Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mitochondria-targeted half-sandwich iridium(III)-Cp*-arylimidazophenanthroline complexes as antiproliferative and bioimaging agents against triple negative breast cancer cells MDA-MB-468†
Ashaparna Mondal‡
 a,
Shanooja Shanavas‡
a,
Shanooja Shanavas‡ b,
Utsav Sen
b,
Utsav Sen b,
Utpal Das
b,
Utpal Das a,
Nilmadhab Roy
a,
Nilmadhab Roy a,
Bipasha Bose
a,
Bipasha Bose *b and
Priyankar Paira
*b and
Priyankar Paira *a
*a
aDepartment of Chemistry, School of Advanced Sciences, Vellore Institute of Technology, Vellore-632014, Tamilnadu, India. E-mail: priyankar.paira@vit.ac.in
bDepartment Stem Cells and Regenerative Medicine Centre, Institution Yenepoya Research Centre, Yenepoya University, University Road, Derlakatte, Mangalore 575018, Karnataka, India. E-mail: bipasha.bose@gmail.com
First published on 19th April 2022
Abstract
To reduce the side effects of marketed cancer drugs against triple negative breast cancer cells we have reported mitochondria targeting half-sandwich iridium(III)-Cp*-arylimidazophenanthroline complexes for MDA-MB-468 cell therapy and diagnosis. Out of five Ir(III) complexes (IrL1–IrL5), [iridium(III)-Cp*-2-(naphthalen-1-yl)-1H-imidazo[4,5-f][1,10]phenanthroline]PF6 (IrL1) has exhibited the best cytoselectivity against MDA-MB-468 cells compared to normal HaCaT cells along with excellent binding efficacy with DNA as well as serum albumin. The subcellular localization study of the complex revealed the localization of the compound in cytoplasm thereby pointing to a possible mitochondrial localization and consequent mitochondrial dysfunction via MMP alteration and ROS generation. Moreover, the IrL1 complex facilitated a substantial G1 phase cell-cycle arrest of MDA-MB-468 cells at the highest tested concentration of 5 μM. The study verdicts support the prospective therapeutic potential of the IrL1 complex in the treatment and eradication of triple negative breast cancer cells. These results validate that these types of scaffolds will be fairly able to exert great potential for tumor diagnosis as well as therapy in the near future.
Introduction
Nowadays, breast cancer prevails over most of the deadly life threats towards women from all over the world. 11.7% of new cases of cancer diagnosed in 2020 were female breast cancer which is the highest number of cases among all types of cancer reported.1 This disease is heterogenous at the molecular level but, as a consequence of diligent research over past decades, chances of healing have been increased by 70–80% in breast cancer patients when the cancer is non-metastatic and treated in the early stage. Over the years of research a few categorisations of tumours were refined based on the alterations at the molecular level.2 Current clinical practices involve an alternate classification of 5 types that includes triple negative breast cancer (TNBC) without expression of ER, PR or HER2. TNBC is adenoid cystic and metaplastic in nature with a poor prognosis profile.2 A thorough investigation of medical literature reveals that the term, “triple negative breast cancer”, was first mentioned in 2005 by Brenton and coworkers.3 However, in the present situation 12–17% of all reported breast cancer cases have been recognised under this class.It has been challenging to create an effective treatment regime for TNBC patients as this aggressive subtype does not respond to the HER2 targeting drugs or hormonal therapy. Consequently, chemotherapy remains as the main systemic treatment option even though TNBC develops resistance easily to existing targeted medicines like trastuzumab. Potential molecular targets for TNBC may include EGFR (a surface receptor), PARP1 (poly ADP-ribose polymerase 1) and DNA. The phenotypic similarity of TNBC to BRCA-1 associated malignancy helped researchers to develop few targeted cytotoxic agents currently which may lead to a new horizon of TNBC therapeutics. In 2020 FDA approved trodelvy (sacituzumab govitecan-hziy) which is a topoisomerase inhibitor conjugate antibody directed to Trop-2 receptor for metastatic TNBC and for the first-time improved progression of overall survival was witnessed. Triple negative breast cancer caused by BRCA1 and BRCA2 mutation can be a potential target of DNA damaging chemotherapeutics and a number of clinical data leads to the suggestion that involving platinum-based chemotherapeutics may be use in standard treatment regime of early stage as well as advanced TNBC.4 As reported in earlier scientific research, platinum salts like cisplatin, carboplatin or oxaliplatin are able to initiate a platinum–DNA adduct formation followed by DNA damage in cancer cells leading towards apoptosis of the targeted cancer cells. BRCA1 deficient breast cancer showed sensitivity towards platinum and gemcitabine neoadjuvant treatment but with poor chance of progression free survival.5 A clinical study, reported by von Minckwitz in 2013 involving 315 early stage TNBC patients, let out the fact that a neoadjuvant treatment regime comprising doxorubicin and carboplatin achieved 59% pathologic complete response rate.6 However, a combination therapy of gemcitabine/carboplatin given to advanced metastatic TNBC patients in a trial scored 34% objective response rate and progression free survival of 5.1 months.7 Although cisplatin and its analogical platinum complexes showed slight prospect in TNBC treatment they have exerted some major drawbacks like toxicity, chemoresistance and narrow activity window.8 This fact motivated broader area of investigation