10 μM was added to the cells. After 30 min, fluorescence imaging was carried out using a fluorescence microscope with a 100× objective lens. Then, add 50 μM Fe3+ solution and incubate for 30 min. Before each step, the cells were rinsed with PBS three times. White light and fluorescence images were then obtained using the methods described previously.3. Results and discussion
3.1 Design of probe BODIPY-CL
In this paper, using BODIPY as fluorophore, we synthesized a fluorescent probe (BODIPY-CL) for the determination of ferric ion (Fe3+). The BODIPY derivative with one p-chloromethylbenzene unit can be complexed with ferric ion (Fe3+) to cause fluorescence quenching. Scheme 1 showed the synthetic route for BODIPY-CL. Details of the experimental procedure and characteristics of BODIPY-CL can be found in the Experimental section and ESI (Fig. S1–S3†).3.2 Fluorescence and UV-vis response of BODIPY-CL to Fe3+
UV-vis and fluorescence spectra of BODIPY-CL were determined in CH3CN/PBS buffer (v/v = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8). Fig. 1 displays the typical optical properties of the BODIPY chromophore, with a maximum absorption wavelength of 498 nm (Fig. 1a) and a maximum emission wavelength of 516 nm (Fig. 1b). Response times of BODIPY-CL to Fe3+ were also evaluated using fluorescence spectra (see Fig. 2a). The magneton was placed in a quartz cuvette containing the probe solution to achieve the mixing of the probe solution and the iron ion solution. The emission intensity at 516 nm decreases drastically within less than 20 s after Fe3+ was added. The response time of BODIPY-CL for Fe3+ detection was quite fast.
8). Fig. 1 displays the typical optical properties of the BODIPY chromophore, with a maximum absorption wavelength of 498 nm (Fig. 1a) and a maximum emission wavelength of 516 nm (Fig. 1b). Response times of BODIPY-CL to Fe3+ were also evaluated using fluorescence spectra (see Fig. 2a). The magneton was placed in a quartz cuvette containing the probe solution to achieve the mixing of the probe solution and the iron ion solution. The emission intensity at 516 nm decreases drastically within less than 20 s after Fe3+ was added. The response time of BODIPY-CL for Fe3+ detection was quite fast.
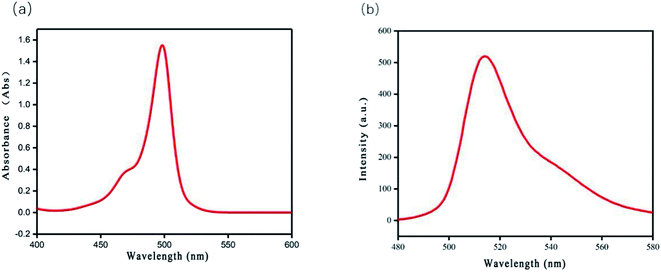 | ||
Fig. 1 (a) UV-vis absorbance spectra and (b) fluorescence emission spectra (λex = 371 nm) of BODIPY-CL (10 μM) in PBS/acetonitrile (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v, pH = 7.4) buffer solution. 2, v/v, pH = 7.4) buffer solution. | ||
The stability of fluorescence intensity was also investigated. Fig. 2b shows the changes of fluorescence intensity over time with or without the addition of Fe3+ in BODIPY-CL solution under continuous UV irradiation. The BODIPY-CL probe exhibited a stable fluorescence emission intensity under UV light irradiation for 2 h in the absence of Fe3+, suggesting the high photochemical stability of the prob. While, when Fe3+ was added to BODIPY-CL solution, the fluorescence emission at 516 nm decreased and the luminescence property was also stable. The quenching efficiency was calculated according to the equation: [1 − (F/F0)] × 100%. The relationship of BODIPY-CL fluorescence intensity to Fe3+ addition concentration was investigated (in Fig. 2c). The intensity decreases with the increase of Fe3+ concentration. Meanwhile, the intensity shows a negative linear relationship with the Fe3+ concentration in the range of 0 to 400.0 μM. And the linear correlation coefficient (R2) was 0.995 (Fig. 2d). The limits of detection (LOD) of the method was also calculated as 2.9 μM. The detection limit and response time of BODIPY-CL are tabulated and compared with previous literature (Table 1).65–69
3.3 Selectivity study
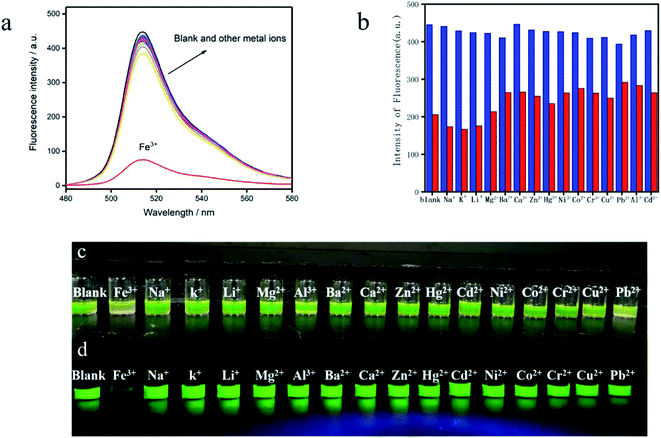
In order to evaluate the selectivity, the emission intensity of the BODIPY-CL, BODIPY-CL with Fe3+ and BODIPY-CL with the addition of others metal ions (Na+, K+, Li+, Mg2+, Al3+, Ba2+, Ca2+, Zn2+, Hg2+, Cd2+, Ni2+, Co2+, Cr2+, Cu2+ and Pb2+) was measured (see Fig. 3a and b). The colorimetric and fluorescence changes of BODIPY-CL for Fe3+ and 16 metal ions are observed by naked eyes. Only with the addition of Fe3+ and Pb2+ ions, the color of BODIPY-CL probe solution changed from yellow-green to light yellow. While, the addition of other metal ions does not change the color of the solution (Fig. 3c). Interesting, the fluorescence response immediately weakens and almost completely disappears with the addition of Fe3+ ion (Fig. 3d) under the UV-light irradiation (λ = 365 nm). In contrast, when other metal ions, including Pb2+ ion, were added to the probe solution, the luminescence intensity did not change obviously. According to the above results, Fe3+ and Pb2+ ions can interact with the probe molecules. While, the other ions do not interact with the probe. At the same time, only the interaction of Fe3+ ion can cause fluorescence quenching of BODIPY-CL probe. It is indicated that the probe has good fluorescence quenching selectivity for Fe3+.3.4 Effect of co-existing metal ions on Fe3+ detection
The BODIPY-CL probe was incubated with other ions (Na+, K+, Li+, Mg2+, Ba2+, Ca2+, Zn2+, Hg2+, Ni2+, Co2+, Cr3+, Cu2+, Pb2+, Al3+, and Cd2+) in buffer, and the fluorescence intensity was detected. As mentioned above, the fluorescence intensity changes little in the presence of other ions compared to the absence of ions (blue columns in Fig. 3b). Then, Fe3+ was added to the above solution with the probe and co-existing metal ion, the fluorescence emission intensity is obviously reduced (see the red columns in Fig. 3b). In general, the quenching response of BODIPY-CL to Fe3+ is not affected by most other metal ions.3.5 Effect of pH
The capability of BODIPY-CL to detect Fe3+ in various pH buffer solutions was investigated in detail (Fig. S4†). In the pH range of 1.0 to 10.0, the recognition ability of BODIPY-CL to Fe3+ is nearly invariant. It is indicated that the probe may determine Fe3+ ion in a wide range of pH values, expanding the probe's ability to detect Fe3+ ion in cells.3.6 Binding mode study
In order to investigate whether REDOX reaction occurred between the probe and iron ion, o-phenanthroline was added into the mixed solution of probe and iron ion. There was no significant discoloration in the solution, indicating that no Fe2+ ions were formed and the REDOX reaction did not occur.The binding stoichiometric ratio of BODIPY-CL to Fe3+ was investigated by fluorescence titration experiments.70 The total concentration of the BODIPY-CL and Fe3+ was fixed at 10−4 M. The Job's plot in Fig. 4 showed that the maximum relative intensity appears at [Fe3+]/([Fe3+] + [BODIPY-CL]) = 0.66, which shows that BODIPY-CL and Fe3+ form a complex with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.71
2.71
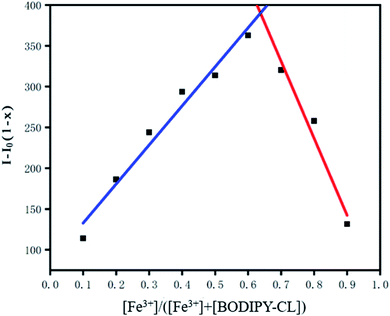 | ||
Fig. 4 Job's plot for BODIPY-CL titrated with Fe3+ indicating the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes. The total [Fe3+] + [BODIPY-CL] = 1 × 10–4 M. (x = [Fe3+]/[Fe3+] + [BODIPY-CL]). 1 complexes. The total [Fe3+] + [BODIPY-CL] = 1 × 10–4 M. (x = [Fe3+]/[Fe3+] + [BODIPY-CL]). | ||
Additionally, the probe forms the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex with ferric ion, which was also confirmed by the positive ion mode mass spectrometry data of electrospray ionization mass spectrometry (ESI-MS). A peak in ESI-MS at m/z = 481.29 (Fig. S5†) exactly corresponds to [BODIPY-CL + 2Fe3+]+ (the complexation of BODIPY-CL with two Fe3+ ions loss of four hydrogen ions).