on efficacy of other metal complexes to target metastasis and defective cell proliferation. A handful of gold, iron, copper, ruthenium, rhodium and iridium complexes happened to be synthesized and tasted against breast cancer cell lines in vitro or in vivo and found to manifest excellent outcomes.9 Of late, Ir(III) based metal complexes have been fascinating the mind of the investigators for being reconnoitered as highly active anticancer agents with their excellent photoluminescence property possessing high photostabilities, long-lived excited triplet states after quick singlet to triplet intersystem crossing (ISC), larger Stokes' shift associated with high quantum yields.10,11 Furthermore, the higher oxidation state of iridium metal, ability to display a wide range of ligand substitution, considerable redox properties, flexible structural features and stability towards cancer cell environments has enabled them to be used in preparing anticancer metal complexes.10,11 Instead, bioactivities of metal complexes are also reliant on the structure of the ligands. Arylimidazophenthroline compounds are well known as pertinent probes of DNA structure having the capability of disentangling the double stranded DNA through strong intercalative interaction.12 Encouraged by our previous work with ruthenium and arylimidazophenthroline herein we have intended to design five iridium(III)-Cp* N^N metal complexes with planar and π-extended arylimidazophenanthroline moiety as ligand having strong metal binding capacity (Fig. 1).13–18 and evaluated their antiproliferative activity against MDA-MB-468 TNBC cells that are EGFR +ve, TGF alpha +ve, high in Ki67. These complexes exhibited dual properties like (i) killing of cancer cells via DNA damage (ii) mitochondrial dysfunction by ROS production.19 The intrinsic phosphorescence property of these Ir(III) complexes are also helpful for cellular imaging and tracking of drug accumulation in subcellular organelles.20
Results and discussion
Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio in methanol and stirred at room temperature for 2 h. After a change in colour from light yellow to orange, 2.5 equivalents of NH4PF6 were added and stirred again for 90 min (Scheme 1). The complexes [Cp*Ir(L1–L5)Cl]PF6 labelled as IrL1–IrL5 were obtained in good yield (92–95%). The structures of all the complexes (IrL1–IrL5) were analysed via 1H, 13C, 19F and 31P NMR, and mass spectroscopy. The complex IrL1 displayed a characteristic singlet peak at 1.74 ppm, corresponding to the five methyl groups of pentamethylcyclopentadiene. The protons of complex IrL1 experienced a considerable downfield effect upon attachment to the iridium Cp* precursor. In the 13C NMR spectrum, the ligand carbons appeared at around δ 125.8–153.2 ppm. The aliphatic CH3 carbons peaks were observed at δ 8.7 ppm and aromatic CH carbons peaks were observed at δ 89.6 ppm. In the 19F NMR spectrum, characteristic peaks of six fluorines appeared at δ −69.2 and −71.08 ppm. The characteristic septate of phosphorous was observed in the range of δ −135.41 to −157.37 ppm in the 31P NMR spectrum. The ESI-MS peak at m/z: 709.17 [M]+ and isotopic pattern of iridium confirmed the formation of complex IrL1. Similarly, clear differences in peak values in the NMR, FT-IR and ESI-MS between the other complexes (IrL2–IrL5) were observed.
2 ratio in methanol and stirred at room temperature for 2 h. After a change in colour from light yellow to orange, 2.5 equivalents of NH4PF6 were added and stirred again for 90 min (Scheme 1). The complexes [Cp*Ir(L1–L5)Cl]PF6 labelled as IrL1–IrL5 were obtained in good yield (92–95%). The structures of all the complexes (IrL1–IrL5) were analysed via 1H, 13C, 19F and 31P NMR, and mass spectroscopy. The complex IrL1 displayed a characteristic singlet peak at 1.74 ppm, corresponding to the five methyl groups of pentamethylcyclopentadiene. The protons of complex IrL1 experienced a considerable downfield effect upon attachment to the iridium Cp* precursor. In the 13C NMR spectrum, the ligand carbons appeared at around δ 125.8–153.2 ppm. The aliphatic CH3 carbons peaks were observed at δ 8.7 ppm and aromatic CH carbons peaks were observed at δ 89.6 ppm. In the 19F NMR spectrum, characteristic peaks of six fluorines appeared at δ −69.2 and −71.08 ppm. The characteristic septate of phosphorous was observed in the range of δ −135.41 to −157.37 ppm in the 31P NMR spectrum. The ESI-MS peak at m/z: 709.17 [M]+ and isotopic pattern of iridium confirmed the formation of complex IrL1. Similarly, clear differences in peak values in the NMR, FT-IR and ESI-MS between the other complexes (IrL2–IrL5) were observed.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solvent system, as shown in Fig. 2. The photophysical data is summarized in Table 1. The characteristic intraligand (π–π*) transitions (N^N ligands) appeared at 250–350 nm and metal to ligand charge transfer (1MLCT) at 360–400 nm.22,23 Among the complexes, we observed the maximum absorption in the 1MLCT region for the benzothiazole derivative (IrL5). In the emission spectra, we observed the MLCT emission of all the complexes in the range of 350–525 nm (Fig. 2). Similar to the absorption spectra, the emission for the anthracene derivative is the most intense because of its strong π conjugation. Using the emission spectral data, the quantum yield of these complexes was calculated. These complexes didn't show remarkable quantum yield, complex IrL1 showed moderate quantum yield (0.002) though for the MLCT transition (Table 1).