2 complex with ferric ion, which was also confirmed by the positive ion mode mass spectrometry data of electrospray ionization mass spectrometry (ESI-MS). A peak in ESI-MS at m/z = 481.29 (Fig. S5†) exactly corresponds to [BODIPY-CL + 2Fe3+]+ (the complexation of BODIPY-CL with two Fe3+ ions loss of four hydrogen ions).
3.7 Theoretical calculations
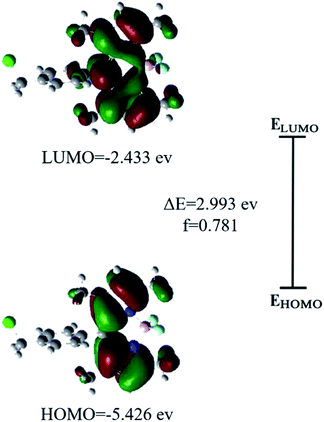
To better understand the photophysical properties of chemosensors, theoretical computational methods are often used.72 Optimized structures (in Fig. S6†) of BODIPY-CL were obtained by using DFT calculations. Energies of relevant orbitals of the BODIPY-CL were listed in Table S1.† According to the frontier orbital analysis, the distribution of electrons in HOMO and LUMO orbitals shows that electrons are localized on the BODIPY group during the transition (see Fig. 5). Through time density functional theory studies, it is found that the electronic transition at the maximum absorption wavelength occurs in the HOMO–LUMO transition. Thus, the BODIPY-CL probe exhibited strong fluorescent behavior. Meanwhile, the absorption spectra (λmax = 422 nm) and emission spectra (λmax = 484 nm) of the BODIPY-CL chemosensor obtained by DFT theoretical calculation were in Fig. S7 and S8.† The experimental results were red-shifted compared with the theoretical calculations, which may be due to the influence of the solvent, because the solvent in the actual detection was a mixed solution of ethanol and PBS.3.8 Detection of Fe3+ in water samples
To further evaluate the feasibility of BODIPY-CL for rapid and sensitive detection of Fe3+ in practical applications, tap water was detected by fluorescence spectroscopy. Detection results are collected in Table 2. No Fe3+ was detected in tap water. The spiked experiment showed good recovery rates, which were between 89.5% and 104.0%. It can be seen that the probe is accurate and reliable in detecting Fe3+ in environmental samples.3.9 Live-cell imaging
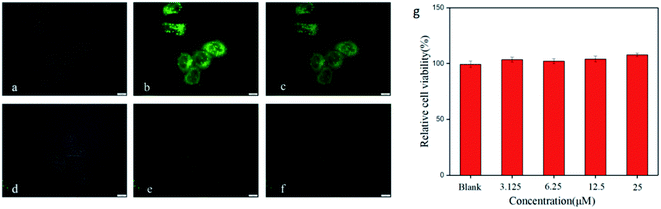
Fluorescence imaging of living HeLa cells was shown in Fig. 6. A strong green fluorescence signal in the cell was observed (Fig. 6b) when cells were incubated with BODIPY-CL. However, the fluorescence intensity of BODIPY-CL probe loaded cells decreased after Fe3+ was added (see Fig. 6e). In addition, in vitro cytotoxicity studies were performed using HeLa cells, and the results were shown in Fig. 6g that the BODIPY-CL probe has almost no toxicity to living cells under fluorescence cell imaging conditions. Therefore, the BODIPY-CL can penetrate the cell membrane and be used in response to Fe3+ in living cells.4. Conclusions
A fluorescent probe BODIPY-CL was synthesized and applied to detect Fe3+. Based on the quenching effect of Fe3+ on the probe, a method to detect Fe3+ was developed. The probe shows highly sensitive and selective response to Fe3+. The fluorescence quenching response is linear with the Fe3+ concentration over the range of 0 to 400 μM, and the detection limit is 2.9 μM. The stoichiometric ratio of BODIPY-CL to Fe3+ is nearly 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. BODIPY-CL was successfully applied to imaging Fe3+ in cells, which is helpful to provide a promising fluorescent imaging probe for further research on the physiological and pathological effects of Fe3+.
1. BODIPY-CL was successfully applied to imaging Fe3+ in cells, which is helpful to provide a promising fluorescent imaging probe for further research on the physiological and pathological effects of Fe3+.
Conflicts of interest
This research work is not funded by any organization and has no potential conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21705139) and Innovative Training Program for College Students in Shandong Province (No. S202011066025).References
- H. Wang, D.-L. Shi, J. Li, H.-Y. Tang, J. Li and Y. Guo, Sens. Actuators, B, 2018, 256, 600–608 CrossRef CAS.
- M. T. Wilson and B. J. Reeder, Exp. Physiol., 2008, 93, 128–132 CrossRef CAS PubMed.
- A. S. Grillo, A. M. SantaMaria, M. D. Kafina, A. G. Cioffi, N. C. Huston, M. Han, Y. A. Seo, Y. Y. Yien, C. Nardone, A. V. Menon, J. Fan, D. C. Svoboda, J. B. Anderson, J. D. Hong, B. G. Nicolau, K. Subedi, A. A. Gewirth, M. Wessling-Resnick, J. Kim, B. H. Paw and M. D. Burke, Science, 2017, 356, 608 CrossRef CAS PubMed.
- W. L. Miller and H. S. Bose, J. Lipid Res., 2011, 52, 2111–2135 CrossRef CAS PubMed.
- I. A. Pikuleva and M. R. Waterman, J. Biol. Chem., 2013, 288, 17091–17098 CrossRef CAS PubMed.
- W. Sornjai, F. Nguyen Van Long, N. Pion, A. Pasquer, J.-C. Saurin, V. Marcel, J. J. Diaz, H. C. Mertani and D. R. Smith, Chem.-Biol. Interact., 2020, 319, 109021 CrossRef CAS PubMed.
- L. Guo, Y. Liu, R. Kong, G. Chen, Z. Liu, F. Qu, L. Xia and W. Tan, Anal. Chem., 2019, 91, 12453–12460 CrossRef CAS PubMed.
- Y. Wu, S. Zhang, X. Gong, S. Tam, D. Xiao, S. Liu and Y. Tao, Mol. Cancer, 2020, 19, 39 CrossRef PubMed.
- Y. Sun, N. Zhang, Q. L. Guan, C. H. Liu, B. Li, K. Y. Zhang, G. H. Li, Y. H. Xing, F. Y. Bai and L. X. Sun, Cryst. Growth Des., 2019, 19, 7217–7229 CrossRef CAS.
- T. Hirayama, M. Niwa, S. Hirosawa and H. Nagasawa, ACS Sens., 2020, 5, 2950–2958 CrossRef CAS PubMed.
- X. Zhu, Y. Duan, P. Li, H. Fan, T. Han and X. Huang, Anal. Methods, 2019, 11, 642–647 RSC.
- T. Hirayama, M. Inden, H. Tsuboi, M. Niwa, Y. Uchida, Y. Naka, I. Hozumi and H. Nagasawa, Chem. Sci., 2019, 10, 1514–1521 RSC.
- X.-Q. Yi, Y.-F. He, Y.-S. Cao, W.-X. Shen and Y.-Y. Lv, ACS Sens., 2019, 4, 856–864 CrossRef CAS PubMed.