1) solvent system, as shown in Fig. 2. The photophysical data is summarized in Table 1. The characteristic intraligand (π–π*) transitions (N^N ligands) appeared at 250–350 nm and metal to ligand charge transfer (1MLCT) at 360–400 nm.22,23 Among the complexes, we observed the maximum absorption in the 1MLCT region for the benzothiazole derivative (IrL5). In the emission spectra, we observed the MLCT emission of all the complexes in the range of 350–525 nm (Fig. 2). Similar to the absorption spectra, the emission for the anthracene derivative is the most intense because of its strong π conjugation. Using the emission spectral data, the quantum yield of these complexes was calculated. These complexes didn't show remarkable quantum yield, complex IrL1 showed moderate quantum yield (0.002) though for the MLCT transition (Table 1).
| Samples | λmaxa (nm) | λfb (nm) | Stoke's shift | εc (M−1 cm−1) | (ϕf)d | Solubilitye (M) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pf Pf |
ΛMg (μs) | |
|---|---|---|---|---|---|---|---|---|---|
| DMSO | 10% DMSO | ||||||||
a Absorption maxima.b Maximum emission wavelength.c Extinction coefficient.d Quantum yield.e DMSO-10% DMEM medium (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 99 v/v, comparable to cell media).f n-Octanol/water partition coefficients.g Conductance in DMSO and 10% aq. DMSO (IrL1–IrL5; 3 × 10−5 M). 99 v/v, comparable to cell media).f n-Octanol/water partition coefficients.g Conductance in DMSO and 10% aq. DMSO (IrL1–IrL5; 3 × 10−5 M). |
|||||||||
| IrL1 | 284 | 409 | 125 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
0.002 | 0.0007 | 0.27 ± 0.07 | 8 | 22 |
| IrL2 | 375 | 437 | 62 | 6850 | — | 0.0006 | 0.34 ± 0.07 | 6 | 27 |
| IrL3 | 397 | 462 | 65 | 2200 | — | 0.0012 | −0.23 ± 0.2 | 7 | 27 |
| IrL4 | 278 | 360 | 82 | 2200 | — | 0.0009 | 0.02 ± 0.05 | 9 | 30 |
| IrL5 | 336 | 380 | 44 | 2350 | — | 0.0007 | 0.29 ± 0.08 | 9 | 36 |
| Quinine sulfate | 350 | 452 | 102 | — | 0.57 | — | — | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 v/v (comparable to cell media) at 25 °C (Table 1). The lipophilicity of these complexes was determined by performing an n-octanol/water partition coefficient (log
90 v/v (comparable to cell media) at 25 °C (Table 1). The lipophilicity of these complexes was determined by performing an n-octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w, where Po/w = the octanol/water partition coefficient) study using the shake flask method (Table 1).24 The experimental log
Po/w, where Po/w = the octanol/water partition coefficient) study using the shake flask method (Table 1).24 The experimental log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w values of these complexes were determined to be in the range of 0.27–0.34 (Table 1). Complex IrL2 exhibited the highest log
Po/w values of these complexes were determined to be in the range of 0.27–0.34 (Table 1). Complex IrL2 exhibited the highest log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w due to the hydrophobic nature of its anthracene group. The lowest log
Po/w due to the hydrophobic nature of its anthracene group. The lowest log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w value was observed for compound IrL3 because it's hydrophilic chromone group. The iridium complexes IrL1–IrL5 exhibited molar conductance values in the range of ∼7–9 S m2 M−1 in pure DMSO. Furthermore, their molar conductance increased in 10% DMSO (∼22–36 S m2 M−1, Table 1), suggesting their 1
Po/w value was observed for compound IrL3 because it's hydrophilic chromone group. The iridium complexes IrL1–IrL5 exhibited molar conductance values in the range of ∼7–9 S m2 M−1 in pure DMSO. Furthermore, their molar conductance increased in 10% DMSO (∼22–36 S m2 M−1, Table 1), suggesting their 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 electrolytic nature in pure DMSO and 10% DMSO, respectively.25–27 This change in the electrolytic behaviour of the complexes from 1
2 electrolytic nature in pure DMSO and 10% DMSO, respectively.25–27 This change in the electrolytic behaviour of the complexes from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 can be attributed to the dissociation of the Ir–Cl bond and subsequent aquation of the complexes.