- Z. Li, Y. Xu, H. Xu, M. Cui, T. Liu, X. Ren, J. Sun, D. Deng, Y. Gu and P. Wang, Spectrochim. Acta, Part A, 2021, 244, 118819 CrossRef CAS PubMed.
- Y. Wang, C. Xia, Z. Han, Y. Jiao, X. Yao, Z. Lun, S. Fu, H. Zhang, P. Hou and D. Ning, J. Photochem. Photobiol., B, 2019, 199, 111602 CrossRef CAS PubMed.
- Z.-H. Fu, J.-C. Qin, Y.-W. Wang, Y. Peng, Y.-M. Zhang, D.-M. Zhao and Z.-H. Zhang, Dyes Pigm., 2021, 185, 108896 CrossRef CAS.
- X. Yuan, T.-H. Leng, Z.-Q. Guo, C.-Y. Wang, J.-Z. Li, W.-W. Yang and W.-H. Zhu, Dyes Pigm., 2019, 161, 403–410 CrossRef CAS.
- Y. Tang, Y. Huang, Y. Chen, L. Lu, C. Wang, T. Sun, M. Wang, G. Zhu, Y. Yang, L. Zhang and J. Zhu, Spectrochim. Acta, Part A, 2019, 218, 359–365 CrossRef CAS PubMed.
- P. Ravichandiran, V. K. Kaliannagounder, A. P. Bella, A. Boguszewska-Czubara, M. Masłyk, C. S. Kim, C. H. Park, P. M. Johnson, B.-H. Park, M.-K. Han, A. R. Kim and D. J. Yoo, Anal. Chem., 2021, 93, 801–811 CrossRef CAS PubMed.
- W. Xuan, R. Pan, Y. Wei, Y. Cao, H. Li, F.-S. Liang, K.-J. Liu and W. Wang, Bioconjugate Chem., 2016, 27, 302–308 CrossRef CAS PubMed.
- R. Kouser, S. Zehra, R. A. Khan, A. Alsalme, F. Arjmand and S. Tabassum, Spectrochim. Acta, Part A, 2021, 247, 119156 CrossRef CAS PubMed.
- M. Kumar, A. Kumar, M. S. H. Faizi, S. Kumar, M. K. Singh, S. K. Sahu, S. Kishor and R. P. John, Sens. Actuators, B, 2018, 260, 888–899 CrossRef CAS.
- T. Hirayama, Free Radical Biol. Med., 2019, 133, 38–45 CrossRef CAS PubMed.
- A. T. Aron, M. O. Loehr, J. Bogena and C. J. Chang, J. Am. Chem. Soc., 2016, 138, 14338–14346 CrossRef CAS PubMed.
- C.-H. Cai, H.-L. Wang and R.-J. Man, Spectrochim. Acta, Part A, 2021, 255, 119729 CrossRef CAS PubMed.
- M. I. Halawa, F. Wu, A. Nsabimana, B. Lou and G. Xu, Sens. Actuators, B, 2018, 257, 980–987 CrossRef CAS.
- J. Liu, Y. Guo, B. Dong, J. Sun, J. Lyu, L. Sun, S. Hu, L. Xu, X. Bai, W. Xu, S. Mintova and H. Song, Sens. Actuators, B, 2020, 320, 128361 CrossRef CAS.
- Y. Wang, S. Lao, W. Ding, Z. Zhang and S. Liu, Sens. Actuators, B, 2019, 284, 186–192 CrossRef CAS.
- Z. Xie, X. Sun, J. Jiao and X. Xin, Colloids Surf., A, 2017, 529, 38–44 CrossRef CAS.
- X. Zhang, Z. Gou, Y. Zuo and W. Lin, Spectrochim. Acta, Part A, 2020, 228, 117679 CrossRef CAS PubMed.
- S. Kuang, X. Liao, X. Zhang, T. W. Rees, R. Guan, K. Xiong, Y. Chen, L. Ji and H. Chao, Angew. Chem., Int. Ed., 2020, 59, 3315–3321 CrossRef CAS PubMed.
- M. Karmakar, S. R. Bhatta, S. Giri and A. Thakur, Inorg. Chem., 2020, 59, 4493–4507 CrossRef CAS PubMed.
- M. Wang, Y.-M. Zhang, Q.-Y. Zhao, Z.-H. Fu and Z.-H. Zhang, Chem. Phys., 2019, 527, 110470 CrossRef CAS.
- B. K. Rani and S. A. John, J. Photochem. Photobiol., A, 2020, 392, 112426 CrossRef CAS.
- Y.-K. Chiang, Y. R. Bhorge, R. D. Divate, Y.-C. Chung and Y.-P. Yen, J. Fluoresc., 2016, 26, 1699–1708 CrossRef CAS PubMed.
- A. Goel, S. Umar, P. Nag, A. Sharma, L. Kumar, Shamsuzzama, Z. Hossain, J. R. Gayen and A. Nazir, Chem. Commun., 2015, 51, 5001–5004 RSC.
- J.-S. Lee, S. D. Warkad, P. B. Shinde, A. Kuwar and S. B. Nimse, Arabian J. Chem., 2020, 13, 8697–8707 CrossRef CAS.
- Q. Wang, D. Zheng, Q. Cao, K. Huang and D. Qin, Spectrochim. Acta, Part A, 2021, 261, 120061 CrossRef CAS PubMed.
- B. Li, X. Gu, M. Wang, X. Liu and K. Xu, Dyes Pigm., 2021, 194, 109637 CrossRef CAS.
- X.-G. Chen, Y. Mei, H. Li and Q.-H. Song, Sens. Actuators, B, 2022, 354, 131202 CrossRef CAS.
- B. Yin, S. Zhang, H. Chen and L. Yan, Sens. Actuators, B, 2021, 344, 130225 CrossRef CAS.
- A. Grzelakowska, J. Modrzejewska, J. Kolińska, M. Szala, M. Zielonka, K. Dębowska, M. Zakłos-Szyda, A. Sikora, J. Zielonka and R. Podsiadły, Free Radical Biol. Med., 2022, 179, 34–46 CrossRef CAS PubMed.
- Y. Zhang, X.-F. Zhang, Q. Chen, X.-Q. Cao and S.-L. Shen, Sens. Actuators, B, 2022, 353, 131145 CrossRef CAS.
- F. Fan, L. Zhang, F. Mu and G. Shi, ACS Sens., 2021, 6, 1400–1406 CrossRef CAS PubMed.
- T. Mukherjee, S. Kanvah, A. S. Klymchenko and M. Collot, ACS Appl. Mater. Interfaces, 2021, 13, 40315–40324 CrossRef PubMed.
- G. Fei, S. Ma, C. Wang, T. Chen, Y. Li, Y. Liu, B. Tang, T. D. James and G. Chen, Coord. Chem. Rev., 2021, 447, 214134 CrossRef CAS.
- Q. Cheng, Y. Tian, H. Dang, C. Teng, K. Xie, D. Yin and L. Yan, Adv. Healthcare Mater., 2022, 11, 2101697 CrossRef CAS PubMed.
- X. Wang, H. N. Chan, N. Desbois, C. P. Gros, F. Bolze, Y. Li, H. W. Li and M. S. Wong, ACS Appl. Mater. Interfaces, 2021, 13, 18525–18532 CrossRef CAS PubMed.
- P. Wu, Y. Zhu, S. Liu and H. Xiong, ACS Cent. Sci., 2021, 7, 2039–2048 CrossRef CAS PubMed.
- J. M. Merkes, A. Hasenbach, F. Kiessling, S. Hermann and S. Banala, ACS Sens., 2021, 6, 4379–4388 CrossRef CAS PubMed.
- M. Priessner, P. A. Summers, B. W. Lewis, M. Sastre, L. Ying, M. K. Kuimova and R. Vilar, Angew. Chem., Int. Ed., 2021, 60, 23148–23153 CrossRef CAS PubMed.
- Z. Zhang, Q. Yuan, M. Li, B. Bao and Y. Tang, Small, 2021, 17, 2104581 CrossRef CAS PubMed.
- C. Geraghty, C. Wynne and R. B. P. Elmes, Coord. Chem. Rev., 2021, 437, 213713 CrossRef CAS.
- J. Bañuelos, Chem. Rec., 2016, 16, 335–348 CrossRef PubMed.