2 can be attributed to the dissociation of the Ir–Cl bond and subsequent aquation of the complexes.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO = 9
DMSO = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
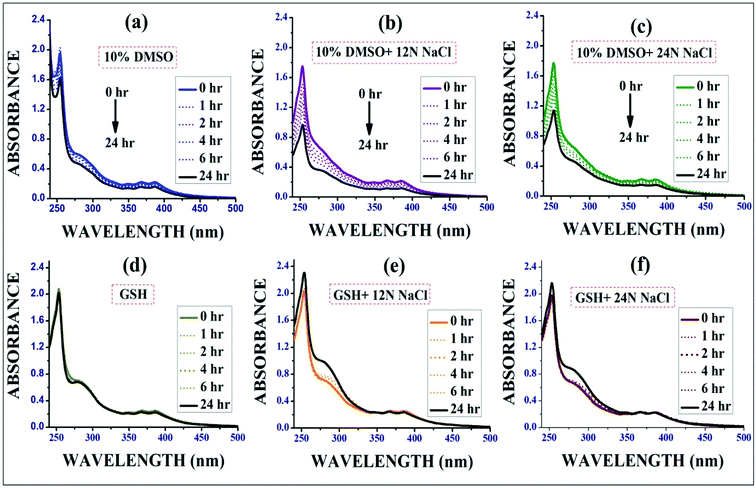
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and aqueous GSH medium (Fig. 3) respectively in presence or absence of different concentrations of NaCl. It is essential that the complexes remain stable in the biological environments of cells, and thus the stability studies were performed. The obvious change in absorbance (∼15–30% decrease after 24 h) with time in aqueous DMSO clearly revealed the moderate dissociation of the –Cl ligand from the Ir(III) complexes followed by aqua complex formation, which was also quantitatively determined based on the observed molar conductivity of the complexes (IrL1–IrL5) in aqueous DMSO and it favours DNA covalent binding as well. It has been reported that many cancer cells become resistant to various drugs by increasing their cellular glutathione level.28 Hence, to determine the effect of GSH on the reported complexes, a stability study was performed in the presence of excess (10 eq.) glutathione (GSH) via time-dependent UV spectroscopy. However, GSH didn't have much impact on the stability of metal complex and hence resistivity may not be induced against this complex in cancer cells containing high level of GSH.
1) and aqueous GSH medium (Fig. 3) respectively in presence or absence of different concentrations of NaCl. It is essential that the complexes remain stable in the biological environments of cells, and thus the stability studies were performed. The obvious change in absorbance (∼15–30% decrease after 24 h) with time in aqueous DMSO clearly revealed the moderate dissociation of the –Cl ligand from the Ir(III) complexes followed by aqua complex formation, which was also quantitatively determined based on the observed molar conductivity of the complexes (IrL1–IrL5) in aqueous DMSO and it favours DNA covalent binding as well. It has been reported that many cancer cells become resistant to various drugs by increasing their cellular glutathione level.28 Hence, to determine the effect of GSH on the reported complexes, a stability study was performed in the presence of excess (10 eq.) glutathione (GSH) via time-dependent UV spectroscopy. However, GSH didn't have much impact on the stability of metal complex and hence resistivity may not be induced against this complex in cancer cells containing high level of GSH.
Biology
All the complexes exhibited significant cytotoxicity with IC50 values of 3.673 μM, 20.35 μM, 20.27 μM, 5.202 μM, 4.412 μM for IrL1, IrL2, IrL3, IrL4 and IrL5 respectively (Fig. 4). The dose dependent cytotoxic effects of these complexes were evident on MDA-MB-468 with IrL1 exhibiting the most potent effects thereby exhibiting the lowest IC50 value of 3.67 μM (Fig. 5). Consequently, the IrL1 complex doses of 1 μM, 3 μM and 5 μM were selected for further experimental analysis. Furthermore, the possible cytotoxic effects of IrL1 on normal cell line were also examined on the immortalized human keratinocyte cell line HaCaT. The IC50 value of IrL1 on HaCaT is found to be 11.42 μM, which is significantly ∼3 times higher than the IC50 value of the complex on the breast cancer cell line MDA-MB-468, suggesting the low cytotoxic effects of the same on normal cells and safety implications to use the compound for selective therapeutics at lower concentrations (Fig. 4).
DNA binding studies
| Complex | λmax (nm) | Change in absorbance | Δεa (%) | Kbb (M−1) | Ksvc (M−1) | Kappd (M−1) |
|---|---|---|---|---|---|---|
| a % change in hypochromism or hyperchromism.b Kb, intrinsic DNA binding constant from UV-visible absorption titration.c Ksv, Stern–Volmer quenching constant.d Kapp, apparent DNA binding constant from competitive displacement. | ||||||
| IrL1 | 250 | Hyperchromism | 60 | — | — | — |
| 305 | Hypochromism | 25 | 0.11 × 106 | 2.2 × 104 | 2.5 × 103 | |
The higher value of binding constant and lower values of Stern–Volmer's quenching constant and apparent binding constant is evidence of the fact that IrL1 complex interacts with DNA via covalent bond and not via intercalation mode.
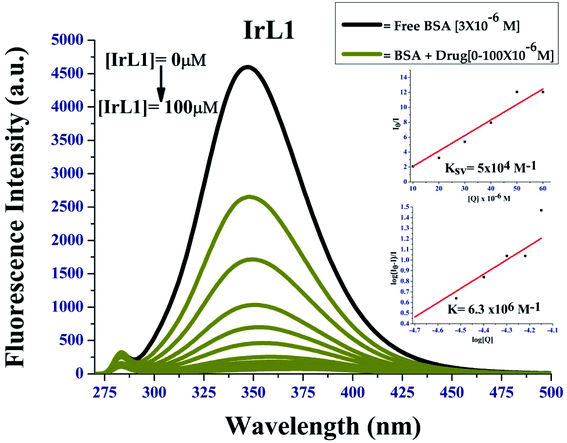 | ||
| Fig. 8 Fluorescence quenching of BSA by IrL1; inset: I0/I vs. [complex] linear plots for stern Volmer's constant and Scatchard plot of log([I0 − I]/I) vs. log[complex] for BSA. | ||
The complexes showed strong binding propensity with BSA, which is required for the transport of protein-bound complexes in biological systems. KBSA for complex IrL1 was found to be 5 × 104 M−1 the K value was 6.3 × 106 M−1. The value of bimolecular quenching constant (kq) calculated from KSV and τ0 (1 × 10−8 s) was observed to be 5 × 1012 M−1 S−1. These values are higher than the maximum possible value for dynamic quenching (2.0 × 1010 L mol−1 s−1),29,30 suggesting the involvement of static quenching mechanism by the present iridium(III) complexes. The non-linearity of the Stern–Volmer plot of IrL1 is due to the formation of a ground state complex between the Ir(III) complex and BSA.
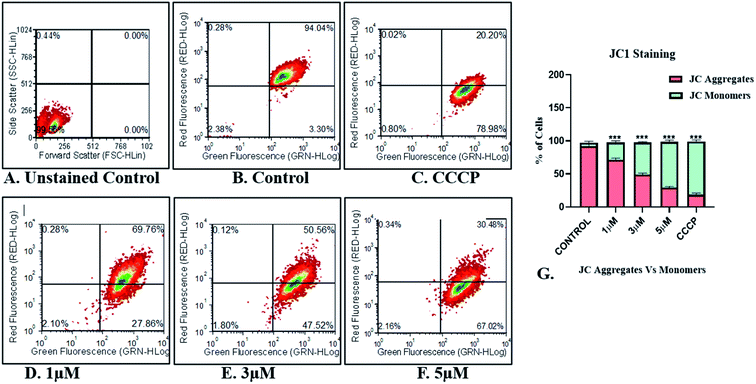
The flow cytometric quantification of JC-1-stained cells revealed a normal mitochondrial function in control cells devoid of any treatment exhibiting ∼94% JC-1 aggregates (red+ green fluorescing healthy mitochondria). Impaired mitochondrial function was observed in only 3.30% of control cells with JC-1 monomers indicating green fluorescence (Fig. 10B). Unstained control represented in Fig. 10A has been used for gating purposes. Conversely, the positive control with CCCP treatment, displayed a high percentage of cells (∼79%) with damaged mitochondria indicating the presence of green fluorescing JC-1 monomers against ∼20% cells retaining healthy mitochondria with red + green fluorescing JC-1 aggregates (Fig. 10C).
Treatment of the MDA-MB-468 cells with IrL1 has ensued a dose dependent increase in JC-1 monomer expressing/green fluorescing cells indicating damaged mitochondria (Fig. 10D–F). IrL1 complex at 1 μM concentration resulted in damaged mitochondria in 27.86% cells (Fig. 10D). However, the higher concentrations of the IrL1 complex at 3 and 5 μM induced alterations of mitochondrial membrane potential and associated damage in high percentage ∼47.52% and 67.02% respectively in MDA-MB-468 breast cancer cells (Fig. 10E and F). The proportion of green fluorescing JC-1 monomers induced by high concentrations of the complex at 3 and 5 μM thereby indicates alteration in the MMP and mitochondrial depolarization and associated mitochondrial dysfunction leading to a dose dependent possible cell death meditated by the IrL1 complex comparable to the CCCP treated positive control cells (Fig. 10C).
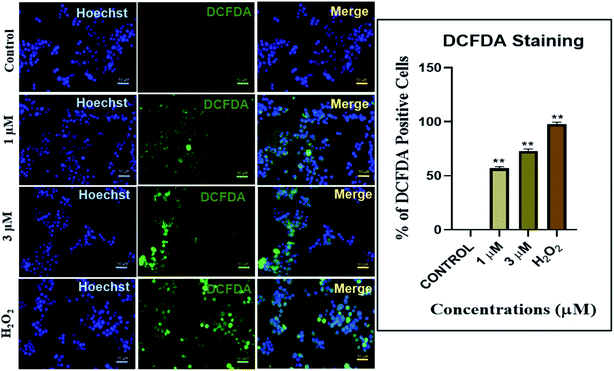
Our results indicated a dose dependent increase in ROS production in the MDA-MB-468 cells treated with IrL1 concentrations 1 μM and 3 μM. The extent of ROS production is indicated by the 2′,7′-dichlorofluorescein (DCF) formed by the deacetylation and subsequent oxidation of 2′,7′-dichlorofluorescin diacetate (DCFDA) through cellular esterases and ROS. 1 μM concentration of the complex has resulted in 55.01% DCF positive cells which has increased up to 70.67% DCF positive cells at 3 μM concentrations. ROS production was absent in untreated control. However, in the positive control, treated with H2O2, ROS production was high which is ∼99% (Fig. 11).