- J. M. Franke, B. K. Raliski, S. C. Boggess, D. V. Natesan, E. T. Koretsky, P. Zhang, R. U. Kulkarni, P. E. Deal and E. W. Miller, J. Am. Chem. Soc., 2019, 141, 12824–12831 CrossRef CAS PubMed.
- M. Poddar, P. Gautam, Y. Rout and R. Misra, Dyes Pigm., 2017, 146, 368–373 CrossRef CAS.
- T. Zhang, C. Ma, T. Sun and Z. Xie, Coord. Chem. Rev., 2019, 390, 76–85 CrossRef CAS.
- P. Kaur and K. Singh, J. Mater. Chem. C, 2019, 7, 11361–11405 RSC.
- P. E. Kesavan, V. Pandey, M. K. Raza, S. Mori and I. Gupta, Bioorg. Chem., 2019, 91, 103139 CrossRef CAS PubMed.
- S. M. Yuriy, V. S. Alexey, S. T. Alexander and V. R. Evgeniy, Curr. Med. Chem., 2017, 24, 2745–2772 Search PubMed.
- P. Guo, S. Yan, J. Hu, X. Xing, C. Wang, X. Xu, X. Qiu, W. Ma, C. Lu, X. Weng and X. Zhou, Org. Lett., 2013, 15, 3266–3269 CrossRef CAS PubMed.
- G. L. Long and J. D. Winefordner, Anal. Chem., 1983, 55, 712A–724A CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- G. W. T. M. J. Frisch, H. B. Schlegel, G. E. Scuseria, J. R. C. M. A. Robb, G. Scalmani, V. Barone, B. Mennucci, H. N. G. A. Petersson, M. Caricato, X. Li, H. P. Hratchian, J. B. A. F. Izmaylov, G. Zheng, J. L. Sonnenberg, M. Hada, K. T. M. Ehara, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, O. K. Y. Honda, H. Nakai, T. Vreven, J. A. Montgomery Jr, F. O. J. E. Peralta, M. Bearpark, J. J. Heyd, E. Brothers, V. N. S. K. N. Kudin, T. Keith, R. Kobayashi, J. Normand, A. R. K. Raghavachari, J. C. Burant, S. S. Iyengar, J. Tomasi, N. R. M. Cossi, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, C. A. V. Bakken, J. Jaramillo, R. Gomperts, R. E. Stratmann, A. J. A. O. Yazyev, R. Cammi, C. Pomelli, J. W. Ochterski, K. M. R. L. Martin, V. G. Zakrzewski, G. A. Voth, J. J. D. P. Salvador, S. Dapprich, A. D. Daniels, J. B. F. O. Farkas, J. V. Ortiz, J. Cioslowski and a. D. J. Fox, Gaussian 09 programme packages was used toperform all the electronic structure calculations and visualizations, Wallingford CT, 2010 Search PubMed.
- L. Xiao, C. Li, X. He and X. Cheng, SN Appl. Sci., 2021, 3, 591 CrossRef CAS.
- S. He, L. Xiao, L. Marin, Y. Bai and X. Cheng, Eur. Polym. J., 2021, 150, 110428 CrossRef CAS.
- M. Oguz, A. N. Kursunlu and M. Yilmaz, Dyes Pigm., 2020, 173, 107974 CrossRef CAS.
- J. Ji, S. S. Chereddy, Y. Ren, X. Chen, D. Su, Z. Zhong, T. Mori, Y. Inoue, W. Wu and C. Yang, J. Photochem. Photobiol., A, 2018, 355, 78–83 CrossRef CAS.
- J. Nootem, C. Sattayanon, R. Daengngern, A. Kamkaew, W. Wattanathana, S. Wannapaiboon, P. Rashatasakhon and K. Chansaenpak, Chemosensors, 2021, 9, 165 CrossRef CAS.
- H. Wang, D. Wang, Q. Wang, X. Li and C. A. Schalley, Org. Biomol. Chem., 2010, 8, 1017–1026 RSC.
- L.-m. Liu and Z.-y. Yang, Inorg. Chim. Acta, 2018, 469, 588–592 CrossRef CAS.
- T. Keawwangchai, N. Morakot and B. Wanno, J. Mol. Model., 2013, 19, 1435–1444 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional supporting information may be found in the online version of the publisher's website. See https://doi.org/10.1039/d2ra00818a |
| This journal is © The Royal Society of Chemistry 2022 |