Experimental section
Materials and methods
In all the experiments, the reagents and solvents used were of the highest grade and best commercial quality. All organic solvents used throughout the chemical synthesis and chromatography procedures were of analytical grade and used without further purification as received from E. Merck (India). Pentamethylcyclopentadienyl iridium(III) chloride dimer, 1,10-phenanthroline-5,6-dione, α-naphthaldehyde, 9-anthraldehyde, chromone-3-carboxaldehyde, indole-3-carboxaldehyde and benzothiazole-2-carboxaldehyde were procured from Sigma Aldrich Chemical Ltd, Merck and Spectrochem. Thin layer chromatography was performed on pre-coated silica gel 60 F254 aluminium sheets (E. Merck, Germany) and the solvent system was an ethyl–acetate–methanol mixture.1H NMR, 13C NMR, 19F NMR and 31P NMR spectra were recorded on a 400 MHz Advance Bruker DPX spectrometer with tetramethylsilane (TMS) as the internal standard. The chemical shifts were reported in ppm units. Abbreviations are as follows: s, singlet; d, doublet; dd, double doublet; t, triplet; and m, multiplet. The melting points of the complexes were measured on an Elchem Microprocessor-based DT apparatus using an open capillary tube. The mass spectra of the synthesized compounds were recorded on a Shimadzu ESI-mass spectrometer having a 4000 triple quadrupole MS, using methanol as the solvent. UV-visible spectra were recorded on a JASCO V-730 spectrometer using a 1 cm quartz cell and fluorescence spectra on Hitachi F7000 fluorescence spectrophotometer equipped with a xenon lamp. A PerkinElmer instrument was used for the elemental analysis. The conductivity and viscosity study were performed using a conductivity-TDS meter-307 and Ostwald viscometer, respectively.
For the cytotoxicity (MTT) assay and imaging study, an Elisa reader, 96-well plate and Olympus CX41 fluorescence microscope were used. Bovine serum albumin (BSA) was purchased from Sigma Aldrich Chemical Limited. The MDA-MB-468 and HaCaT cell lines were purchased from NCCS, Pune.
Synthetic procedure
Synthesis of [Ir(III)-Cp*-(arylimidazophenanthroline)Cl]PF6 complexes [IrL1–IrL5]: 30 mg (0.037 mmole, 1 eq.) of pentamethylcyclopentadienyl iridium dichloride dimer, [(C5(CH3)5IrCl2)]2, was dissolved in about 10 ml of methanol in a 50 ml round bottomed flask and was stirred continuously for 5–10 min to dissolve the reactant. To the completely dissolved solution, 2.1 equivalents of the previously synthesized ligands (L1–L5) were added and stirred at room temperature. After 90 min, 2.5 equivalents of ammonium hexafluorophosphate (NH4PF6) (13.3 mg, 0.082 mmol) was added as ligand exchange salt in order to increase crystallinity and hence, purity of the product and again the reaction mixture was stirred for 90 min more at room temperature. The progress of reaction was confirmed by TLC. After complete conversion of the starting materials to the desired product, the solvent was evaporated under reduced pressure. The crude product was washed with hexane and further recrystallized from diethyl ether/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solvent system. Finally, the complexes (IrL1–IrL5) were obtained as light brown crystals with high yield (90–92%).
1) solvent system. Finally, the complexes (IrL1–IrL5) were obtained as light brown crystals with high yield (90–92%).
Characterization data for complexes (IrL1–IrL5)
Cytotoxicity studies
Cytotoxicity study of all the synthesized compounds against triple negative breast cancer cells MDA-MB-468 and immortalized human keratinocyte cell line HaCaT were done with a drug incubation period of 48 h by following standard procedure that were mentioned in ESI.† Preliminary studies were done to understand the mode of action of the complex to induce cytotoxicity using flow cytometric methods.Mitochondrial membrane dysfunction assay
Alterations in mitochondrial membrane potential (MMP, ΔΨm) and associated mitochondrial dysfunction was demonstrated by JC-1, a cationic carbocyanine dye. The cells were treated with the mitochondrial uncoupler CCCP (carbonyl cyanide m-chlorophenylhydrazone), which mediates the dissipation of mitochondrial membrane potential, served as positive control for the detection of mitochondrial dysfunction. The flow cytometric quantification of JC-1 stained cells revealed a normal mitochondrial function in control cells and the results obtained from the quantified JC-1 stained cells among IrL1 treated cells were compared with the same of the control.Imaging studies
Colocalization study and ROS generation studies were performed using live cell lines in their log phase.Statistical analysis
As the study had more than one group, one way ANOVA was used for statistical analysis. The p value <0.05 was considered as significant.Conclusions
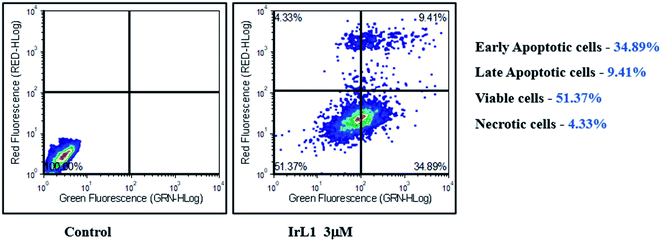
Five Ir(III)-imidazophenanthroline complexes were synthesized and characterized successfully. Our study has demonstrated the cytotoxic potential of IrL1 complex on MDA-MB-468 breast cancer cells in a dose dependent manner. The subcellular localization study of the complex revealed the localization of the compound in cytoplasm thereby pointing to a possible mitochondrial localization and consequent mitochondrial dysfunction. Subsequent analysis to demonstrate the alterations in MMP and ROS generation mediated by the IrL1 complex has revealed a significant increase in mitochondrial dysfunction and a resultant increase in ROS production suggestive of the mitochondrial targeting potential of the IrL1 complex. The Annexin V-FITC/PI assay has validated the cytotoxic potential, along with the IC50 dose of the complex on MDA-MB-468 breast cancer cells by initiating apoptotic pathway probably due to the cellular energetic stress triggered by elevated ROS levels. Furthermore, the IrL1 complex mediated a substantial G1 phase cell-cycle arrest of MDA-MB-468 cells at the highest tested concentration of 5 μM. The study findings support the prospective therapeutic potential of the IrL1 complex in the treatment and eradication of triple negative breast cancer cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors are grateful to Department of Science and Technology, Government of India for supporting the work through the DST-SERB CRG project grant (CRG/2021/002267) and ICMR-SRF fellowship and contingency grant. The authors are grateful to VIT University for providing VIT SEED funding. We acknowledge DST, New Delhi, India for DST-FIST project.Notes and references
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, Ca-Cancer J. Clin., 2021, 71(3), 209–249 CrossRef PubMed
.
- N. Harbeck, F. Penault-Llorca, J. Cortes, M. Gnant, N. Houssami, P. Poortmans, K. Ruddy, J. Tsang and F. Cardoso, Breast Cancer, 2019, 5, 66 Search PubMed
.
- J. D. Brenton, L. A. Carey, A. A. Ahmed and C. Caldas, Molecular classification and molecular forecasting of breast cancer: ready for clinical application?, J. Clin. Oncol., 2005, 23(29), 7350–7360 CrossRef CAS PubMed
.
- M. Telli, Optimizing chemotherapy in triple-negative breast cancer: the role of platinum, Am. Soc. Clin. Oncol. Educ. Book, 2014, 34(1), e37–e42 CrossRef PubMed
.
- E. Alli, V. B. Sharma, A. R. Hartman, P. S. Lin, L. McPherson and J. M. Ford, Enhanced sensitivity to cisplatin and gemcitabine in Brca1-deficient murine mammary epithelial cells, BMC Pharmacol., 2011, 11(1), 1–12 Search PubMed
.
- G. Von Minckwitz, A. Schneeweiss, C. Salat, M. Rezai, D. M. Zahm, P. Klare, J. U. Blohmer, H. Tesch, F. Khandan, S. Jud and C. Jackisch, A randomized phase II trial investigating the addition of carboplatin to neoadjuvant therapy for triple-negative and HER2-positive early breast cancer (GeparSixto), J. Clin. Oncol., 2013, 13(15), 1004 Search PubMed
.
- J. O'shaughnessy, L. S. Schwartzberg, M. A. Danso, H. S. Rugo, K. Miller, D. A. Yardley, R. W. Carlson, R. S. Finn, E. Charpentier, M. Freese and S. Gupta, A randomized phase III study of iniparib (BSI-201) in combination with gemcitabine/carboplatin (G/C) in metastatic triple-negative breast cancer (TNBC), J. Clin. Oncol., 2011, 29(15), 1007 Search PubMed
.
- G. Ciarimboli, Membrane transporters as mediators of cisplatin side-effects, Anticancer Res., 2014, 34(1), 547–550 CAS
.
- E. M. Domínguez-Martís, D. G. Mosteiro-Miguéns, L. Vigo-Gendre, D. López-Ares, M. Freire-Garabal, M. J. Núñez-Iglesias and S. Novío, Non-platinum metal complexes as potential anti-triple negative breast cancer agents, Crystals, 2018, 8(10), 369–393 CrossRef
.
- P.-Y. Ho, C.-L. Ho and W.-Y. Wong, Coord. Chem. Rev., 2020, 413, 213267–213305 CrossRef CAS
.
-
(a) P. Steunenberg, A. Ruggi, N. S. van den Berg, T. Buckle, J. Kuil, F. W. B. van Leeuwen and A. H. Velders, Inorg. Chem., 2012, 51, 2105–2114 CrossRef CAS PubMed
; (b) L. He, Y. Li, C. P. Tan, R. R. Ye, M. H. Chen, J. J. Cao, L. N. Ji and Z. W. Mao, Chem. Sci., 2015, 6, 5409–5418 RSC
.
- A. Bencini and V. Lippolis, 1,10-Phenanthroline: a versatile building block for the construction of ligands for various purposes, Coord. Chem. Rev., 2010, 254(17–18), 2096–2180 CrossRef CAS
.
- B. Kar, U. Das, S. De, S. Pete, S. A. Sharma, N. Roy, S. K. A. Kumar, D. Panda and P. Paira, GSH-resistant and highly cytoselective ruthenium(II)-p-cymene-(imidazo[4,5-f][1,10]phenanthrolin-2-yl) phenol complexes as potential anticancer agents, Dalton Trans., 2021, 50(30), 10369–10373 RSC
.
- N. Roy, U. Sen, S. Ray Chaudhuri, V. Muthukumar, P. Moharana, P. Paira, B. Bose, A. Gauthaman and A. Moorthy, Mitochondria specific highly cytoselective iridium(III)-Cp* dipyridophenazine (dppz) complexes as cancer cell imaging agents, Dalton Trans., 2021, 50(6), 2268–2283 RSC
.
- A. Mondal, U. Sen, N. Roy, V. Muthukumar, S. K. Sahoo, B. Bose and P. Paira, DNA targeting half sandwich Ru(II)-p-cymene-N^N complexes as cancer cell imaging and terminating agents: influence of regioisomers in cytotoxicity, Dalton Trans., 2021, 50(3), 979–997 RSC
.
- N. Roy, U. Sen, Y. Madaan, V. Muthukumar, S. Varddhan, S. K. Sahoo, P. Panda, B. Bose and P. Paira, Mitochondria-Targeting Click-Derived Pyridinyltriazolylmethylquinoxaline-Based Y-Shaped Binuclear Luminescent Ruthenium(II) and Iridium(III) Complexes as Cancer Theranostic Agents, Inorg. Chem., 2020, 59(23), 17689–17711 CrossRef CAS PubMed
.
- B. Sarkar, A. Mondal, Y. Madaan, N. Roy, A. Moorthy, Y.-C. Kuo and P. Paira, Luminescent anticancer ruthenium(II)-p-cymene complexes of extended imidazophenanthroline ligands: synthesis, structure, reactivity, biomolecular interactions and live cell imaging, Dalton Trans., 2019, 48(32), 12257–12271 RSC
.
- S. De, S. Ray Chaudhuri, A. Panda, G. R. Jadhav, R. S. Kumar, P. Manohar, N. Ramesh, A. Mondal, A. Moorthy, S. Banerjee and P. Paira, Synthesis, characterisation, molecular docking, biomolecular interaction and cytotoxicity studies of novel ruthenium(II)-arene-2-heteroarylbenzoxazole complexes, New J. Chem., 2019, 43(8), 3291–3302 RSC
.
- S. Rangasamy, H. Ju, S. Um, D.-C. Oh and J. M. Song, Mitochondria and DNA Targeting of 5,10,15,20-Tetrakis(7-sulfonatobenzo[b]thiophene) Porphyrin-Induced Photodynamic Therapy via Intrinsic and Extrinsic Apoptotic Cell Death, J. Med. Chem., 2015, 58, 6864–6874 CrossRef CAS PubMed
.
- C. Zhang, K. Qiu, C. Liu, H. Huang, T. W. Rees, L. Ji, Q. Zhang and H. Chao, Tracking mitochondrial dynamics during apoptosis with phosphorescent fluorinated iridium(III) complexes, Dalton Trans., 2018, 47, 12907–12913 RSC
.
- A. Mondal and P. Paira, Hypoxia efficient and glutathione-resistant cytoselective ruthenium(II)-p-cymene-arylimidazophenanthroline complexes: biomolecular interaction and live cell imaging, Dalton Trans., 2020, 49(36), 12865–12878 RSC
.
- Y. Chen, L. Qiao, L. Ji and H. Chao, Phosphorescent iridium(III) complexes as multicolor probes for specific mitochondrial imaging and tracking, Biomaterials, 2014, 35(1), 2–13 CrossRef CAS PubMed
.
- C. Jin, J. Liu, Y. Chen, L. Zeng, R. Guan, C. Ouyang, L. Ji and H. Chao, Cyclometalated iridium (III) complexes as two-photon phosphorescent probes for specific mitochondrial dynamics tracking in living cells, Chem.–Eur. J., 2015, 21(34), 12000–12010 CrossRef CAS PubMed
.
- M. Kubanik, H. Holtkamp, T. Sohnel, S. M. Jamieson and C. G. Hartinger, Impact of the halogen substitution pattern on the biological activity of organoruthenium 8-hydroxyquinoline anticancer agents, Organometallics, 2015, 34(23), 5658–5668 CrossRef CAS
.
- M. Sunita, B. Anupama, B. Ushaiah and C. G. Kumari, Synthesis, characterization, DNA binding and cleavage studies of mixed-ligand copper (II) complexes, Arabian J. Chem., 2017, 10, S3367–S3374 CrossRef CAS
.
- W. J. Geary, The use of conductivity measurements in organic solvents for the characterisation of coordination compounds, Coord. Chem. Rev., 1971, 7(1), 81–122 CrossRef CAS
.
- I. Ali, W. A. Wani and K. Saleem, Empirical formulae to molecular structures of metal complexes by molar conductance, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43(9), 1162–1170 CrossRef CAS
.
- B. G. Campling, K. Baer, H. M. Baker, Y. M. Lam and S. P. Cole, Do glutathione and related enzymes play a role in drug resistance in small cell lung cancer cell lines?, Br. J. Cancer, 1993, 68(2), 327–335 CrossRef CAS PubMed
.
- V. G. Sankareswari, D. Vinod, A. Mahalakshmi, M. Alamelu, G. Kumaresan, R. Ramaraj and S. Rajagopal, Interaction of oxovanadium(IV)–salphen complexes with bovine serum albumin and their cytotoxicity against cancer, Dalton Trans., 2014, 43(8), 3260–3272 RSC
.
- S. S. Iqbal, M. W. Mayo, J. G. Bruno, B. V. Bronk, C. A. Batt and J. P. Chambers, A review of molecular recognition technologies for detection of biological threat agents, Biosens. Bioelectron., 2000, 15(11–12), 549–578 CrossRef CAS
.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H, 31P and 19F NMR, LCMS, IR, UV and fluorescence spectra of all compounds. See https://doi.org/10.1039/d2ra01036d |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |